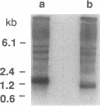
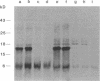
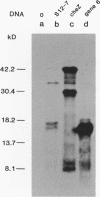
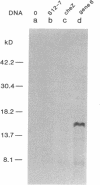
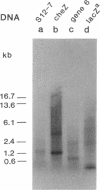
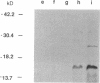
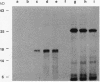
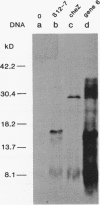
Abstract
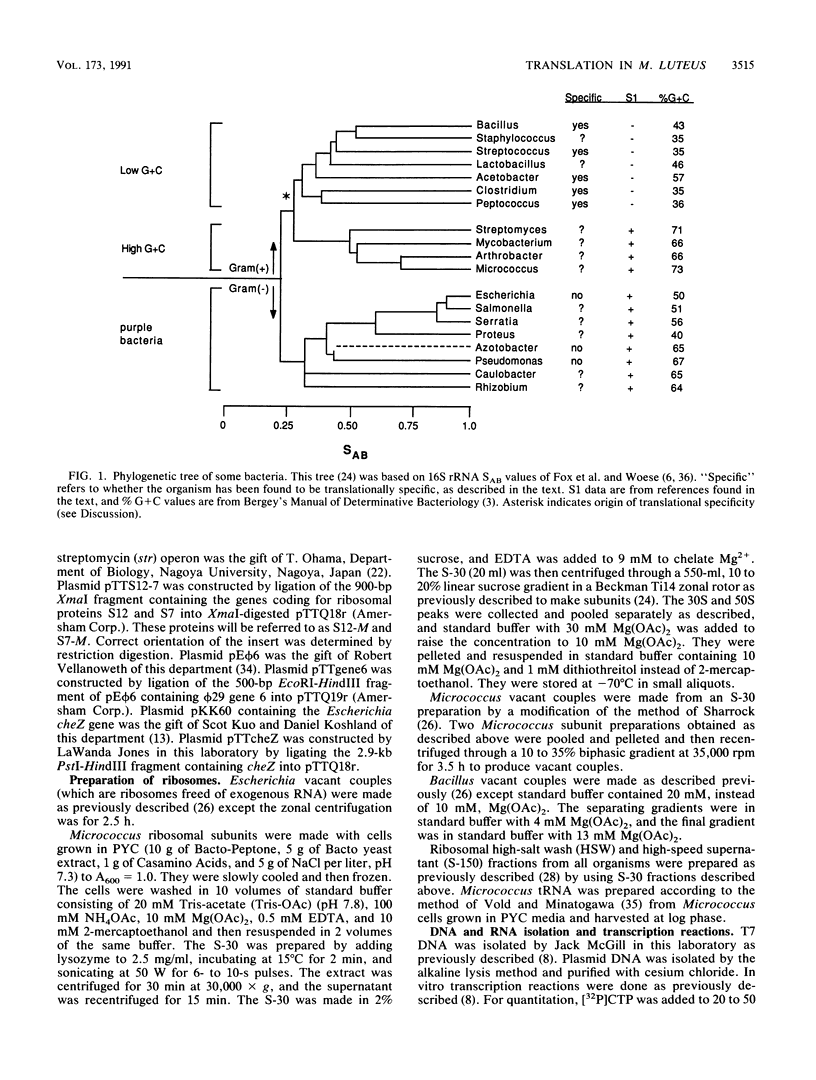
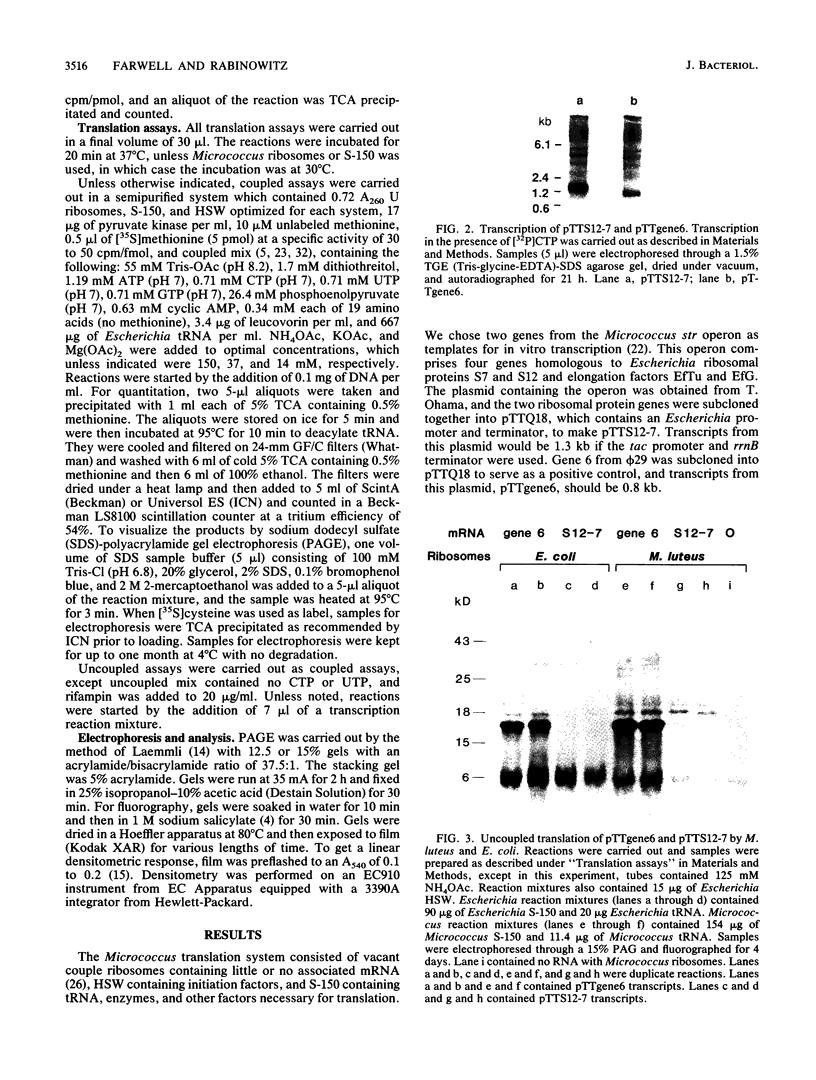
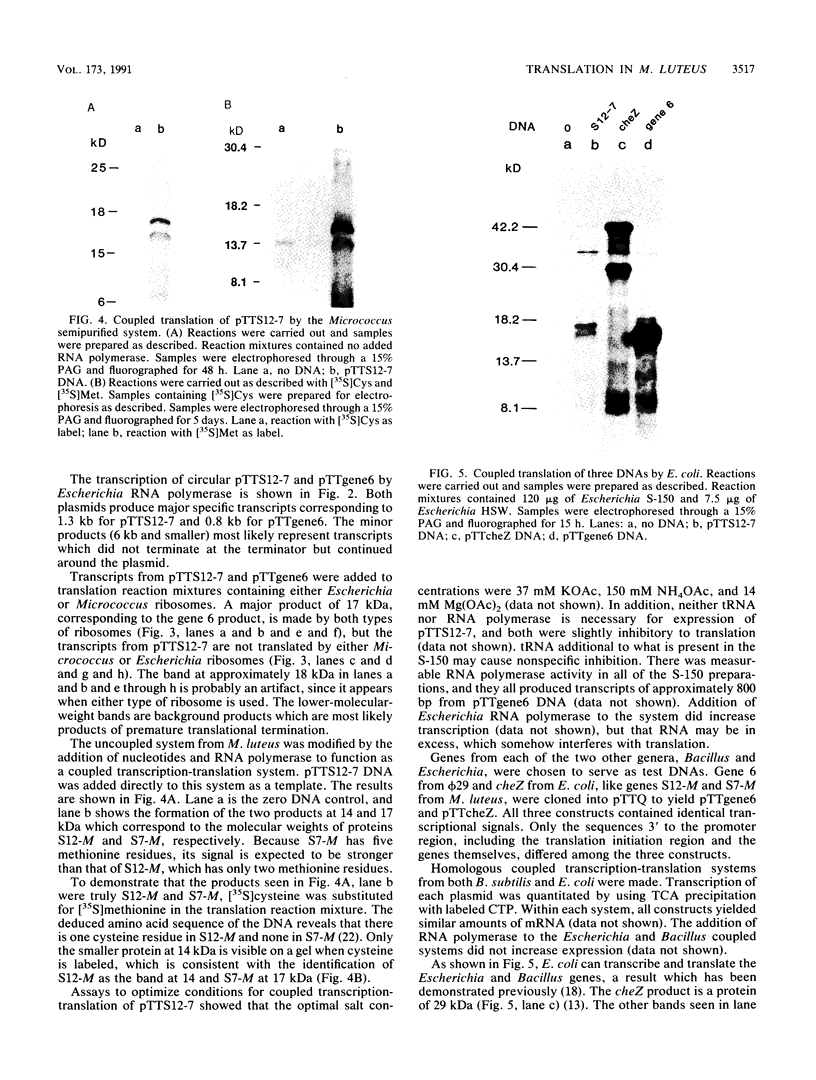
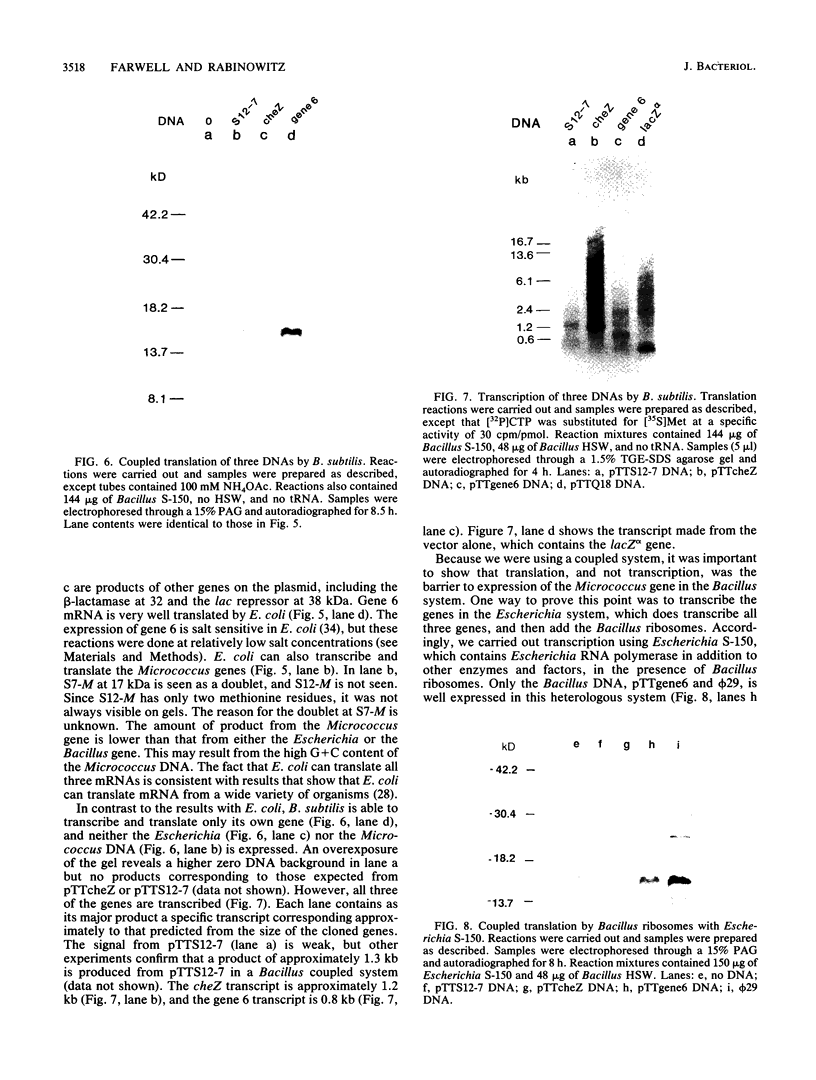
Bacillus subtilis and related gram-positive bacteria which have low to moderate genomic G + C contents are unable to efficiently translate mRNA derived from gram-negative bacteria, whereas Escherichia coli and other gram-negative bacteria are able to translate mRNA from both types of organisms. This phenomenon has been termed translational species specificity. Ribosomes from the low-G + C-content group (low-G + C group) of gram-positive organisms (B. subtilis and relatives) lack an equivalent to Escherichia ribosomal protein S1. The requirement for S1 for translation in E. coli (G. van Dieijen, P. H. van Knippenberg, J. van Duin, B. Koekman, and P. H. Pouwels, Mol. Gen. Genet. 153:75-80, 1977) and its specific role (A.R. Subramanian, Trends Biochem. Sci. 9:491-494, 1984) have been proposed. The group of gram-positive bacteria characterized by high genomic G + C content (formerly Actinomyces species and relatives) contain S1, in contrast to the low-G + C group (K. Mikulik, J. Smardova, A. Jiranova, and P. Branny, Eur. J. Biochem. 155:557-563, 1986). It is not known whether members of the high-G + C group are translationally specific, although there is evidence that one genus, Streptomyces, can express Escherichia genes in vivo (M. J. Bibb and S. N. Cohen, Mol. Gen. Genet. 