Abstract
Absence or presence of glial cells missing (GCM) in cells of the developing nervous system of Drosophila decides over their future fate as neurons or glia with only those cells turning into glia that express GCM. To understand how GCM exerts its function we performed a detailed structure–function analysis. Using fusions between the DNA binding domain of the yeast GAL4 protein and GCM, we detected a transactivation function within the C-terminal part of GCM. In addition to this transactivation domain we mapped a sequence-specific DNA-binding domain within the N-terminal part of the GCM protein in close proximity to a bipartite nuclear localization signal. Binding site selection assays determined the motif 5′-AT(G/A)CGGGT-3′ as the preferred binding site for GCM. Both the lack of homology to known proteins and the novel DNA binding specificity indicate that GCM contained a new type of DNA-binding domain. In transiently transfected cells, GCM also activated transcription from promoters consisting of the newly identified GCM-binding site and a TATA box. Thus, GCM is a novel type of transcription factor involved in early gliogenesis.
Keywords: glia, neurogenesis, differentiation/gene regulation/nuclear localization
Neurons and glia are the two main cell types in the nervous system with neurons being involved in the processing of information and glia providing trophic, structural, and functional support. Despite their distinct functions, neurons and glia originate from a common precursor both in vertebrates and in invertebrates (1–3). What determines the choice between these two different cell fates has become apparent only recently with the isolation of the gene glial cells missing (GCM) in Drosophila (4, 5). In the absence of GCM, presumptive glia develop into additional neurons, whereas overexpression of GCM leads to a conversion of presumptive neurons into surplus glia. Thus, GCM functions as an important switch during early neurogenesis by committing cells to the glial cell fate. Because of its nuclear localization it has been speculated that GCM might activate glia-specific gene expression or repress the expression of neuronal genes in the glial precursor either in a direct or an indirect manner (6). Whether a functionally equivalent protein is present in presumptive neurons or whether differentiation along the neuronal pathway occurs by default remains to be determined. Unfortunately, the primary amino acid sequence of GCM did not reveal any information concerning the protein’s mode of action as its sequence did not exhibit any apparent homology to other known proteins. Therefore we initiated a detailed structure-function analysis of the GCM protein probing for features expected to be present in transcriptional regulators.
EXPERIMENTAL PROCEDURES
Plasmids.
Gcm-N9delta containing the entire GCM coding sequences in pBluescript II SK was generously provided by T. Hosoya and Y. Hotta (University of Tokyo). GCM sequences were retrieved by PCR from this vector as a EcoRI/SalI fragment, and inserted into pGEX-KG, yielding pGEX-GCM for the bacterial expression of fusion proteins between glutathione-S-transferase (GST) and GCM. To obtain a mammalian expression plasmid for GCM, restriction sites were introduced immediately upstream (EcoRI) and downstream (KpnI) of the open reading frame. The resulting GCM fragment was inserted into pCMV5 and pCMV/GAL4, yielding the mammalian expression plasmids pCMV–GCM fl and pCMV/GAL4–GCM fl. pCMV/GAL4 already contained coding sequences for the DNA-binding domain of the yeast GAL4 protein (7), and insertion of GCM sequences led to an in-frame fusion of both open reading frames with GCM following GAL4 sequences. A stop codon and KpnI site were introduced by PCR-directed mutagenesis downstream of codons 171, 253, 338, or 421 to obtain the C-terminal GCM deletion mutants GCM 171, GCM 253, GCM 338, and GCM 421. For N-terminal GCM deletion mutants, EcoRI site and eukaryotic translation initiation consensus (including start ATG) were introduced immediately in front of codon 86, 169, 248, 344, or 419 of GCM, thus generating GCM Δ85, GCM Δ168, GCM Δ247, GCM Δ343, and GCM Δ418. All deletion mutants were inserted as EcoRI/KpnI fragments into pCMV/GAL4 in a manner analogous to full-length GCM. C-terminal mutants were also inserted into pCMV5 whereas out of the N-terminal mutants only GCM Δ168 and GCM Δ247 were cloned into pCMV5 after additional introduction of a T7-tag (Novagen).
