Abstract
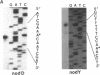
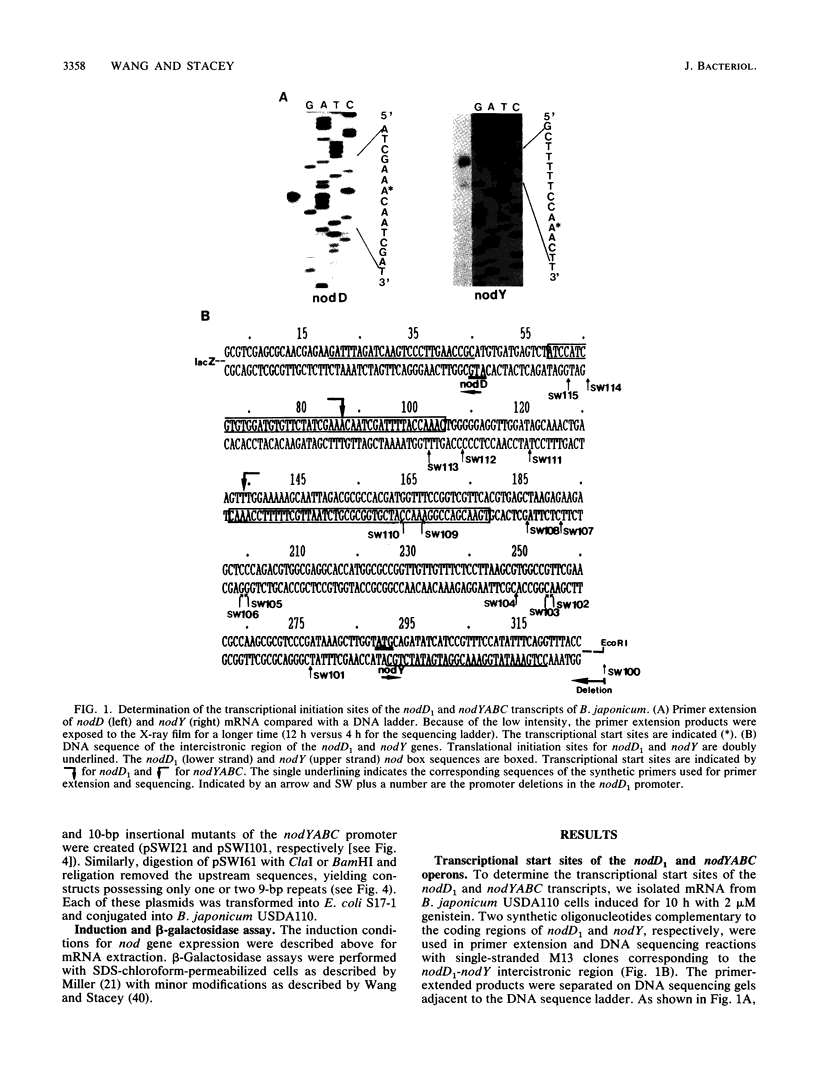
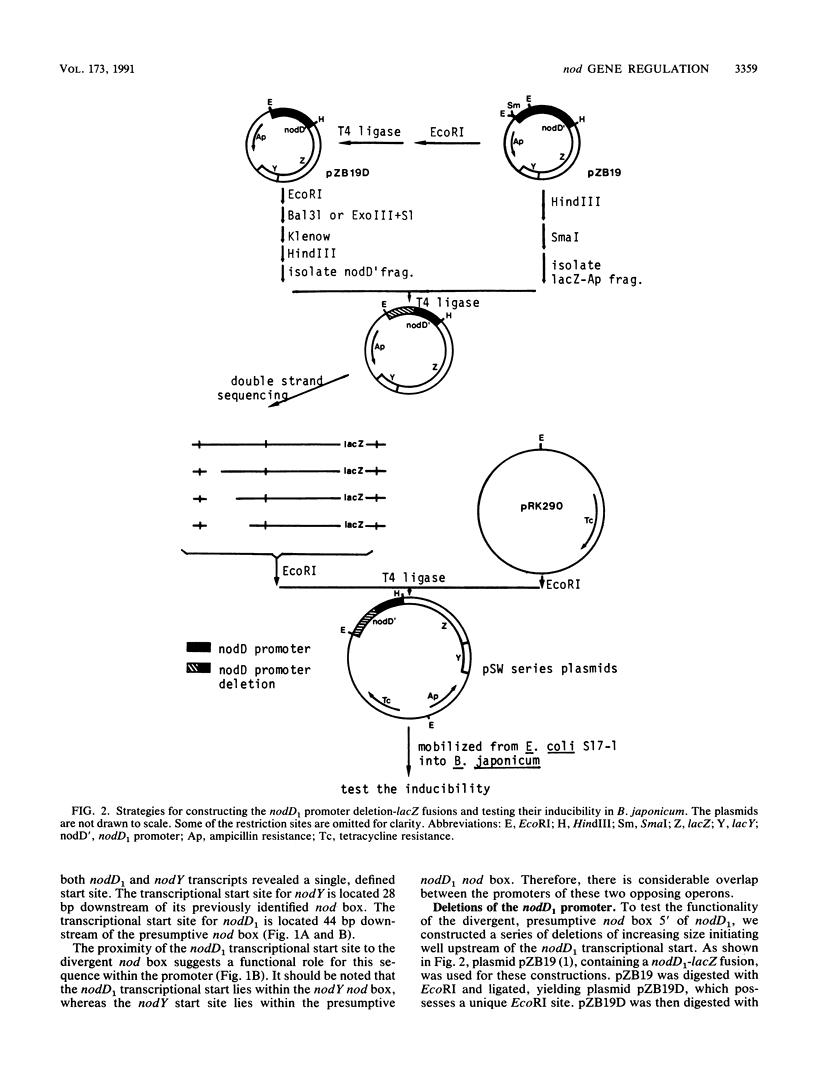
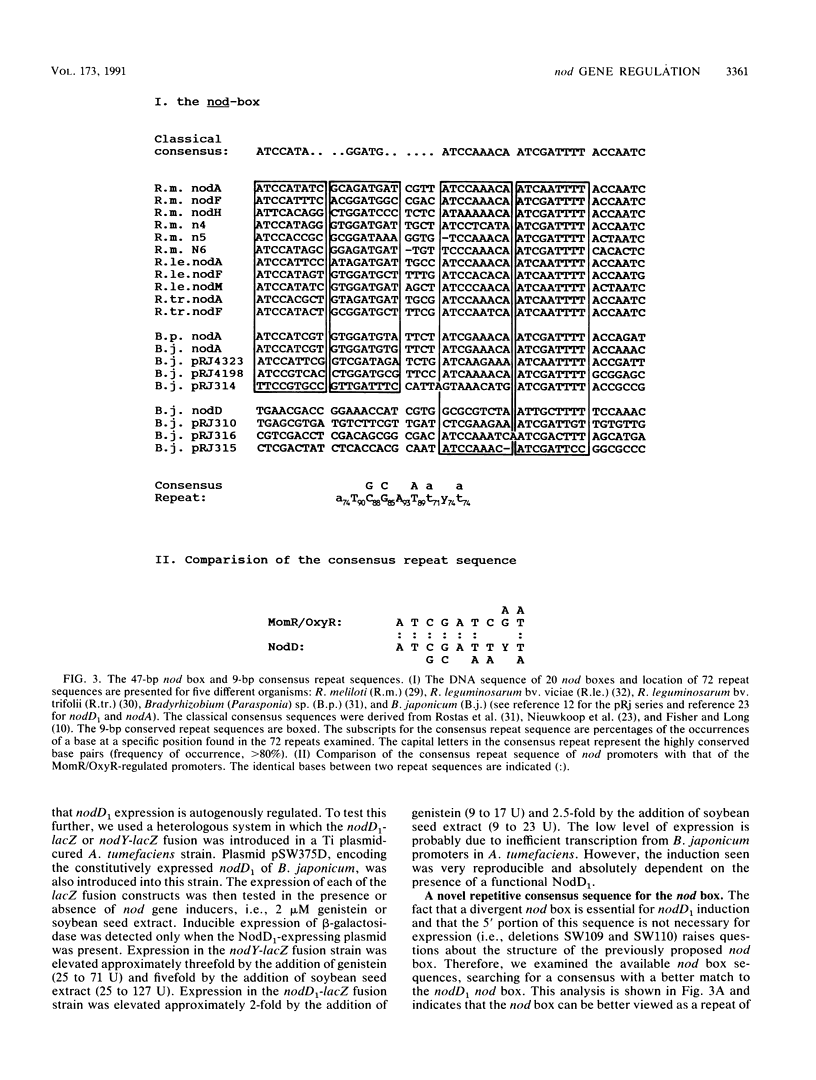
Induction of nod genes in Rhizobium and Bradyrhizobium species is dependent on the presence of plant-produced flavonoids, the NodD protein, and the cis-acting nod box promoter sequence. Although the nodD (nodD1) gene in Rhizobium species is constitutively expressed, nodD1 expression in Bradyrhizobium japonicum is inducible by isoflavones in a manner similar to that of the nodYABC operon. A consensus nod box sequence is found 5' of the nodYABC operon, whereas a presumptive, nod box-like sequence is found 5' of the nodD1 gene. As an initial step toward examining the nodD1 promoter, the transcriptional start sites of the nodD1 and nodYABC operons were determined and found to be 44 and 28 bp, respectively, downstream of their respective nod box sequences. A series of deletions of the nodD1 promoter were constructed and fused to the lacZ gene. Analysis of the activity of these deletions clearly showed that the divergent nod box sequence was essential for nodD1 induction by isoflavones or soybean seed extract. The induction of nodD1 expression requires NodD1, as tested in B. japonicum and in a heterologous system, Agrobacterium tumefaciens. On the basis of these data, we analyzed the published nod box sequences and propose a new consensus sequence composed of paired 9-bp repeats. Analysis of the nodD1 nod box and synthetic constructs of the nocYABC nod box indicate that at least two 9-bp repeats are required for NodD1-mediated induction. Furthermore, insertions between the paired repeats of the nodYABC nod box suggest that orientation of the repeats on opposite faces of the DNA helix is essential for maximum nod gene expression.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banfalvi Z., Nieuwkoop A., Schell M., Besl L., Stacey G. Regulation of nod gene expression in Bradyrhizobium japonicum. Mol Gen Genet. 1988 Nov;214(3):420–424. doi: 10.1007/BF00330475. [DOI] [PubMed] [Google Scholar]