187:265-277, 1985; J. L. Schottel, M. J. Bibb, and S. N. Cohen, J. Bacteriol. 146:360-368, 1981). In order to determine whether the organisms of this group are translationally specific, we examined the in vitro translational characteristics of a member of the high-G + C group, Micrococcus luteus, whose genomic G + C content is 73%. A semipurified coupled transcription-translation system of M. luteus translates Escherichia mRNA as well as Bacillus and Micrococcus mRNA. Therefore, M. luteus is translationally nonspecific and resembles E. coli rather than B. subtilis in its translational characteristics.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams C. W., Fornwald J. A., Schmidt F. J., Rosenberg M., Brawner M. E. Gene organization and structure of the Streptomyces lividans gal operon. J Bacteriol. 1988 Jan;170(1):203–212. doi: 10.1128/jb.170.1.203-212.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M. J., Cohen S. N. Gene expression in Streptomyces: construction and application of promoter-probe plasmid vectors in Streptomyces lividans. Mol Gen Genet. 1982;187(2):265–277. doi: 10.1007/BF00331128. [DOI] [PubMed] [Google Scholar]
- Chamberlain J. P. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979 Sep 15;98(1):132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- Collins J. Cell-free synthesis of proteins coding for mobilisation functions of ColE1 and transposition functions of Tn3. Gene. 1979 May;6(1):29–42. doi: 10.1016/0378-1119(79)90083-0. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Stackebrandt E., Hespell R. B., Gibson J., Maniloff J., Dyer T. A., Wolfe R. S., Balch W. E., Tanner R. S., Magrum L. J. The phylogeny of prokaryotes. Science. 1980 Jul 25;209(4455):457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- Hager P. W., Rabinowitz J. C. Inefficient translation of T7 late mRNA by Bacillus subtilis ribosomes. Implications for species-specific translation. J Biol Chem. 1985 Dec 5;260(28):15163–15167. [PubMed] [Google Scholar]
