Abstract
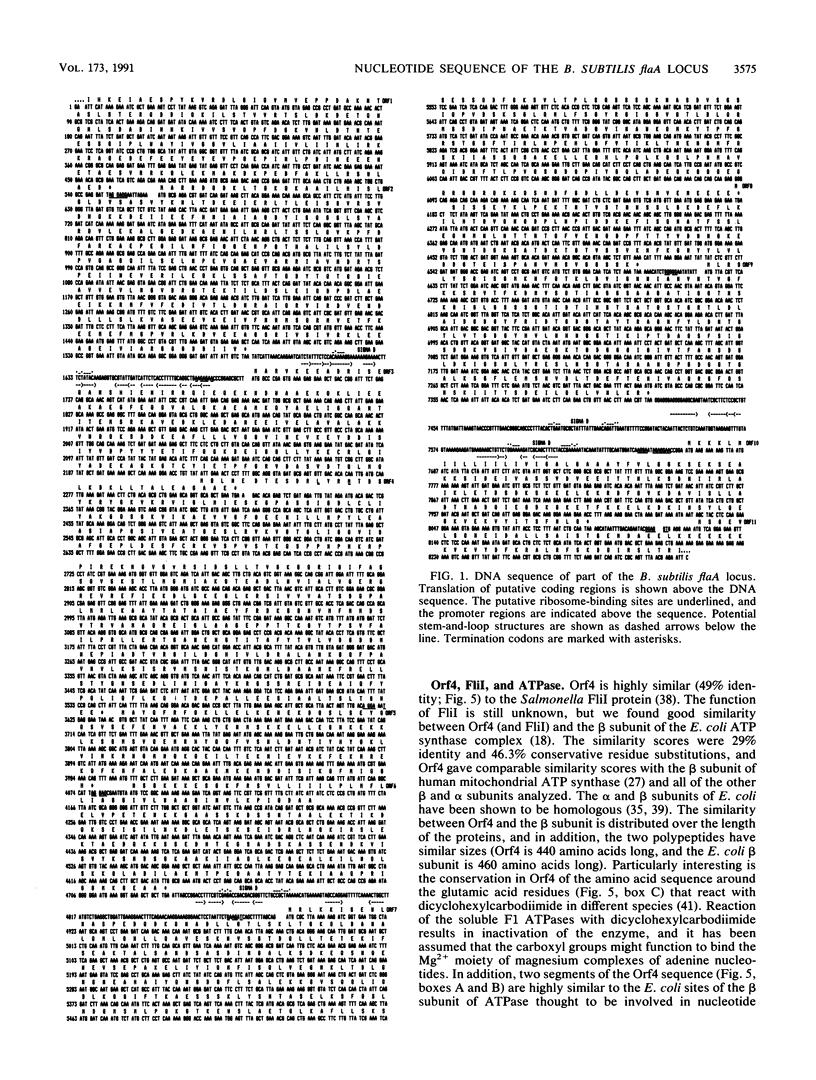
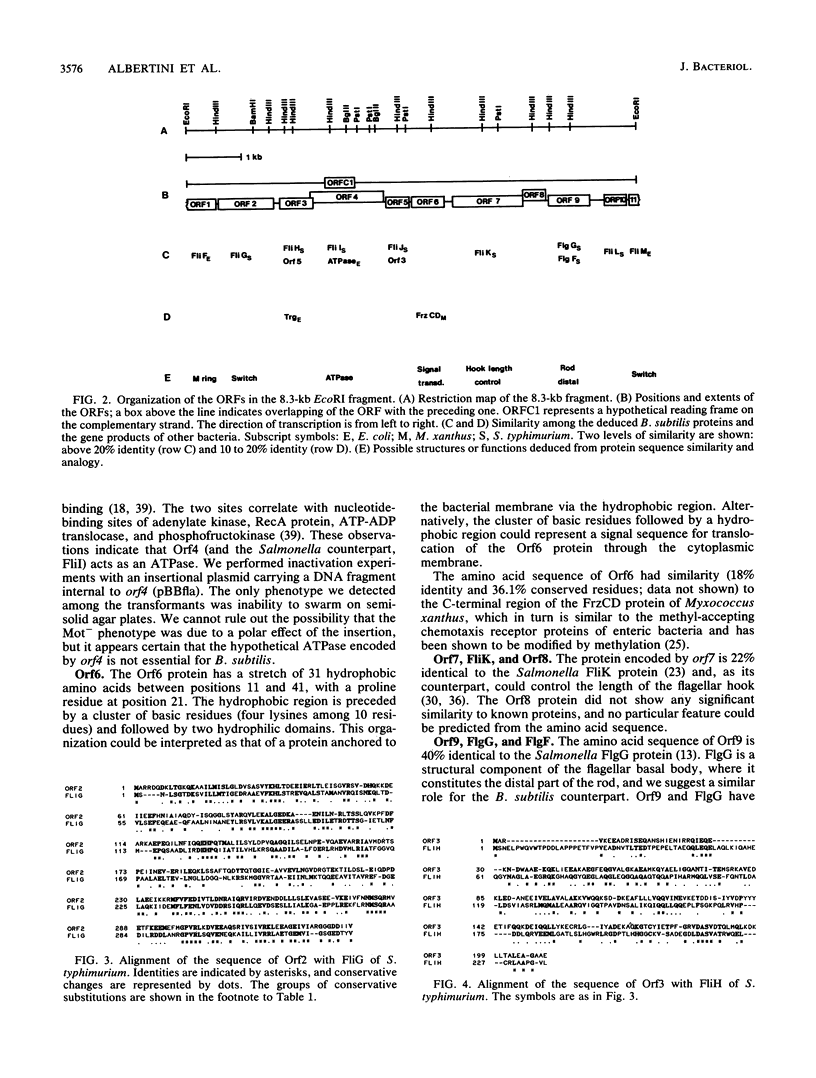
We cloned and sequenced 8.3 kb of Bacillus subtilis DNA corresponding to the flaA locus involved in flagellar biosynthesis, motility, and chemotaxis. The DNA sequence revealed the presence of 10 complete and 2 incomplete open reading frames. Comparison of the deduced amino acid sequences to data banks showed similarities of nine of the deduced products to a number of proteins of Escherichia coli and Salmonella typhimurium for which a role in flagellar functioning has been directly demonstrated. In particular, the sequence data suggest that the flaA operon codes for the M-ring protein, components of the motor switch, and the distal part of the basal-body rod. The gene order is remarkably similar to that described for region III of the enterobacterial flagellar regulon. One of the open reading frames was translated into a protein with 48% amino acid identity to S. typhimurium FliI and 29% identity to the beta subunit of E. coli ATP synthase.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bollinger J., Park C., Harayama S., Hazelbauer G. L. Structure of the Trg protein: Homologies with and differences from other sensory transducers of Escherichia coli. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3287–3291. doi: 10.1073/pnas.81.11.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Emerson S. U., Tokuyasu K., Simon M. I. Bacterial flagella: polarity of elongation. Science. 1970 Jul 10;169(3941):190–192. doi: 10.1126/science.169.3941.190. [DOI] [PubMed] [Google Scholar]
- Ferrari E., Henner D. J., Hoch J. A. Isolation of Bacillus subtilis genes from a charon 4A library. J Bacteriol. 1981 Apr;146(1):430–432. doi: 10.1128/jb.146.1.430-432.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Gianni M., Galizzi A. Isolation of genes preferentially expressed during Bacillis subtilis spore outgrowth. J Bacteriol. 1986 Jan;165(1):123–132. doi: 10.1128/jb.165.1.123-132.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant G. F., Simon M. I. Synthesis of bacterial flagella. II. PBS1 transduction of flagella-specific markers in Bacillus subtilis. J Bacteriol. 1969 Jul;99(1):116–124. doi: 10.1128/jb.99.1.116-124.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser P. M., Crabb W. D., Fiora M. G., Scoffone F., Galizzi A. Genetic analysis of the flaA locus of Bacillus subtilis. J Bacteriol. 1991 Jun;173(11):3580–3583. doi: 10.1128/jb.173.11.3580-3583.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988 Dec 15;73(1):237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Homma M., Iino T., Macnab R. M. Identification and characterization of the products of six region III flagellar genes (flaAII.3 through flaQII) of Salmonella typhimurium. J Bacteriol. 1988 May;170(5):2221–2228. doi: 10.1128/jb.170.5.2221-2228.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., Kutsukake K., Hasebe M., Iino T., Macnab R. M. FlgB, FlgC, FlgF and FlgG. A family of structurally related proteins in the flagellar basal body of Salmonella typhimurium. J Mol Biol. 1990 Jan 20;211(2):465–477. doi: 10.1016/0022-2836(90)90365-S. [DOI] [PubMed] [Google Scholar]