- Bölker M., Kahmann R. The Escherichia coli regulatory protein OxyR discriminates between methylated and unmethylated states of the phage Mu mom promoter. EMBO J. 1989 Aug;8(8):2403–2410. doi: 10.1002/j.1460-2075.1989.tb08370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmane N., Stacey G. Identification of Bradyrhizobium nod genes involved in host-specific nodulation. J Bacteriol. 1989 Jun;171(6):3324–3330. doi: 10.1128/jb.171.6.3324-3330.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelhoff T. T., Fisher R. F., Jacobs T. W., Mulligan J. T., Long S. R. Nucleotide sequence of Rhizobium meliloti 1021 nodulation genes: nodD is read divergently from nodABC. DNA. 1985 Jun;4(3):241–248. doi: 10.1089/dna.1985.4.241. [DOI] [PubMed] [Google Scholar]
- Fisher R. F., Egelhoff T. T., Mulligan J. T., Long S. R. Specific binding of proteins from Rhizobium meliloti cell-free extracts containing NodD to DNA sequences upstream of inducible nodulation genes. Genes Dev. 1988 Mar;2(3):282–293. doi: 10.1101/gad.2.3.282. [DOI] [PubMed] [Google Scholar]
- Fisher R. F., Long S. R. DNA footprint analysis of the transcriptional activator proteins NodD1 and NodD3 on inducible nod gene promoters. J Bacteriol. 1989 Oct;171(10):5492–5502. doi: 10.1128/jb.171.10.5492-5502.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttfert M., Lamb J. W., Gasser R., Semenza J., Hennecke H. Mutational analysis of the Bradyrhizobium japonicum common nod genes and further nod box-linked genomic DNA regions. Mol Gen Genet. 1989 Feb;215(3):407–415. doi: 10.1007/BF00427037. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Haughn G. W., Calvo J. M., Wallace J. C. A large family of bacterial activator proteins. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6602–6606. doi: 10.1073/pnas.85.18.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong G. F., Burn J. E., Johnston A. W. Evidence that DNA involved in the expression of nodulation (nod) genes in Rhizobium binds to the product of the regulatory gene nodD. Nucleic Acids Res. 1987 Dec 10;15(23):9677–9690. doi: 10.1093/nar/15.23.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Geiduschek E. P. Bacteriophage T4 late promoters: mapping 5' ends of T4 gene 23 mRNAs. EMBO J. 1982;1(1):107–114. doi: 10.1002/j.1460-2075.1982.tb01132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S. R. Rhizobium-legume nodulation: life together in the underground. Cell. 1989 Jan 27;56(2):203–214. doi: 10.1016/0092-8674(89)90893-3. [DOI] [PubMed] [Google Scholar]
