Abstract
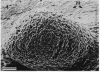
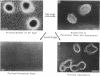
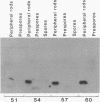
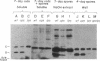
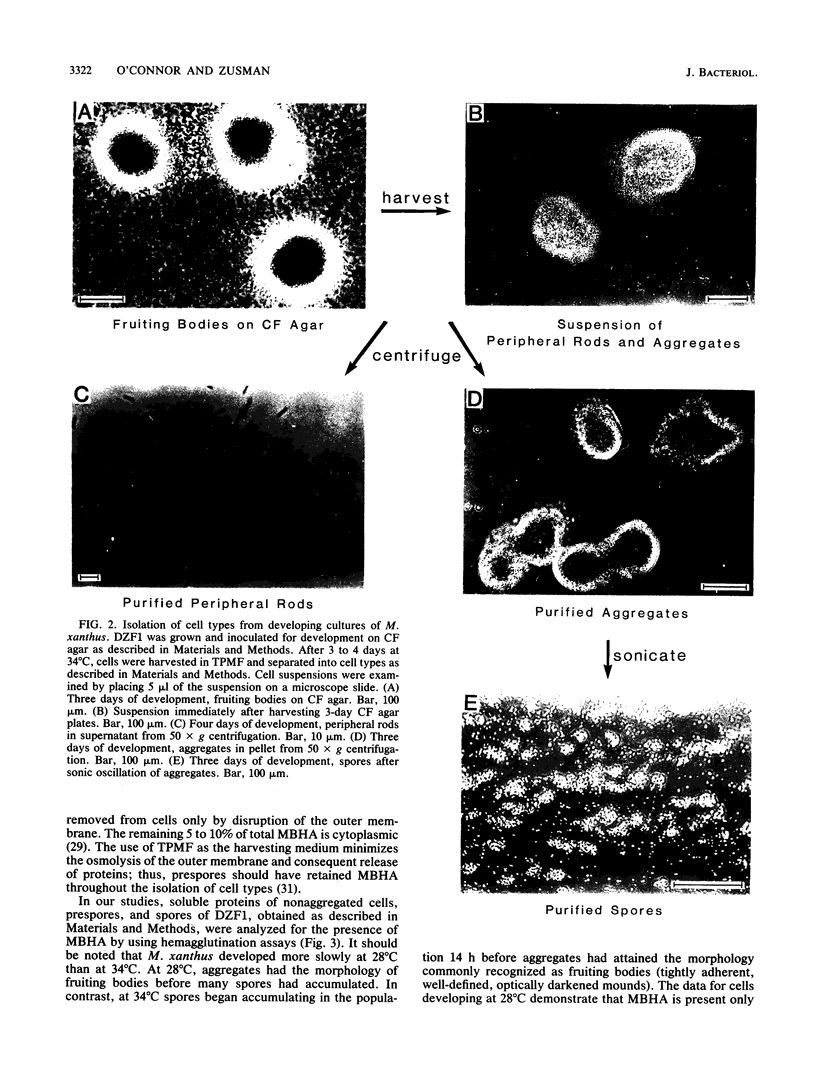
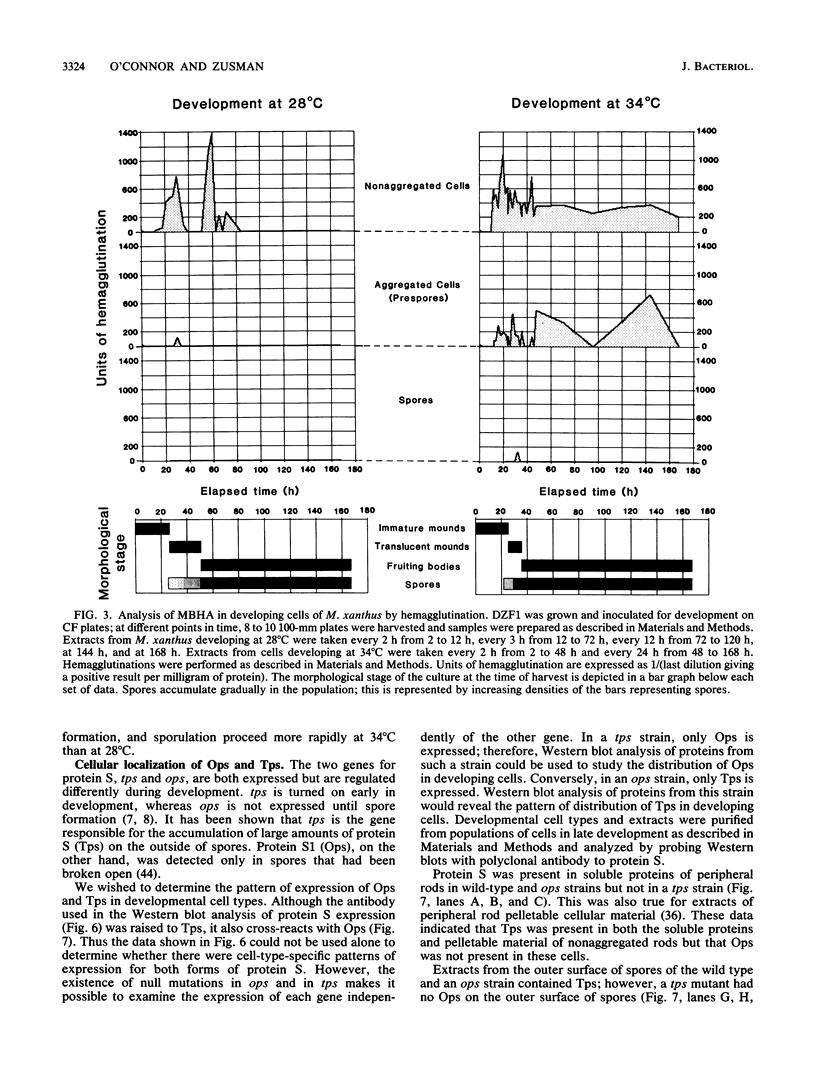
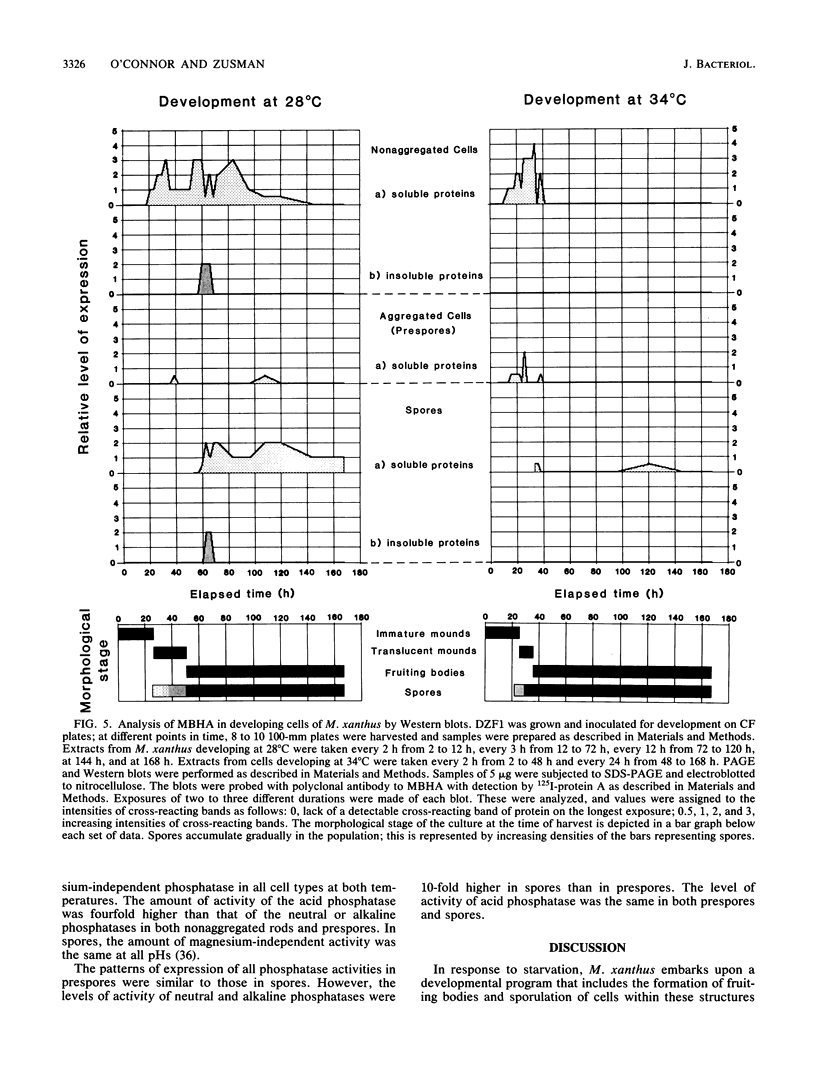
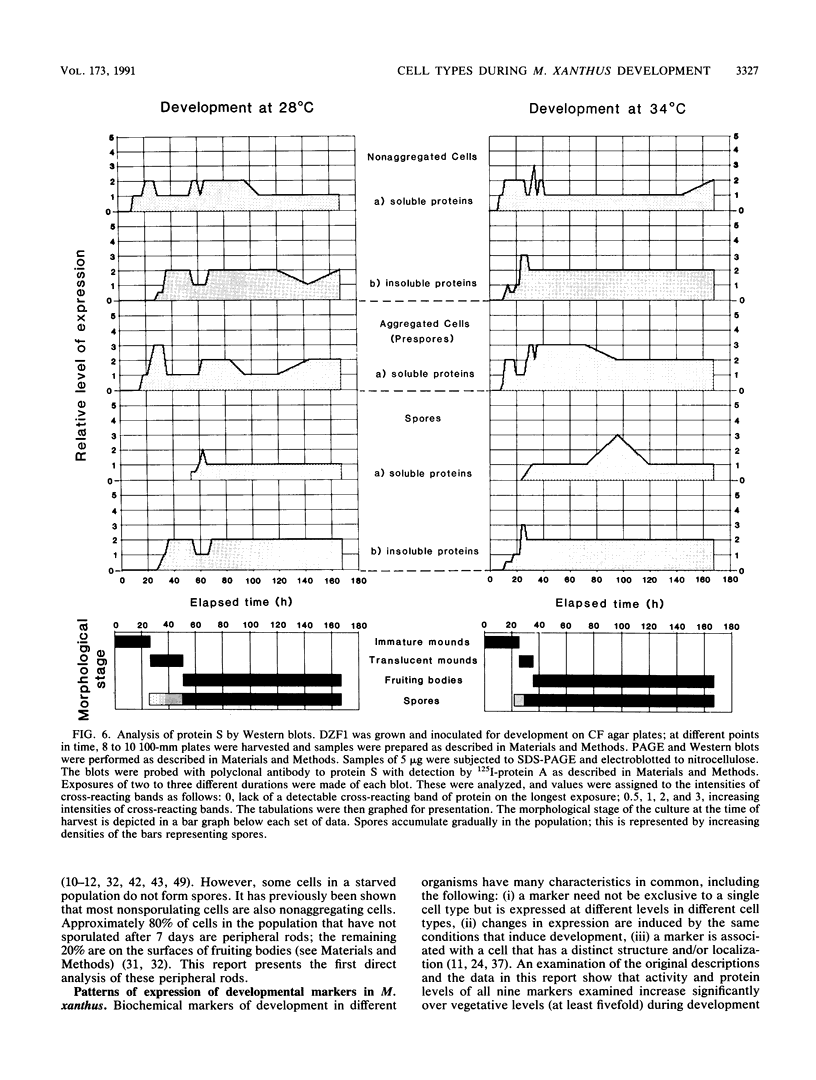
Myxococcus xanthus, a gram-negative bacterium, has a complex life cycle. In response to starvation, most cells in a population participate in the formation of multicellular aggregates (i.e., fruiting bodies) in which cells differentiate into spores. However, some cells do not enter aggregates. In this and the two accompanying reports, the biology and physiology of these nonaggregated cells is examined. A technique to separate aggregated cells from nonaggregated cells was developed; then differentiating cells at stages throughout the course of development were isolated. In this report we (i) describe peripheral rods, those cells which remain outside aggregates after aggregation has ceased in the rest of the population; (ii) document the occurrence of peripheral rods in several wild-type strains; and (iii) characterize the expression of developmentally regulated genes in both aggregated and nonaggregated cells. These studies have shown that myxobacterial hemagglutinin, protein S (Tps), protein S1 (Ops), protein C, and several phosphatase activities are expressed in cell-type-specific patterns. These data demonstrate that peripheral rods constitute a cell type distinct from either vegetatively growing cells or spores. The description of a second, late developmental cell type (in addition to spores) opens an entirely new line of investigation in M. xanthus, i.e., the regulation of the differentiation of vegetatively growing cells into two cell types that differ significantly in biology, shape, and localization within the population.