- Iino T., Komeda Y., Kutsukake K., Macnab R. M., Matsumura P., Parkinson J. S., Simon M. I., Yamaguchi S. New unified nomenclature for the flagellar genes of Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1988 Dec;52(4):533–535. doi: 10.1128/mr.52.4.533-535.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino T. Polarity of flagellar growth in salmonella. J Gen Microbiol. 1969 May;56(2):227–239. doi: 10.1099/00221287-56-2-227. [DOI] [PubMed] [Google Scholar]
- Jones C. J., Homma M., Macnab R. M. L-, P-, and M-ring proteins of the flagellar basal body of Salmonella typhimurium: gene sequences and deduced protein sequences. J Bacteriol. 1989 Jul;171(7):3890–3900. doi: 10.1128/jb.171.7.3890-3900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. J., Macnab R. M. Flagellar assembly in Salmonella typhimurium: analysis with temperature-sensitive mutants. J Bacteriol. 1990 Mar;172(3):1327–1339. doi: 10.1128/jb.172.3.1327-1339.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa H., Kayano T., Kiyasu T., Futai M. Nucleotide sequence of the genes for beta and epsilon subunits of proton-translocating ATPase from Escherichia coli. Biochem Biophys Res Commun. 1982 Apr 29;105(4):1257–1264. doi: 10.1016/0006-291x(82)90922-6. [DOI] [PubMed] [Google Scholar]
- Kihara M., Homma M., Kutsukake K., Macnab R. M. Flagellar switch of Salmonella typhimurium: gene sequences and deduced protein sequences. J Bacteriol. 1989 Jun;171(6):3247–3257. doi: 10.1128/jb.171.6.3247-3257.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo S. C., Koshland D. E., Jr Sequence of the flaA (cheC) locus of Escherichia coli and discovery of a new gene. J Bacteriol. 1986 Jun;166(3):1007–1012. doi: 10.1128/jb.166.3.1007-1012.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malakooti J., Komeda Y., Matsumura P. DNA sequence analysis, gene product identification, and localization of flagellar motor components of Escherichia coli. J Bacteriol. 1989 May;171(5):2728–2734. doi: 10.1128/jb.171.5.2728-2734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleary W. R., McBride M. J., Zusman D. R. Developmental sensory transduction in Myxococcus xanthus involves methylation and demethylation of FrzCD. J Bacteriol. 1990 Sep;172(9):4877–4887. doi: 10.1128/jb.172.9.4877-4887.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirel D. B., Chamberlin M. J. The Bacillus subtilis flagellin gene (hag) is transcribed by the sigma 28 form of RNA polymerase. J Bacteriol. 1989 Jun;171(6):3095–3101. doi: 10.1128/jb.171.6.3095-3101.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta S., Tomura H., Matsuda K., Kagawa Y. Gene structure of the human mitochondrial adenosine triphosphate synthase beta subunit. J Biol Chem. 1988 Aug 15;263(23):11257–11262. [PubMed] [Google Scholar]