- Himes R. H., Stallcup M. R., Rabinowitz J. C. Translation of synthetic and endogenous messenger ribonucleic acid in vitro by ribosomes and polyribosomes from Clostridium pasteurianum. J Bacteriol. 1972 Dec;112(3):1057–1069. doi: 10.1128/jb.112.3.1057-1069.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Furuya K., Nishiyama M., Suzuki H., Beppu T. Nucleotide sequence of the streptothricin acetyltransferase gene from Streptomyces lavendulae and its expression in heterologous hosts. J Bacteriol. 1987 May;169(5):1929–1937. doi: 10.1128/jb.169.5.1929-1937.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo S. C., Koshland D. E., Jr Roles of cheY and cheZ gene products in controlling flagellar rotation in bacterial chemotaxis of Escherichia coli. J Bacteriol. 1987 Mar;169(3):1307–1314. doi: 10.1128/jb.169.3.1307-1314.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur J Biochem. 1975 Aug 15;56(2):335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- Legault-Demare L., Chambliss G. H. Natural messenger ribonucleic acid-directed cell-free protein-synthesizing system of Bacillus subtilis. J Bacteriol. 1974 Dec;120(3):1300–1307. doi: 10.1128/jb.120.3.1300-1307.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H. F. Secondary structure of bacteriophage f2 ribonucleic acid and the initiation of in vitro protein biosynthesis. J Mol Biol. 1970 Jun 28;50(3):689–702. doi: 10.1016/0022-2836(70)90093-8. [DOI] [PubMed] [Google Scholar]
- Mikulík K., Smardová J., Jiránová A., Branny P. Molecular and functional properties of protein SS1 from small ribosomal subunits of Streptomyces aureofaciens. Eur J Biochem. 1986 Mar 17;155(3):557–563. doi: 10.1111/j.1432-1033.1986.tb09524.x. [DOI] [PubMed] [Google Scholar]