Full text
PDF






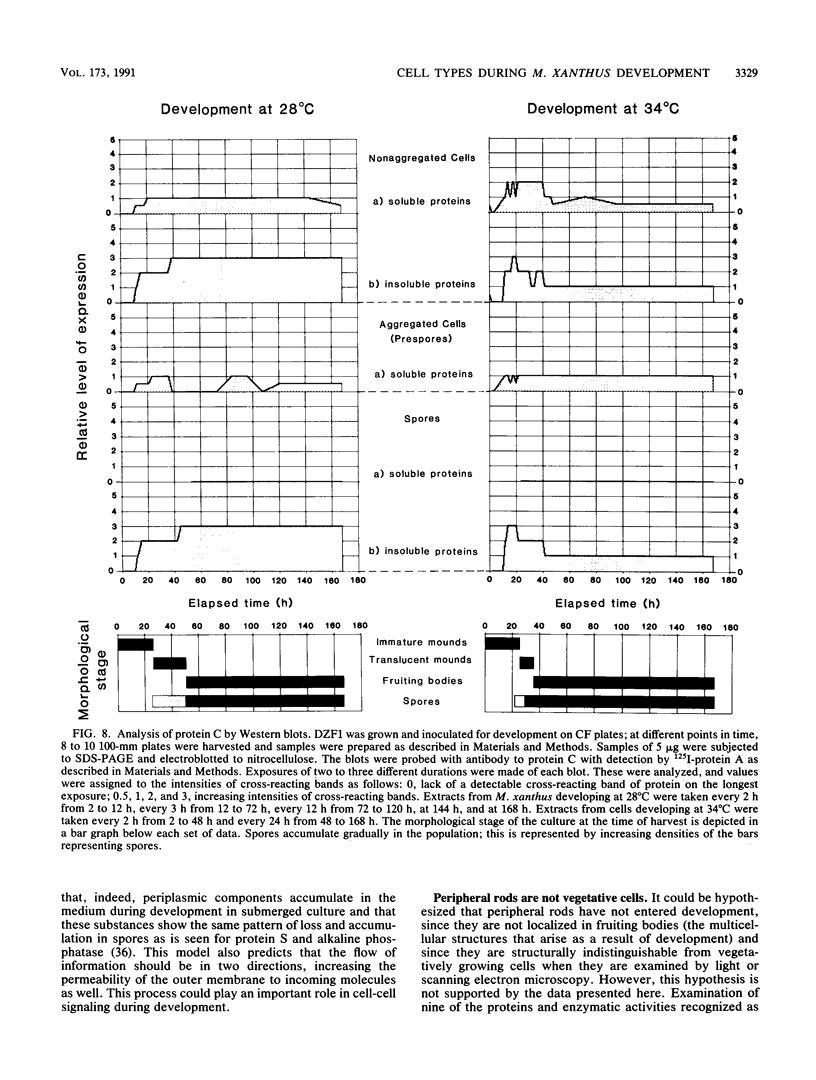
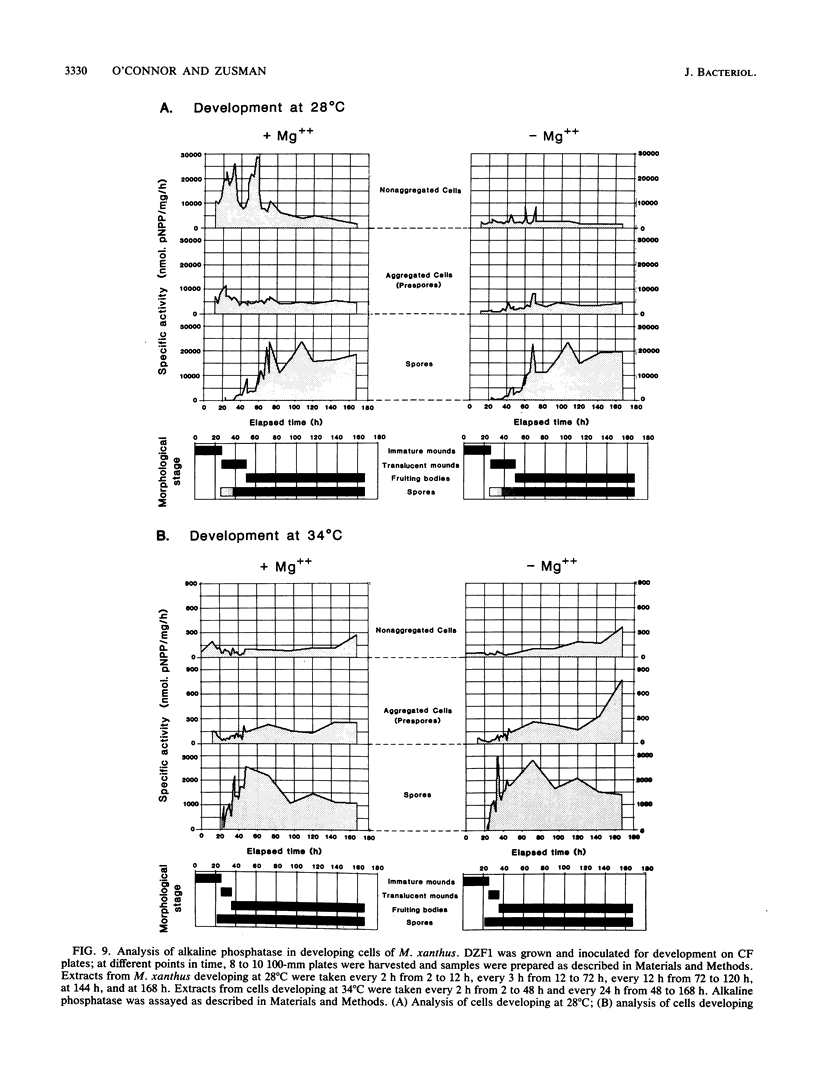



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blackhart B. D., Zusman D. R. "Frizzy" genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8767–8770. doi: 10.1073/pnas.82.24.8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Campos J. M., Zusman D. R. Regulation of development in Myxococcus xanthus: effect of 3':5'-cyclic AMP, ADP, and nutrition. Proc Natl Acad Sci U S A. 1975 Feb;72(2):518–522. doi: 10.1073/pnas.72.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumsky M., Zusman D. R. Myxobacterial hemagglutinin: a development-specific lectin of Myxococcus xanthus. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5505–5509. doi: 10.1073/pnas.76.11.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DWORKIN M. Nutritional requirements for vegetative growth of Myxococcus xanthus. J Bacteriol. 1962 Aug;84:250–257. doi: 10.1128/jb.84.2.250-257.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downard J. S., Kupfer D., Zusman D. R. Gene expression during development of Myxococcus xanthus. Analysis of the genes for protein S. J Mol Biol. 1984 Jun 5;175(4):469–492. doi: 10.1016/0022-2836(84)90180-3. [DOI] [PubMed] [Google Scholar]
- Downard J. S., Zusman D. R. Differential expression of protein S genes during Myxococcus xanthus development. J Bacteriol. 1985 Mar;161(3):1146–1155. doi: 10.1128/jb.161.3.1146-1155.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin M., Kaiser D. Cell interactions in myxobacterial growth and development. Science. 1985 Oct 4;230(4721):18–24. doi: 10.1126/science.3929384. [DOI] [PubMed] [Google Scholar]
- Hagen D. C., Bretscher A. P., Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol. 1978 Jun;64(2):284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- Huang J. H., Schell M. A. DNA sequence analysis of pglA and mechanism of export of its polygalacturonase product from Pseudomonas solanacearum. J Bacteriol. 1990 Jul;172(7):3879–3887. doi: 10.1128/jb.172.7.3879-3887.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M., Inouye S., Zusman D. R. Biosynthesis and self-assembly of protein S, a development-specific protein of Myxococcus xanthus. Proc Natl Acad Sci U S A. 1979 Jan;76(1):209–213. doi: 10.1073/pnas.76.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M., Inouye S., Zusman D. R. Gene expression during development of Myxococcus xanthus: pattern of protein synthesis. Dev Biol. 1979 Feb;68(2):579–591. doi: 10.1016/0012-1606(79)90228-8. [DOI] [PubMed] [Google Scholar]
