Abstract
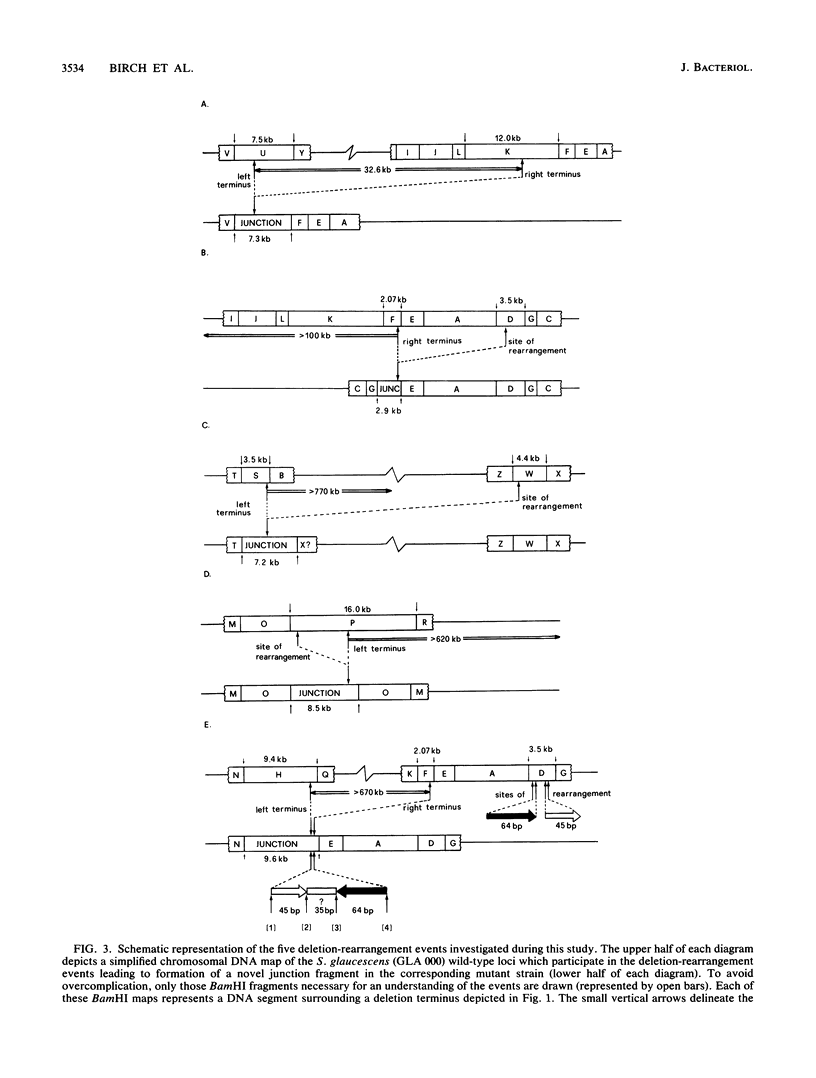
The Streptomyces glaucescens genome frequently undergoes gross genomic rearrangement events which result in the deletion of extremely large segments of chromosomal DNA. The structure and origin of the DNA forming the novel junctions arising from five of these deletion events are described. Only one junction proved to be the result of a relatively simple event; the remainder were more complex, with one involving DNA which originated from at least five distinct loci. In three of the investigated cases, DNA sequences present in the junctions appeared to have resulted from the duplication of previously unique sequences, suggesting that duplication of chromosomal segments may be an important factor in genetic instability. The nucleotide sequences surrounding these junctions and their respective wild-type termini were determined.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Betzler M., Dyson P., Schrempf H. Relationship of an unstable argG gene to a 5.7-kilobase amplifiable DNA sequence in Streptomyces lividans 66. J Bacteriol. 1987 Oct;169(10):4804–4810. doi: 10.1128/jb.169.10.4804-4810.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch A., Häusler A., Hütter R. Genome rearrangement and genetic instability in Streptomyces spp. J Bacteriol. 1990 Aug;172(8):4138–4142. doi: 10.1128/jb.172.8.4138-4142.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch A., Häusler A., Vögtli M., Krek W., Hütter R. Extremely large chromosomal deletions are intimately involved in genetic instability and genomic rearrangements in Streptomyces glaucescens. Mol Gen Genet. 1989 Jun;217(2-3):447–458. doi: 10.1007/BF02464916. [DOI] [PubMed] [Google Scholar]
- Boccard F., Smokvina T., Pernodet J. L., Friedmann A., Guérineau M. Structural analysis of loci involved in pSAM2 site-specific integration in Streptomyces. Plasmid. 1989 Jan;21(1):59–70. doi: 10.1016/0147-619x(89)90087-5. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- DasGupta U., Weston-Hafer K., Berg D. E. Local DNA sequence control of deletion formation in Escherichia coli plasmid pBR322. Genetics. 1987 Jan;115(1):41–49. doi: 10.1093/genetics/115.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson P., Schrempf H. Genetic instability and DNA amplification in Streptomyces lividans 66. J Bacteriol. 1987 Oct;169(10):4796–4803. doi: 10.1128/jb.169.10.4796-4803.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P. J., Schmeissner U., Hofer M., Miller J. H. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 1978 Dec 25;126(4):847–857. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Flett F., Cullum J. DNA deletions in spontaneous chloramphenicol-sensitive mutants of Streptomyces coelicolor A 3(2) and Streptomyces lividans 66. Mol Gen Genet. 1987 May;207(2-3):499–502. doi: 10.1007/BF00331621. [DOI] [PubMed] [Google Scholar]
- Frischauf A. M., Lehrach H., Poustka A., Murray N. Lambda replacement vectors carrying polylinker sequences. J Mol Biol. 1983 Nov 15;170(4):827–842. doi: 10.1016/s0022-2836(83)80190-9. [DOI] [PubMed] [Google Scholar]
- Glickman B. W., Ripley L. S. Structural intermediates of deletion mutagenesis: a role for palindromic DNA. Proc Natl Acad Sci U S A. 1984 Jan;81(2):512–516. doi: 10.1073/pnas.81.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M., Hintermann G., Simonet J. M., Crameri R., Piret J., Hütter R. Certain chromosomal regions in Streptomyces glaucescens tend to carry amplifications and deletions. Mol Gen Genet. 1985;200(3):375–384. doi: 10.1007/BF00425720. [DOI] [PubMed] [Google Scholar]
- Hintermann G., Crameri R., Vögtli M., Hütter R. Streptomycin-sensitivity in Streptomyces glaucescens is due to deletions comprising the structural gene coding for a specific phosphotransferase. Mol Gen Genet. 1984;196(3):513–520. doi: 10.1007/BF00436201. [DOI] [PubMed] [Google Scholar]