The GAL4-responsive luciferase reporter was created from p36luc (8) by inserting three tandem copies of a previously characterized GAL4 binding site (9) in front of the rat prolactin minimal promoter. siteA-luc has been described (8, 10). GCM-responsive reporter plasmids were constructed by inserting one, three, or six copies of the GCM-binding site 5′-GATCCCGATGCGGGTGCAGATC-3′ into pTATAluc, which carried the luciferase gene under the control of the β-globin minimal promoter. 6xoct luc contained six copies of the DNA binding element for POU-domain proteins 5′-GATCCGAGAATATGCAAATCAATTGGAGATC-3′ in pTATAluc.
All β-galactosidase (β-gal) expression vectors were based on pCMVlacZ, which together with pCMVlacZ/Tst-1 nuclear localization signal (NLS) has been described (11). Plasmid pCMVlacZ/GCM NLS and pCMV/MESKRRRlacZ were obtained by inserting a short sequence corresponding to amino acids 218–233 of GCM (KRQAKTQSIQESKRRR) or amino acids 228–233 of GCM (ESKRRR) between NcoI and NruI sites of pCMVlacZ directly behind the start methionine.
Antibodies.
An antiserum against Drosophila GCM was raised in rabbit using standard protocols. Purified recombinant GCM protein produced in bacteria with an N-terminal polyhistidine tag was used as antigen in a partially denatured state. The antiserum recognized primarily N-terminal epitopes of GCM.
Cell Culture, Transfections, Luciferase Assays, Immunofluorescence, and Histochemistry.
U138 human glioblastoma cells were propagated in RPMI 1640 medium supplemented with 10% fetal calf serum, whereas CV1 and COS cells were kept in Dulbecco’s modified Eagle’s medium containing 10% fetal calf serum. U138 and CV1 cells were transfected by the calcium phosphate technique as described (11). COS cells were transfected using DEAE-dextran (12). For luciferase assays, U138 cells were transfected with 2 μg of luciferase reporter plasmid and 2 μg of expression plasmid per 60-mm plate. The total amount of plasmid was kept constant. Cells were harvested 48 h after transfection, and extracts were assayed for luciferase activity (8).
For immunofluorescence and β-gal histochemistry, CV1 cells were transfected on chamber slides (Lab-Tec; Nunc). Forty-eight hours posttransfection, cells were fixed and either stained for β-gal activity in PBS containing 1 mg/ml 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal), 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, and 2 mM MgCl2 or labeled with primary antibodies for immunofluorescence studies (11). Depending on the primary antibody, Cy3-conjugated goat anti-rabbit antibodies or goat anti-mouse antibodies (Dianova, Hamburg, Germany) were used as secondaries. All antibodies were used at a dilution of 1:500.
Preparation of Whole Cell Extracts and Recombinant Proteins.
Transiently transfected COS cells were lysed in the presence of 2 μg/μl leupeptin and aprotinin each in ice-cold 10 mM Hepes (pH 7.9), 0.2 mM EDTA, 2 mM DTT, and 1% Nonidet P-40. Immediately after lysis, NaCl was added to a final concentration of 400 mM. After incubation for 15 min under constant rotation, cell debris was removed from the extract by centrifugation. GST-fusion proteins were expressed in bacteria and affinity-purified as described (10, 13).
Western Blot Analysis.
A total of 15 μl of nuclear extract (≈4 mg/ml) were size fractionated on SDS/12% polyacrylamide gels and transferred to nitrocellulose membranes. Nitrocellulose filters were blocked and incubated consecutively with primary antibody and horseradish-peroxidase-coupled secondary antibody as described (11). The rabbit antiserum against GCM or a mouse monoclonal against the DNA-binding domain of GAL4 (CLONTECH) served as primary antibodies, each at a dilution of 1:3000. The enhanced chemiluminescence system (Amersham) was used for detection.
DNA–Protein Binding Assays.