- Inouye S., Franceschini T., Inouye M. Structural similarities between the development-specific protein S from a gram-negative bacterium, Myxococcus xanthus, and calmodulin. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6829–6833. doi: 10.1073/pnas.80.22.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen G. R., Wireman J. W., Dworkin M. Effect of temperature on the growth of Myxococcus xanthus. J Bacteriol. 1977 Apr;130(1):561–562. doi: 10.1128/jb.130.1.561-562.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroos L., Kuspa A., Kaiser D. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev Biol. 1986 Sep;117(1):252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Legocki R. P., Verma D. P. Multiple immunoreplica Technique: screening for specific proteins with a series of different antibodies using one polyacrylamide gel. Anal Biochem. 1981 Mar 1;111(2):385–392. doi: 10.1016/0003-2697(81)90577-7. [DOI] [PubMed] [Google Scholar]
- Matin A., Auger E. A., Blum P. H., Schultz J. E. Genetic basis of starvation survival in nondifferentiating bacteria. Annu Rev Microbiol. 1989;43:293–316. doi: 10.1146/annurev.mi.43.100189.001453. [DOI] [PubMed] [Google Scholar]
- McBride M. J., Zusman D. R. Trehalose accumulation in vegetative cells and spores of Myxococcus xanthus. J Bacteriol. 1989 Nov;171(11):6383–6386. doi: 10.1128/jb.171.11.6383-6386.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleary W. R., Esmon B., Zusman D. R. Myxococcus xanthus protein C is a major spore surface protein. J Bacteriol. 1991 Mar;173(6):2141–2145. doi: 10.1128/jb.173.6.2141-2145.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. R., Cumsky M. G., Zusman D. R. Localization of myxobacterial hemagglutinin in the periplasmic space and on the cell surface of Myxococcus xanthus during developmental aggregation. J Biol Chem. 1981 Dec 10;256(23):12589–12595. [PubMed] [Google Scholar]
