Abstract
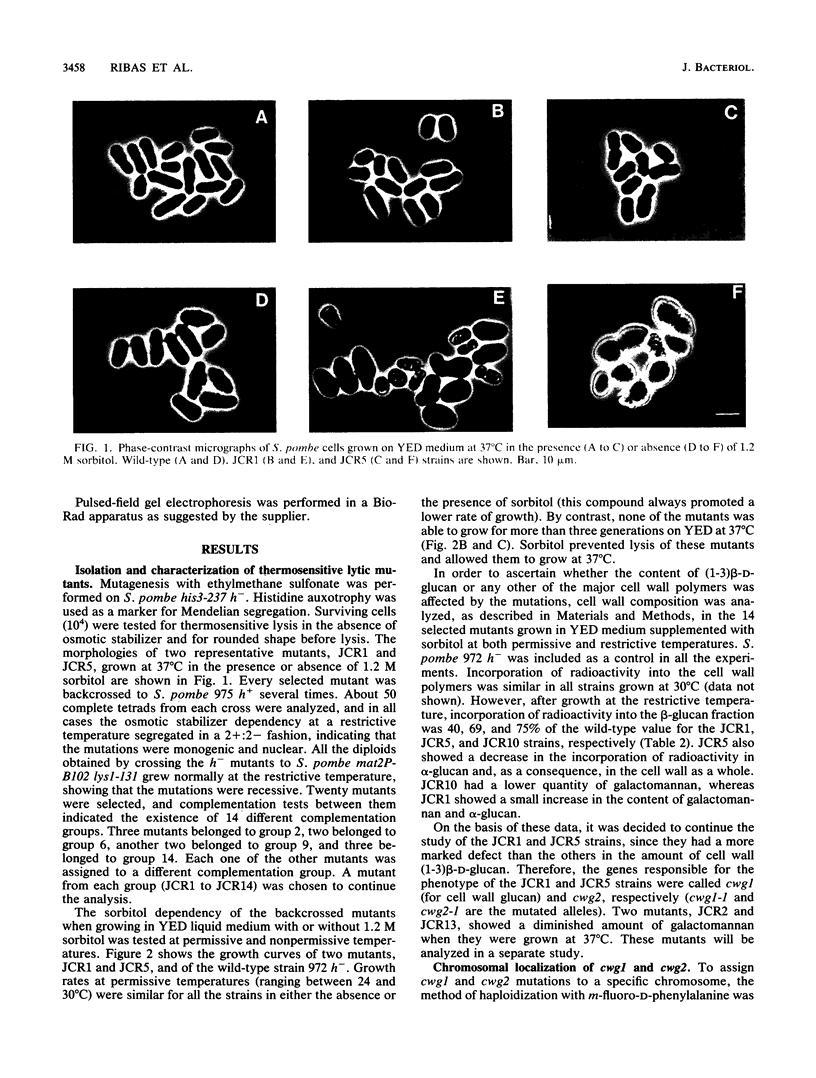
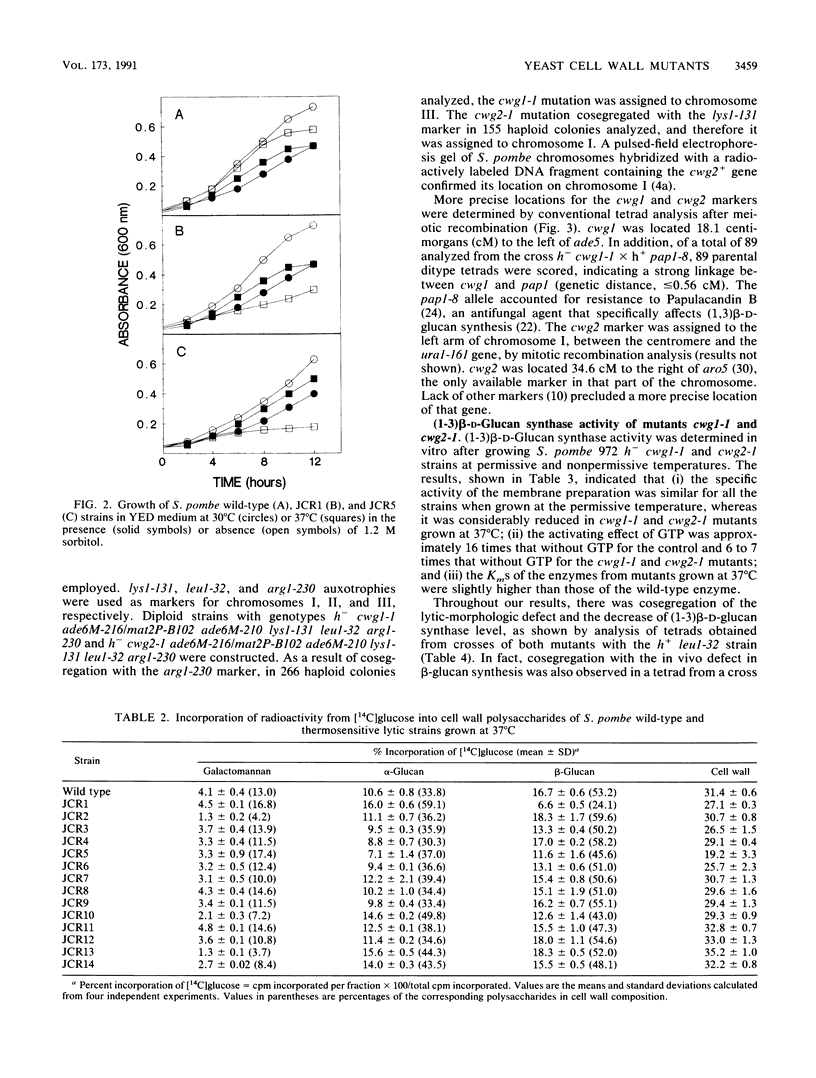
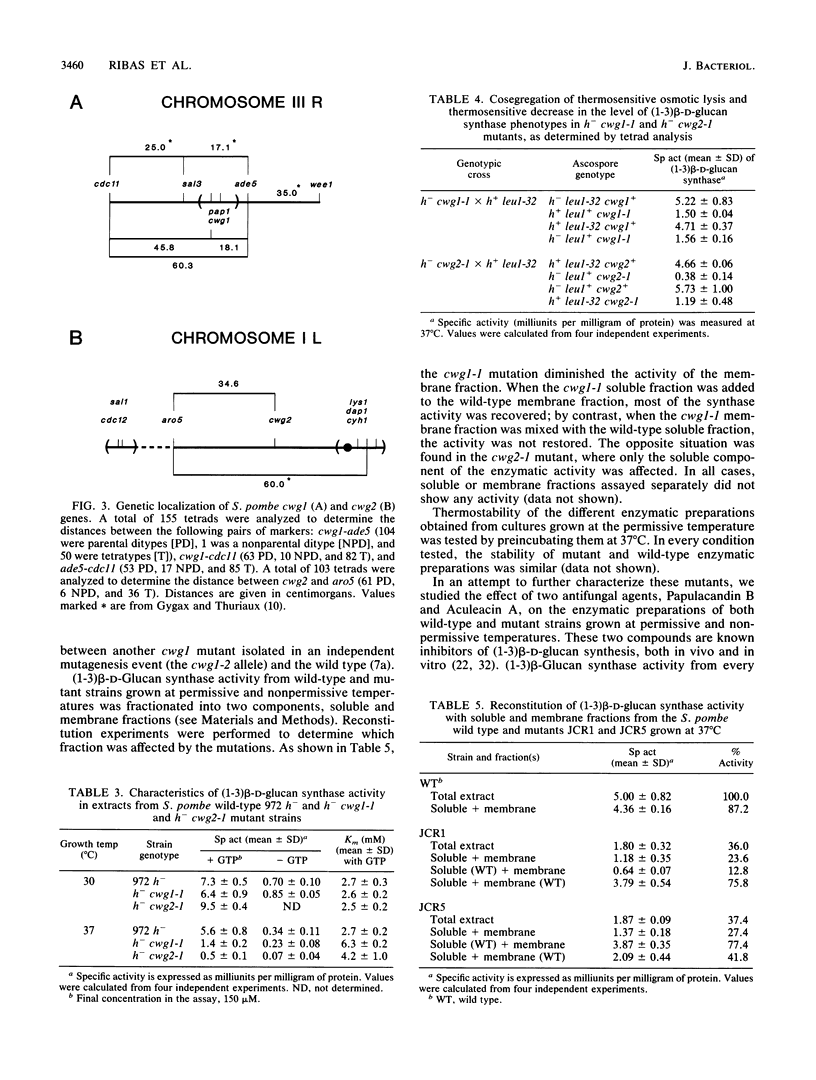
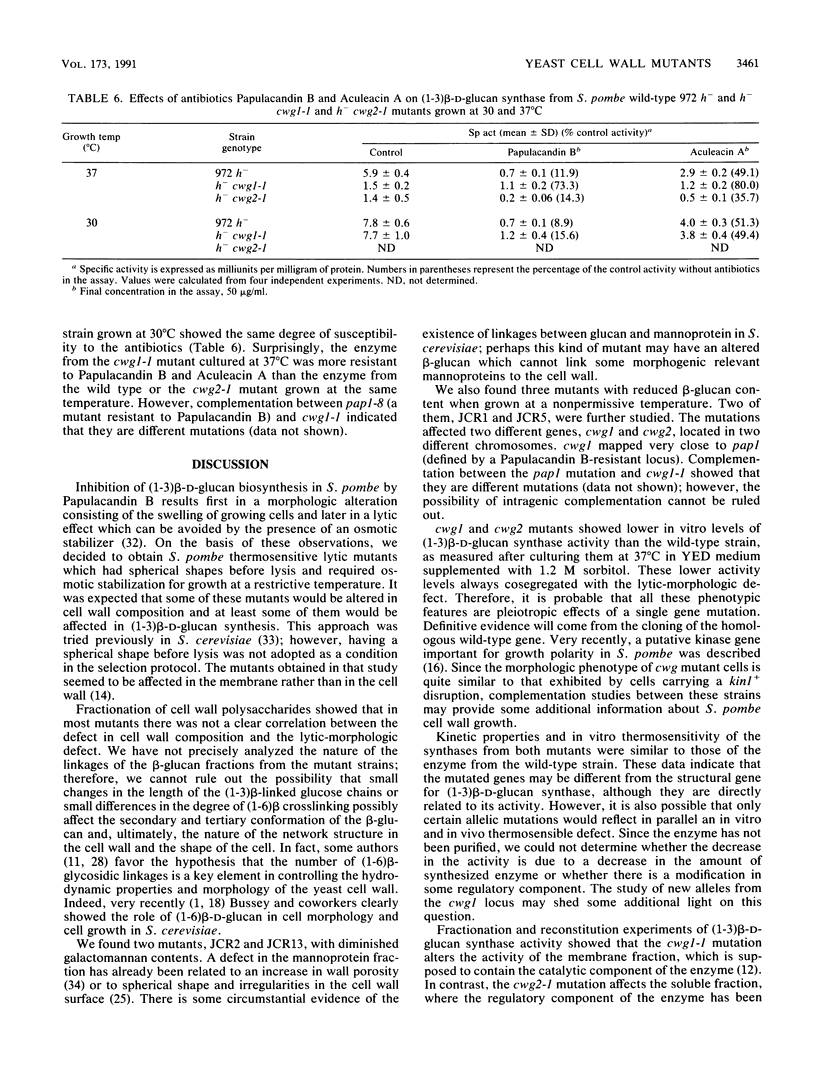
Schizosaccharomyces pombe thermosensitive mutants requiring the presence of an osmotic stabilizer to survive and grow at a nonpermissive temperature were isolated. The mutants were genetically and biochemically characterized. In all of them, the phenotype segregated in Mendelian fashion as a single gene which coded for a recessive character. Fourteen loci were defined by complementation analysis. Studies of cell wall composition showed a reduction in the amount of cell wall beta-glucan in three strains (JCR1, JCR5, and JCR10) when growing at 37 degrees C. Galactomannan was diminished in two others. Strains JCR1 and JCR5, with mutant alleles cwg1-1 and cwg2-1, respectively, were further studied. The cwg1 locus was mapped on the right arm of chromosome III, 18.06 centimorgans (cM) to the left of the ade5 marker; cwg2 was located on the left arm of chromosome I, 34.6 cM away from the aro5 marker. (1-3)beta-D-Glucan synthase activities from cwg1-1 and cwg2-1 mutant strains grown at 37 degrees C were diminished, as measured in vitro, compared with the wild-type strain; however, Km values and activation by GTP were similar to the wild-type values. Mutant synthases behaved like the wild-type enzyme in terms of thermostability. Analyses of round shape, lytic behavior, and low (1-3)beta-D-glucan