Electrophoretic mobility shift experiments were performed as described (8, 13) using either 50 ng of purified GST fusion proteins or 4 μg of whole COS cell extracts as a protein source. Oligonucleotides contained either of two versions of the GCM-binding site (5′-GATCCCGATGCGGGTGCAGATC-3′ or 5′-GATCCCATACGGGTGAGAAGATC-3′). Identification of the preferred GCM-binding site from a pool of random oligonucleotides was performed as a selected and amplified binding site (SAAB) assay (14) using 5′-GGATCCATTCCTAAGCGCAT(N)8GAGCTCAGATCAGATCT-3′ as template and GST–GCM as protein source. Protein-bound template was identified by elelctrophoretic mobility shift analysis and amplified by PCR in 34 cycles with 5′-GGATCCATTCCTAAG-3′ and 5′-AGATCTGATCTGAGC-3′ as primers. Throughout all six rounds of selection conditions were as previously described for the binding site selection of Sprm-1 (15).
RESULTS AND DISCUSSION
Transcription factors all share a number of characteristics, including a NLS, a DNA-binding domain, and a transactivation domain (16). The presence of all these features in GCM would therefore be strongly indicative of its function as a transcription factor.
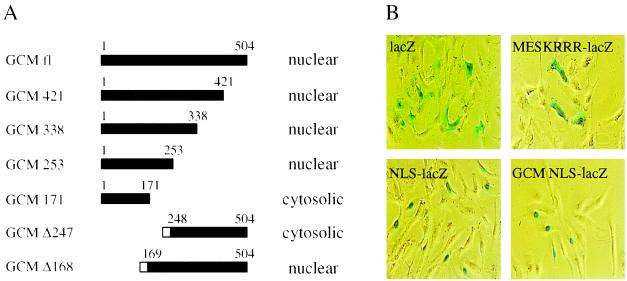
GCM had already been shown to be a nuclear protein and inspection of the primary amino acid sequence had predicted the presence of a nuclear localization function within the central region of the protein (4, 5). To test this prediction, we generated a set of GCM mutants that were successively shortened from the C-terminal end. Using a polyclonal antiserum directed against the N-terminal half of the GCM protein, we analyzed the cellular distribution of these mutants in transiently transfected CV1 cells using immunofluorescence studies (Fig. 1A). As previously reported (4, 5), full-length GCM protein localized to the nucleus. No difference could be detected in cellular distribution for the larger GCM mutants as deletion of all sequences C-terminal to amino acid 253 did not affect nuclear localization. A mutant GCM protein, however, which contained only the N-terminal 171 amino acids, was predominantly found in the cytoplasm of transfected cells, clearly indicating that the NLS was present between amino acids 171 and 253 of GCM. To corroborate this conclusion, we generated two additional GCM mutants with deletions in the N-terminal part of the protein. As these proteins were not recognized by the anti-GCM antiserum, we tagged them with an epitope. As indicated in Fig. 1A, a mutant GCM protein lacking only the N-terminal 168 amino acids localized correctly to the nucleus of transfected cells, whereas a mutant without the N-terminal 247 amino acids remained cytosolic. Thus, our immunofluorescence studies mapped the nuclear localization function to the region between amino acids 171 and 247 predicted to contain the NLS (4, 5). This putative NLS (amino acids 218–233, KR-N10-KRRR) conforms to the consensus for bipartite NLSs consisting of a cluster of basic amino acids preceded by two additional basic amino acids with a spacing of 10–12 amino acids (17).
Figure 1.
Nuclear localization of GCM. (A) Cellular distribution of full-length GCM and various mutants as detected in immunofluorescence studies on transiently transfected CV1 cells. Mutants GCM 421, GCM 338, GCM 253, and GCM 171 as well as full-length GCM were detected using rabbit anti-GCM antiserum, whereas the tagged mutants GCM Δ247 and GCM Δ168 were visualized using a mAb directed against the T7-tag (Novagen). Amino acids present within each mutant are indicated by the filled bar; the open box represents the T7-tag. (B) Localization of β-gal by histochemical X-gal staining in CV1 cells transiently transfected with pCMVlacZ (lacZ), pCMVlacZ/Tst-1 NLS (NLS-lacZ), pCMV/MESKRRRlacZ (MESKRRR-lacZ), or pCMVlacZ/GCM NLS (GCM NLS-lacZ).