- Muralikrishna P., Suryanarayana T. Comparison of ribosomes from gram-positive and gram-negative bacteria with respect to the presence of protein S1. Biochem Int. 1985 Nov;11(5):691–699. [PubMed] [Google Scholar]
- Ohama T., Muto A., Osawa S. Spectinomycin operon of Micrococcus luteus: evolutionary implications of organization and novel codon usage. J Mol Evol. 1989 Nov;29(5):381–395. doi: 10.1007/BF02602908. [DOI] [PubMed] [Google Scholar]
- Ohama T., Yamao F., Muto A., Osawa S. Organization and codon usage of the streptomycin operon in Micrococcus luteus, a bacterium with a high genomic G + C content. J Bacteriol. 1987 Oct;169(10):4770–4777. doi: 10.1128/jb.169.10.4770-4777.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt J. M., Boulnois G. J., Darby V., Orr E., Wahle E., Holland I. B. Identification of gene products programmed by restriction endonuclease DNA fragments using an E. coli in vitro system. Nucleic Acids Res. 1981 Sep 25;9(18):4459–4474. doi: 10.1093/nar/9.18.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M. W., Rabinowitz J. C. The effect of Escherichia coli ribosomal protein S1 on the translational specificity of bacterial ribosomes. J Biol Chem. 1989 Feb 5;264(4):2228–2235. [PubMed] [Google Scholar]
- Schottel J. L., Bibb M. J., Cohen S. N. Cloning and expression in streptomyces lividans of antibiotic resistance genes derived from Escherichia coli. J Bacteriol. 1981 Apr;146(1):360–368. doi: 10.1128/jb.146.1.360-368.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock W. J., Rabinowitz J. C. Protein synthesis in Bacillus subtilis. I. Hydrodynamics and in vitro functional properties of ribosomes from B. subtilis W168. J Mol Biol. 1979 Dec 15;135(3):611–626. doi: 10.1016/0022-2836(79)90167-0. [DOI] [PubMed] [Google Scholar]