synthase activity in cultures derived from ascospores of the same tetrad showed cosegregation of all these characters. Detergent dissociation of (1-3)beta-D-glucan synthase into soluble and particulate fractions and subsequent reconstitution demonstrated that the cwg1-1 mutant was affected in the particulate fraction of the enzymatic activity while cwg2-1 was affected in the soluble component. The antifungal agents Papulacandin B and Aculeacin A had similar effects on the enzymatic activities of the wild type and the cwg2-1 mutant strain, whereas the cwg1-1 mutant, when growing at 37 degrees C, had a more inhibitor-resistant (1,3)beta-D-glucan synthase. It is concluded that the cwg1+ and cwg2+ genes are related to (1,3)beta-D-glucan biosynthesis.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boone C., Sommer S. S., Hensel A., Bussey H. Yeast KRE genes provide evidence for a pathway of cell wall beta-glucan assembly. J Cell Biol. 1990 May;110(5):1833–1843. doi: 10.1083/jcb.110.5.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulawa C. E., Slater M., Cabib E., Au-Young J., Sburlati A., Adair W. L., Jr, Robbins P. W. The S. cerevisiae structural gene for chitin synthase is not required for chitin synthesis in vivo. Cell. 1986 Jul 18;46(2):213–225. doi: 10.1016/0092-8674(86)90738-5. [DOI] [PubMed] [Google Scholar]
- Cabib E., Roberts R., Bowers B. Synthesis of the yeast cell wall and its regulation. Annu Rev Biochem. 1982;51:763–793. doi: 10.1146/annurev.bi.51.070182.003555. [DOI] [PubMed] [Google Scholar]