- Nelson D. R., Zusman D. R. Transport and localization of protein S, a spore coat protein, during fruiting body formation by Myxococcus xanthus. J Bacteriol. 1983 May;154(2):547–553. doi: 10.1128/jb.154.2.547-553.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor K. A., Zusman D. R. Analysis of Myxococcus xanthus cell types by two-dimensional polyacrylamide gel electrophoresis. J Bacteriol. 1991 Jun;173(11):3334–3341. doi: 10.1128/jb.173.11.3334-3341.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor K. A., Zusman D. R. Behavior of peripheral rods and their role in the life cycle of Myxococcus xanthus. J Bacteriol. 1991 Jun;173(11):3342–3355. doi: 10.1128/jb.173.11.3342-3355.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor K. A., Zusman D. R. Patterns of cellular interactions during fruiting-body formation in Myxococcus xanthus. J Bacteriol. 1989 Nov;171(11):6013–6024. doi: 10.1128/jb.171.11.6013-6024.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor K. A., Zusman D. R. Reexamination of the role of autolysis in the development of Myxococcus xanthus. J Bacteriol. 1988 Sep;170(9):4103–4112. doi: 10.1128/jb.170.9.4103-4112.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo J. M., Zusman D. R. Cloning of the gene for myxobacterial hemagglutinin and isolation and analysis of structural gene mutations. J Bacteriol. 1987 Aug;169(8):3801–3808. doi: 10.1128/jb.169.8.3801-3808.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets L. J. Social and developmental biology of the myxobacteria. Microbiol Rev. 1990 Dec;54(4):473–501. doi: 10.1128/mr.54.4.473-501.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teintze M., Thomas R., Furuichi T., Inouye M., Inouye S. Two homologous genes coding for spore-specific proteins are expressed at different times during development of Myxococcus xanthus. J Bacteriol. 1985 Jul;163(1):121–125. doi: 10.1128/jb.163.1.121-125.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelz H., Ortigoza R. O. Cytochemistry of phosphatases in Myxococcus xanthus. J Bacteriol. 1968 Oct;96(4):1357–1365. doi: 10.1128/jb.96.4.1357-1365.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg R. A., Zusman D. R. Alkaline, acid, and neutral phosphatase activities are induced during development in Myxococcus xanthus. J Bacteriol. 1990 May;172(5):2294–2302. doi: 10.1128/jb.172.5.2294-2302.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wireman J. W., Dworkin M. Morphogenesis and developmental interactions in myxobacteria. Science. 1975 Aug 15;189(4202):516–523. doi: 10.1126/science.806967. [DOI] [PubMed] [Google Scholar]