To investigate whether this amino acid sequence would indeed be sufficient for nuclear localization, we carried out transfer experiments with β-gal (Fig. 1B). When expressed in eukaryotic cells, β-gal was localized to the cytoplasm (Fig. 1B). As shown before (11), N-terminal addition of a short stretch of amino acids that represented the NLS of the POU-domain transcription factor Tst-1/Oct6, redistributed the resulting β-gal fusion to the cell nucleus. Now we transferred amino acids 218–233 corresponding to the putative bipartite signal of GCM in an analogous manner to β-gal. Interestingly, this region of GCM was as efficient in targeting β-gal to the cell nucleus as the NLS of Tst-1/Oct6. In contrast, nuclear localization was not achieved by fusing only a shortened version corresponding to amino acids 228–233 to β-gal, clearly showing that the bipartite signal is required in full to mediate nuclear localization. It has been previously noted by us and others that certain NLSs are found in close proximity to phosphorylation sites that in some cases have been shown to regulate the access of the respective protein to the nucleus (11, 18, 19). In this context, it is interesting to note that the NLS of GCM also overlaps with potential phosphorylation sites for protein kinase C and casein kinase II.
Having mapped the nuclear localization domain of GCM, we next turned to the question whether GCM contained a transactivation domain. We constructed a chimera between the well-characterized DNA-binding domain of the yeast GAL4 protein and GCM, and compared its transactivation potential to that of the isolated GAL4 DNA-binding domain in transient transfection experiments using a GAL4-responsive reporter. Fusing full-length GCM to the GAL4 DNA-binding domain led to an 11-fold stimulation of promoter activity when compared with the GAL4 DNA-binding domain alone. This result already indicated that GCM had the capacity to transactivate.
In an attempt to localize the region within GCM responsible for transactivation, we generated two series of GCM mutants by either successively shortening the protein from the N terminus or from the C terminus and fused these mutants in frame with the GAL4 DNA-binding domain. Using a mAb directed against the GAL4 part of the fusion, we confirmed by Western blot analysis on whole cell extracts that chimeric proteins of correct size were produced in transfected cells (Fig. 2). In general, the amount of fusion protein in transfected cells correlated inversely with its size, with the full-length GCM fusion being least, and the shortest GCM fusion being most abundant.
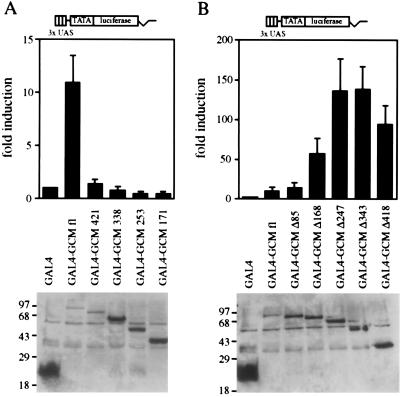
Figure 2.
Transactivation potential of GCM. A GAL4-responsive luciferase reporter (3xUAS TATA luciferase) was transfected into U138 glioblastoma cells together with expression plasmids for the GAL4 DNA-binding domain (GAL4) or for fusions of the GAL4 DNA-binding domain with full-length GCM (GAL4–GCM fl) or mutant versions thereof. (A) Fusions containing C-terminal deletion mutants of GCM: GAL4–GCM 421, GAL4–GCM 338, GAL4–GCM 253, GAL4–GCM 171. (B) Fusions containing N-terminal deletion mutants of GCM: GAL4–GCM Δ85, GAL4–GCM Δ168, GAL4–GCM Δ247, GAL4–GCM Δ343, GAL4–GCM Δ418. (Upper) Luciferase activities in extracts from transfected cells were determined in four independent experiments, each performed in duplicates. Data are presented for each GAL4–GCM fusion as fold induction above the level of luciferase activity obtained in transfections with an expression plasmid for the GAL4 DNA-binding domain, which was given an arbitrary value of 1. (Lower) Expression of GAL4 fusion proteins in transfected cells was confirmed by Western blot analyses of whole cell extracts. Numbers on left indicate size of molecular weight markers in kDa.