- Deshaies R. J., Kepes F., Böhni P. C. Genetic dissection of the early stages of protein secretion in yeast. Trends Genet. 1989 Mar;5(3):87–93. doi: 10.1016/0168-9525(89)90032-2. [DOI] [PubMed] [Google Scholar]
- Gutz H. Induction of mitotic segregation with p-fluorophenylalanine in Schizosaccharomyces pombe. J Bacteriol. 1966 Nov;92(5):1567–1568. doi: 10.1128/jb.92.5.1567-1568.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M. S., Cabib E. Regulation of fungal cell wall growth: a guanine nucleotide-binding, proteinaceous component required for activity of (1----3)-beta-D-glucan synthase. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5808–5812. doi: 10.1073/pnas.83.16.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli J., Hottinger H., Munz P., Strauss A., Thuriaux P. Genetic Mapping in SCHIZOSACCHAROMYCES POMBE by Mitotic and Meiotic Analysis and Induced Haploidization. Genetics. 1977 Nov;87(3):471–489. doi: 10.1093/genetics/87.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotyk A., Venkov P., Dvoráková M. Membrane transport in an osmotically fragile mutant of Saccharomyces cerevisiae. Yeast. 1988 Dec;4(4):241–247. doi: 10.1002/yea.320040402. [DOI] [PubMed] [Google Scholar]
- Kukuruzinska M. A., Bergh M. L., Jackson B. J. Protein glycosylation in yeast. Annu Rev Biochem. 1987;56:915–944. doi: 10.1146/annurev.bi.56.070187.004411. [DOI] [PubMed] [Google Scholar]