- Ordal G. W., Nettleton D. O., Hoch J. A. Genetics of Bacillus subtilis chemotaxis: isolation and mapping of mutations and cloning of chemotaxis genes. J Bacteriol. 1983 Jun;154(3):1088–1097. doi: 10.1128/jb.154.3.1088-1097.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordal G. W., Parker H. M., Kirby J. R. Complementation and characterization of chemotaxis mutants of Bacillus subtilis. J Bacteriol. 1985 Nov;164(2):802–810. doi: 10.1128/jb.164.2.802-810.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson-Delafield J., Martinez R. J., Stocker B. A., Yamaguchi S. A new fla gene in Salmonella typhimurium--flaR--and its mutant phenotype-superhooks. Arch Mikrobiol. 1973 Mar 26;90(2):107–120. doi: 10.1007/BF00414513. [DOI] [PubMed] [Google Scholar]
- Pooley H. M., Karamata D. Genetic analysis of autolysin-deficient and flagellaless mutants of Bacillus subtilis. J Bacteriol. 1984 Dec;160(3):1123–1129. doi: 10.1128/jb.160.3.1123-1129.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste M., Gay N. J., Eberle A., Runswick M. J., Walker J. E. The atp operon: nucleotide sequence of the genes for the gamma, beta, and epsilon subunits of Escherichia coli ATP synthase. Nucleic Acids Res. 1981 Oct 24;9(20):5287–5296. doi: 10.1093/nar/9.20.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Iino T. Role of the flaR gene in flagellar hook formation in Salmonella spp. J Bacteriol. 1981 Dec;148(3):973–979. doi: 10.1128/jb.148.3.973-979.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoelke M. S., Casper J. M., Ordal G. W. Methyl group turnover on methyl-accepting chemotaxis proteins during chemotaxis by Bacillus subtilis. J Biol Chem. 1990 Feb 5;265(4):1928–1932. [PubMed] [Google Scholar]
- Vogler A. P., Homma M., Irikura V. M., Macnab R. M. Salmonella typhimurium mutants defective in flagellar filament regrowth and sequence similarity of FliI to F0F1, vacuolar, and archaebacterial ATPase subunits. J Bacteriol. 1991 Jun;173(11):3564–3572. doi: 10.1128/jb.173.11.3564-3572.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying C. W., Scoffone F., Albertini A. M., Galizzi A., Ordal G. W. Properties of the Bacillus subtilis chemotaxis protein CheF, a homolog of the Salmonella typhimurium flagellar protein FliJ. J Bacteriol. 1991 Jun;173(11):3584–3586. doi: 10.1128/jb.173.11.3584-3586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Allison W. S., Esch F. S., Futai M. The specificity of carboxyl group modification during the inactivation of the Escherichia coli F1-ATPase with dicyclohexyl[14C]carbodiimide. J Biol Chem. 1982 Sep 10;257(17):10033–10037. [PubMed] [Google Scholar]
- Zuberi A. R., Bischoff D. S., Ordal G. W. Nucleotide sequence and characterization of a Bacillus subtilis gene encoding a flagellar switch protein. J Bacteriol. 1991 Jan;173(2):710–719. doi: 10.1128/jb.173.2.710-719.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuberi A. R., Ying C. W., Weinreich M. R., Ordal G. W. Transcriptional organization of a cloned chemotaxis locus of Bacillus subtilis. J Bacteriol. 1990 Apr;172(4):1870–1876. doi: 10.1128/jb.172.4.1870-1876.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]