First, we tested the set of chimeras containing the C-terminal deletion mutants of GCM in transient transfections for their ability to transactivate (Fig. 2A). By removing merely 83 amino acids from the C terminus of the GAL4–GCM fusion, transactivation was almost fully obliterated. Removal of additional amino acids did not lead to a recovery of the transactivation potential.
To extend these studies, we then turned to the series of GAL4 fusions with N-terminal GCM deletion mutants (Fig. 2B). Removal of the first 85 amino acids remained without effect on the transactivation potential of the GAL4–GCM fusion. However, deletion of the N-terminal 168 amino acids caused a substantial increase in the transactivation potential (from an 11-fold induction for the fusion between GAL4 and the full-length GCM protein to a 58-fold induction). Deletion of the first 168 amino acids thus seemed to disrupt a structure that masked part of the transactivation potential of GCM. As shown later, this structure turned out to be the DNA-binding domain. Even higher activity was detected for the mutant that lacked the first 247 amino acids (136-fold stimulation). The smallest fusion of this series, which only contained the 85 C-terminal amino acids, still exhibited a remarkable 94-fold activation of the reporter. Taken together, these data prove that GCM possesses transactivation potential, most of which is localized to a domain within the extreme C-terminal part of the protein.
To analyze whether GCM contains a DNA-binding domain, we produced a recombinant GST–GCM fusion protein in bacteria and purified it using affinity chromatography. The purified GST–GCM protein was then used in a binding site selection assay, in which the GST–GCM protein was incubated with a pool of oligonucleotides that exhibited randomized sequence at 8 bp in the center (15). GST–GCM bound to such an oligonucleotide (see Fig. 3A). A GST fusion with the non-DNA binding N-terminal part of Brn-2 (20), on the other hand, did not bind to the same oligonucleotide, making it unlikely that the observed complex results from a minor bacterial contaminant of the protein preparation.
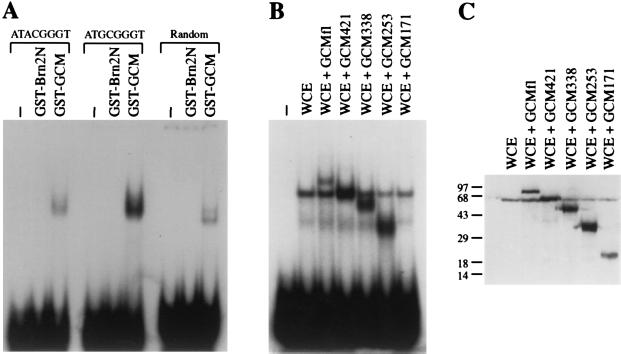
Figure 3.
DNA-binding of GCM. (A) Purified GST–GCM and GST–Brn2N protein were analyzed in electrophoretic mobility shift assays for their ability to bind to radiolabeled oligonucleotides containing either the sequence 5′-ATGCGGGT-3′ or 5′-ATACGGGT-3′, or to a random oligonucleotide which had undergone three of six selection cycles for an optimal GCM-binding site. (B) The radiolabeled oligonucleotide containing the 5′-ATGCGGGT-3′ motif was used to detect complex formation with whole cell extracts from COS cells that were either mock-transfected (WCE), transfected with full length GCM (WCE + GCM fl) or transfected with various GCM mutants (WCE + GCM 421, WCE + GCM 338, WCE + GCM 253, WCE + GCM 171). (C) Western blot analyses of cell extracts used in B. Numbers on left indicate size of molecular weight markers in kDa.
We amplified the preferred binding site from the pool of random oligonucleotides by six selection cycles and determined its sequence as 5′-AT(G/A)CGGGT-3′. To verify the ability of GST–GCM to bind to this sequence, we used oligonucleotides with the determined sequence in their center. Indeed, both 5′-ATACGGGT-3′ and 5′-ATGCGGGT-3′ were bound by GST–GCM (Fig. 3A). However, binding of GST–GCM to 5′-ATACGGGT-3′ was consistently weaker than binding to 5′-ATGCGGGT-3′. This might indicate, that a G at the third position is preferred over an A. The GCM-binding site is related to the octamer motif (5′-ATGCAAAT-3′) that is recognized by all members of the POU family of transcription factors (21–23). Compared with the octamer motif, three consecutive G’s are present in the GCM-binding site instead of three A’s. Despite this similarity, GCM did not recognize the octamer motif just as POU domain proteins did not recognize the GCM-binding site (data not shown). Binding to the GCM-binding site was not only detected for the recombinant GST–GCM fusion from bacteria, but also for a GCM protein produced in transiently transfected COS cells (Fig. 3B).