- Levin D. E., Bishop J. M. A putative protein kinase gene (kin1+) is important for growth polarity in Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8272–8276. doi: 10.1073/pnas.87.21.8272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaden P., Hill K., Wagner J., Slipetz D., Sommer S. S., Bussey H. The yeast KRE5 gene encodes a probable endoplasmic reticulum protein required for (1----6)-beta-D-glucan synthesis and normal cell growth. Mol Cell Biol. 1990 Jun;10(6):3013–3019. doi: 10.1128/mcb.10.6.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai S., Mortimer R. K. Studies on the genetic mechanism of radiation-induced mitotic segregation in yeast. Mol Gen Genet. 1969;103(4):329–338. doi: 10.1007/BF00383483. [DOI] [PubMed] [Google Scholar]
- Notario V., Kawai H., Cabib E. Interaction between yeast beta-(1 goes to 3)glucan synthetase and activating phosphorylated compounds. A kinetic study. J Biol Chem. 1982 Feb 25;257(4):1902–1905. [PubMed] [Google Scholar]
- Perkins D. D. Biochemical Mutants in the Smut Fungus Ustilago Maydis. Genetics. 1949 Sep;34(5):607–626. doi: 10.1093/genetics/34.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shematek E. M., Braatz J. A., Cabib E. Biosynthesis of the yeast cell wall. I. Preparation and properties of beta-(1 leads to 3)glucan synthetase. J Biol Chem. 1980 Feb 10;255(3):888–894. [PubMed] [Google Scholar]
- Shematek E. M., Cabib E. Biosynthesis of the yeast cell wall. II. Regulation of beta-(1 leads to 3)glucan synthetase by ATP and GTP. J Biol Chem. 1980 Feb 10;255(3):895–902. [PubMed] [Google Scholar]