- Shiota S., Nakayama H. Evidence for a Micrococcus luteus gene homologous to uvrB of Escherichia coli. Mol Gen Genet. 1988 Jul;213(1):21–29. doi: 10.1007/BF00333393. [DOI] [PubMed] [Google Scholar]
- Stallcup M. R., Rabinowitz J. C. Initiation of protein synthesis in vitro by a clostridial system. I. Specificity in the translation of natural messenger ribonucleic acids. J Biol Chem. 1973 May 10;248(9):3209–3215. [PubMed] [Google Scholar]
- Stallcup M. R., Sharrock W. J., Rabinowitz J. C. Specificity of bacterial ribosomes and messenger ribonucleic acids in protein synthesis reactions in vitro. J Biol Chem. 1976 Apr 25;251(8):2499–2510. [PubMed] [Google Scholar]
- Thompson C. J., Gray G. S. Nucleotide sequence of a streptomycete aminoglycoside phosphotransferase gene and its relationship to phosphotransferases encoded by resistance plasmids. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5190–5194. doi: 10.1073/pnas.80.17.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J., Rae S., Cundliffe E. Coupled transcription--translation in extracts of Streptomyces lividans. Mol Gen Genet. 1984;195(1-2):39–43. doi: 10.1007/BF00332721. [DOI] [PubMed] [Google Scholar]
- Vold B., Minatogawa S. Comparison of procedures for extracting transfer RNA from spores of Bacillus. Arch Biochem Biophys. 1972 Mar;149(1):62–68. doi: 10.1016/0003-9861(72)90299-8. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dieijen G., van Knippenberg P. H., van Duin J., Koekman B., Pouwels P. H. The effect of the ribosomal protein S1 from Escherichia coli on the synthesis in vitro of bacterial-, DNA phage- and RNA phage proteins. Mol Gen Genet. 1977 May 20;153(1):75–80. doi: 10.1007/BF01035998. [DOI] [PubMed] [Google Scholar]