To identify the region within GCM that was responsible for the observed DNA-binding activity, we expressed the same series of successively shortened C-terminal GCM mutants in COS cells which we had already used in the characterization of the NLS. The presence of the respective GCM mutants in COS whole cell extracts was confirmed by Western blot analysis using the anti-GCM antiserum (Fig. 3C). When the same extracts were used in electrophoretic mobility shift experiments, specific protein–DNA complexes were observed not only for full-length GCM, but also for most mutants, except for the smallest (Fig. 3B). Thus, the N-terminal 253 amino acids were still sufficient for DNA binding, whereas the N-terminal 171 amino acids were not. This result clearly indicates that the DNA binding domain of GCM must be localized in the N-terminal part of the GCM protein. As this region does not exhibit any sequence similarities to other DNA-binding domains, Drosophila GCM seems to contain a hitherto unknown DNA-binding domain.
Recently, the sequence of a mouse GCM homolog has been reported and the partial sequence of a human homolog was found in the expressed sequence tag (EST) database (24). When comparing these sequences to Drosophila GCM it became evident that a region of Drosophila GCM corresponding to amino acids 32–188 is highly conserved in its mammalian homologs. Given the strong conservation of DNA-binding domains in other classes of transcription factors, amino acids 32–188 might contain the DNA-binding domain of GCM. As a blast search did not reveal any similarities between amino acids 32–188 of Drosophila GCM and DNA-binding domains represented in EMBL or GenBank databases, this region most likely defines a new class of transcriptional regulators which comprise GCM and its mammalian homologs.
So far, we have shown that GCM contained NLS, transactivation, and DNA-binding domains. In short, it fulfills all the criteria of a transcription factor. This prompted us to investigate whether GCM also functions as a transcription factor in living cells. Therefore, we constructed artificial promoters each consisting of a TATA box and a varying number of GCM-binding sites. These promoters were used to drive the expression of a luciferase reporter, which was cotransfected with an expression plasmid for GCM into mammalian cells. As shown in Fig. 4A, GCM did not exhibit any effect on a control promoter which consisted only of a TATA box or on a promoter which additionally contained a single binding site for POU domain proteins. In the presence of six consecutive binding sites for POU domain proteins within the promoter, GCM even exerted a 4-fold repression—despite the fact that GCM did not bind to these sites.
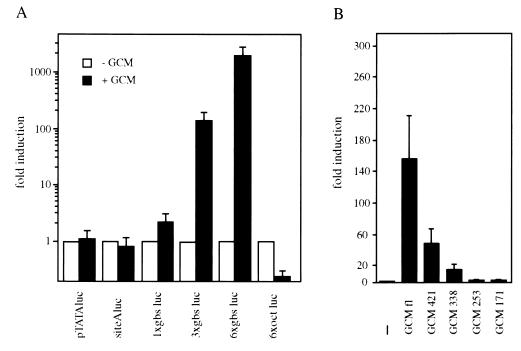
Figure 4.
In vivo function of GCM. (A) Various luciferase reporter plasmids were transfected into U138 glioblastoma cells in the absence (open bars) or presence (filled bars) of cotransfected full-length GCM. pTATAluc contained only a minimal promoter, whereas all other reporter plasmids carried additional binding sites for transcription factors. siteAluc and 6xoct luc contained one or six binding sites for POU-domain proteins; gbs luc constructs had either one, three, or six copies of a GCM-binding site inserted in front of the minimal promoter. (B) The 3xgbs luc reporter was transfected together with empty expression plasmid (−) or various GCM expression plasmids (GCM fl, GCM 421, GCM 338, GCM 253, and GCM 171) into U138 cells. Luciferase activities in extracts from transfected cells were determined in three independent experiments, each performed in duplicate. Data are presented as fold inductions that were calculated for each reporter plasmid by comparing luciferase activities to values from cells that were transfected with reporter plasmid and empty CMV expression plasmid. Note the logarithmic scale of the y axis in A.