- Häusler A., Birch A., Krek W., Piret J., Hütter R. Heterogeneous genomic amplification in Streptomyces glaucescens: structure, location and junction sequence analysis. Mol Gen Genet. 1989 Jun;217(2-3):437–446. doi: 10.1007/BF02464915. [DOI] [PubMed] [Google Scholar]
- Ikeda H., Aoki K., Naito A. Illegitimate recombination mediated in vitro by DNA gyrase of Escherichia coli: structure of recombinant DNA molecules. Proc Natl Acad Sci U S A. 1982 Jun;79(12):3724–3728. doi: 10.1073/pnas.79.12.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H., Kawasaki I., Gellert M. Mechanism of illegitimate recombination: common sites for recombination and cleavage mediated by E. coli DNA gyrase. Mol Gen Genet. 1984;196(3):546–549. doi: 10.1007/BF00436208. [DOI] [PubMed] [Google Scholar]
- Innis M. A., Myambo K. B., Gelfand D. H., Brow M. A. DNA sequencing with Thermus aquaticus DNA polymerase and direct sequencing of polymerase chain reaction-amplified DNA. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9436–9440. doi: 10.1073/pnas.85.24.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall K. J., Cohen S. N. Complete nucleotide sequence of the Streptomyces lividans plasmid pIJ101 and correlation of the sequence with genetic properties. J Bacteriol. 1988 Oct;170(10):4634–4651. doi: 10.1128/jb.170.10.4634-4651.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhstoss S., Richardson M. A., Rao R. N. Site-specific integration in Streptomyces ambofaciens: localization of integration functions in S. ambofaciens plasmid pSAM2. J Bacteriol. 1989 Jan;171(1):16–23. doi: 10.1128/jb.171.1.16-23.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez P., Espinosa M., Greenberg B., Lacks S. A. Generation of deletions in pneumococcal mal genes cloned in Bacillus subtilis. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5189–5193. doi: 10.1073/pnas.81.16.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvo S. L., King S. R., Jaskunas S. R. Role of short regions of homology in intermolecular illegitimate recombination events. Proc Natl Acad Sci U S A. 1983 May;80(9):2452–2456. doi: 10.1073/pnas.80.9.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura-Masuda A., Ikeda H. The DNA gyrase of Escherichia coli participates in the formation of a spontaneous deletion by recA-independent recombination in vivo. Mol Gen Genet. 1990 Feb;220(3):345–352. doi: 10.1007/BF00391737. [DOI] [PubMed] [Google Scholar]
- Murray N. E., Brammar W. J., Murray K. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol Gen Genet. 1977 Jan 7;150(1):53–61. doi: 10.1007/BF02425325. [DOI] [PubMed] [Google Scholar]
- Nakano M. M., Ogawara H., Sekiya T. Recombination between short direct repeats in Streptomyces lavendulae plasmid DNA. J Bacteriol. 1984 Feb;157(2):658–660. doi: 10.1128/jb.157.2.658-660.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Passananti C., Davies B., Ford M., Fried M. Structure of an inverted duplication formed as a first step in a gene amplification event: implications for a model of gene amplification. EMBO J. 1987 Jun;6(6):1697–1703. doi: 10.1002/j.1460-2075.1987.tb02420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair R. B., Bibb M. J. The repressor gene (c) of the Streptomyces temperate phage phi c31: nucleotide sequence, analysis and functional cloning. Mol Gen Genet. 1988 Aug;213(2-3):269–277. doi: 10.1007/BF00339591. [DOI] [PubMed] [Google Scholar]
- Sosio M., Madoń J., Hütter R. Excision of pIJ408 from the chromosome of Streptomyces glaucescens and its transfer into Streptomyces lividans. Mol Gen Genet. 1989 Jul;218(1):169–176. doi: 10.1007/BF00330580. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]