- Shiota M., Nakajima T., Satoh A., Shida M., Matsuda K. Comparison of beta-glucan structures in a cell wall mutant of Saccharomyces cerevisiae and the wild type. J Biochem. 1985 Nov;98(5):1301–1307. doi: 10.1093/oxfordjournals.jbchem.a135397. [DOI] [PubMed] [Google Scholar]
- Silverman S. J., Sburlati A., Slater M. L., Cabib E. Chitin synthase 2 is essential for septum formation and cell division in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4735–4739. doi: 10.1073/pnas.85.13.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szaniszlo P. J., Kang M. S., Cabib E. Stimulation of beta(1----3)glucan synthetase of various fungi by nucleoside triphosphates: generalized regulatory mechanism for cell wall biosynthesis. J Bacteriol. 1985 Mar;161(3):1188–1194. doi: 10.1128/jb.161.3.1188-1194.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkov P. V., Hadjiolov A. A., Battaner E., Schlessinger D. Saccharomyces cerevisiae: sorbitol-dependent fragile mutants. Biochem Biophys Res Commun. 1974 Feb 4;56(3):599–604. doi: 10.1016/0006-291x(74)90646-9. [DOI] [PubMed] [Google Scholar]
- Zlotnik H., Fernandez M. P., Bowers B., Cabib E. Saccharomyces cerevisiae mannoproteins form an external cell wall layer that determines wall porosity. J Bacteriol. 1984 Sep;159(3):1018–1026. doi: 10.1128/jb.159.3.1018-1026.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]