Only with the newly identified GCM-binding site present in front of the TATA box did we observe a GCM-dependent increase of promoter activity. The degree of stimulation was dependent on the number of GCM-binding sites. Single GCM-binding sites were not very effective in increasing promoter activity leading only to a 2.3-fold increase. Three GCM-binding sites in tandem already led to a robust 157-fold stimulation of promoter activity. When six GCM sites were present, promoter activity even increased 2,020-fold over basal activity in the presence of cotransfected GCM. These transient transfections show impressively that GCM can indeed function as a bona fide transcription factor in living cells. The observed difference in the use of single versus multiple binding sites might indicate that GCM is a highly cooperative transcription factor, requiring interaction with other GCM molecules or different transcription factors for optimal function.
Using the promoter construct with three GCM-binding sites, we analyzed the transactivation capacity of the GCM mutants previously used in nuclear localization and DNA binding studies (Fig. 4B). Deletion of 85 amino acids from the C terminus led to a 70% reduction in promoter activity. This result is in good agreement with the one obtained for the corresponding GAL4 chimera with both experiments showing that the major transactivation domain is confined within the C-terminal part of GCM. Deletion of additional amino acids led to further significant losses of activity as mutant GCM 338 only exhibited 10% of the activity of wild-type GCM. No substantial promoter activation was obtained with the mutant that contained the N-terminal 253 amino acids despite the fact that this mutant still exhibited full DNA-binding activity. Thus, DNA binding and transactivation are carried out by two separate domains of GCM as is the case in most other transcription factors.
In summary, we have shown both on the basis of structural as well as functional criteria that Drosophila GCM is a transcription factor with novel DNA binding domain that recognizes the motif AT(G/A)CGGGT. This result should help to elucidate the function of GCM during early gliogenesis and facilitate the search for natural target genes of which the Drosophila homeodomain gene Repo/rk2 is the most obvious one because of its expression pattern in developing glia (25, 26). Given the existence of at least one GCM homolog in mammals, our results should also be instrumental in answering the question whether the function of the GCM protein is as well conserved as its structure (27).
Acknowledgments
We thank T. Hosoya and Y. Hotta for providing the Drosophila GCM cDNA. J. Enderich and D. Feist are acknowledged for expert technical assistance. J.S. is a member of the Graduiertenkolleg Scha692/1-1. This work was supported by a grant from the Deutsche Forschungsgemeinschaft to M.W.
ABBREVIATIONS
- GCM
glial cells missing
- GST
glutathione-S-transferase
- X-Gal
5-bromo-4-chloro-3-indolyl β-d-galactoside
- β-gal
β-galactosidase
- NLS
nuclear localization signal
Note
While this manuscript was under review, Akiyama et al. (28) independently analyzed the DNA binding activity of GCM with very similar results.