- Miller B. E., Kredich N. M. Purification of the cysB protein from Salmonella typhimurium. J Biol Chem. 1987 May 5;262(13):6006–6009. [PubMed] [Google Scholar]
- Mulligan J. T., Long S. R. Induction of Rhizobium meliloti nodC expression by plant exudate requires nodD. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6609–6613. doi: 10.1073/pnas.82.19.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop A. J., Banfalvi Z., Deshmane N., Gerhold D., Schell M. G., Sirotkin K. M., Stacey G. A locus encoding host range is linked to the common nodulation genes of Bradyrhizobium japonicum. J Bacteriol. 1987 Jun;169(6):2631–2638. doi: 10.1128/jb.169.6.2631-2638.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noti J. D., Dudas B., Szalay A. A. Isolation and characterization of nodulation genes from Bradyrhizobium sp. (Vigna) strain IRc 78. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7379–7383. doi: 10.1073/pnas.82.21.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters N. K., Frost J. W., Long S. R. A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science. 1986 Aug 29;233(4767):977–980. doi: 10.1126/science.3738520. [DOI] [PubMed] [Google Scholar]
- Priefer U. B., Simon R., Pühler A. Extension of the host range of Escherichia coli vectors by incorporation of RSF1010 replication and mobilization functions. J Bacteriol. 1985 Jul;163(1):324–330. doi: 10.1128/jb.163.1.324-330.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossen L., Johnston A. W., Downie J. A. DNA sequence of the Rhizobium leguminosarum nodulation genes nodAB and C required for root hair curling. Nucleic Acids Res. 1984 Dec 21;12(24):9497–9508. doi: 10.1093/nar/12.24.9497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossen L., Shearman C. A., Johnston A. W., Downie J. A. The nodD gene of Rhizobium leguminosarum is autoregulatory and in the presence of plant exudate induces the nodA,B,C genes. EMBO J. 1985 Dec 16;4(13A):3369–3373. doi: 10.1002/j.1460-2075.1985.tb04092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostas K., Kondorosi E., Horvath B., Simoncsits A., Kondorosi A. Conservation of extended promoter regions of nodulation genes in Rhizobium. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1757–1761. doi: 10.1073/pnas.83.6.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield P. R., Watson J. M. DNA sequence of Rhizobium trifolii nodulation genes reveals a reiterated and potentially regulatory sequence preceding nodABC and nodFE. Nucleic Acids Res. 1986 Apr 11;14(7):2891–2903. doi: 10.1093/nar/14.7.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K. F. Conserved nodulation genes from the non-legume symbiont Bradyrhizobium sp. (Parasponia). Nucleic Acids Res. 1986 Apr 11;14(7):2905–2919. doi: 10.1093/nar/14.7.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman C. A., Rossen L., Johnston A. W., Downie J. A. The Rhizobium leguminosarum nodulation gene nodF encodes a polypeptide similar to acyl-carrier protein and is regulated by nodD plus a factor in pea root exudate. EMBO J. 1986 Apr;5(4):647–652. doi: 10.1002/j.1460-2075.1986.tb04262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So J. S., Hodgson A. L., Haugland R., Leavitt M., Banfalvi Z., Nieuwkoop A. J., Stacey G. Transposon-induced symbiotic mutants of Bradyrhizobium japonicum: isolation of two gene regions essential for nodulation. Mol Gen Genet. 1987 Apr;207(1):15–23. doi: 10.1007/BF00331485. [DOI] [PubMed] [Google Scholar]
- Török I., Kondorosi E., Stepkowski T., Pósfai J., Kondorosi A. Nucleotide sequence of Rhizobium meliloti nodulation genes. Nucleic Acids Res. 1984 Dec 21;12(24):9509–9524. doi: 10.1093/nar/12.24.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. P., Stacey G. Ammonia regulation of nod genes in Bradyrhizobium japonicum. Mol Gen Genet. 1990 Sep;223(2):329–331. doi: 10.1007/BF00265071. [DOI] [PubMed] [Google Scholar]
- Wek R. C., Hatfield G. W. Transcriptional activation at adjacent operators in the divergent-overlapping ilvY and ilvC promoters of Escherichia coli. J Mol Biol. 1988 Oct 5;203(3):643–663. doi: 10.1016/0022-2836(88)90199-4. [DOI] [PubMed] [Google Scholar]
- de Maagd R. A., Wijfjes A. H., Spaink H. P., Ruiz-Sainz J. E., Wijffelman C. A., Okker R. J., Lugtenberg B. J. nodO, a new nod gene of the Rhizobium leguminosarum biovar viciae sym plasmid pRL1JI, encodes a secreted protein. J Bacteriol. 1989 Dec;171(12):6764–6770. doi: 10.1128/jb.171.12.6764-6770.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]