References
- 1.Anderson D J. Neuron. 1989;3:1–12. doi: 10.1016/0896-6273(89)90110-4. [DOI] [PubMed] [Google Scholar]
- 2.Doe C Q, Technau G M. Trends Neurosci. 1993;16:510–514. doi: 10.1016/0166-2236(93)90195-r. [DOI] [PubMed] [Google Scholar]
- 3.Jan Y N, Jan L Y. Curr Opin Neurobiol. 1994;4:8–13. doi: 10.1016/0959-4388(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 4.Hosoya T, Takizawa K, Nitta K, Hotta Y. Cell. 1995;82:1025–1036. doi: 10.1016/0092-8674(95)90281-3. [DOI] [PubMed] [Google Scholar]
- 5.Jones B W, Fetter R D, Tear G, Goodman C S. Cell. 1995;82:1013–1023. doi: 10.1016/0092-8674(95)90280-5. [DOI] [PubMed] [Google Scholar]
- 6.Anderson D J. Neuron. 1995;15:1219–1222. doi: 10.1016/0896-6273(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 7.Sadowski I, Ptashne M. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wegner M, Drolet D W, Rosenfeld M G. Proc Natl Acad Sci USA. 1993;90:4743–4747. doi: 10.1073/pnas.90.10.4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Webster N, Jin J R, Green S, Hollis M, Chambon P. Cell. 1988;52:169–178. doi: 10.1016/0092-8674(88)90505-3. [DOI] [PubMed] [Google Scholar]
- 10.Renner K, Leger H, Wegner M. Proc Natl Acad Sci USA. 1994;91:6433–6437. doi: 10.1073/pnas.91.14.6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sock E, Enderich J, Rosenfeld M G, Wegner M. J Biol Chem. 1996;271:17512–17518. doi: 10.1074/jbc.271.29.17512. [DOI] [PubMed] [Google Scholar]
- 12.Renner K, Sock E, Bermingham J R, Wegner M. Nucleic Acids Res. 1996;24:4552–4557. doi: 10.1093/nar/24.22.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leger H, Sock E, Renner K, Grummt F, Wegner M. Mol Cell Biol. 1995;15:3738–3747. doi: 10.1128/mcb.15.7.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blackwell T K, Weintraub H. Science. 1990;250:1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- 15.Andersen B, Pearse R V I, Schlegel P N, Cichon Z, Schonemann M D, Bardin W, Rosenfeld M G. Proc Natl Acad Sci USA. 1993;90:11084–11088. doi: 10.1073/pnas.90.23.11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson P F, McKnight S L. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- 17.Dingwall C, Laskey R A. Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 18.Jans D A, Ackerman M, Bischoff R A, Beach D H, Peters R. J Cell Biol. 1991;115:1203–1212. doi: 10.1083/jcb.115.5.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rihs H P, Jans D A, Fan H, Peters R. EMBO J. 1991;10:633–639. doi: 10.1002/j.1460-2075.1991.tb07991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li P, He X, Gerrero M R, Mok M, Aggarwal A, Rosenfeld M G. Genes Dev. 1993;7:2483–2496. doi: 10.1101/gad.7.12b.2483. [DOI] [PubMed] [Google Scholar]
- 21.Herr W, Sturm R A, Clerc R G, Corcoran L M, Baltimore D, Sharp P A, Ingraham H A, Rosenfeld M G, Finney M, Ruvkun G, Horvitz H R. Genes Dev. 1988;2:1513–1516. doi: 10.1101/gad.2.12a.1513. [DOI] [PubMed] [Google Scholar]
- 22.Schöler H R. Trends Genet. 1991;7:323–329. doi: 10.1016/0168-9525(91)90422-m. [DOI] [PubMed] [Google Scholar]
- 23.Wegner M, Drolet D W, Rosenfeld M G. Curr Opin Cell Biol. 1993;5:488–498. doi: 10.1016/0955-0674(93)90015-i. [DOI] [PubMed] [Google Scholar]
- 24.Altshuller Y, Copeland N G, Gilbert D J, Jenkins N A, Frohman M A. FEBS Lett. 1996;393:201–204. doi: 10.1016/0014-5793(96)00890-3. [DOI] [PubMed] [Google Scholar]
- 25.Campbell G, Goering H, Lin T, Spana E, Andersson S, Doe C, Tomlinson A. Development (Cambridge, UK) 1994;120:2957–2966. doi: 10.1242/dev.120.10.2957. [DOI] [PubMed] [Google Scholar]
- 26.Xiong W, Okano H, Patel N, Blendy J, Montell C. Genes Dev. 1994;8:981–994. doi: 10.1101/gad.8.8.981. [DOI] [PubMed] [Google Scholar]
- 27.Pfrieger F W, Barres B A. Cell. 1996;83:671–674. doi: 10.1016/0092-8674(95)90178-7. [DOI] [PubMed] [Google Scholar]
- 28.Akiyama Y, Hosoya T, Poole A M, Hotta Y. Proc Natl Acad Sci USA. 1996;93:14912–14916. doi: 10.1073/pnas.93.25.14912. [DOI] [PMC free article] [PubMed] [Google Scholar]