Abstract
To examine the physiological role of the Fyn tyrosine kinase in neurons, we generated transgenic mice that expressed a fyn cDNA under the control of the calcium/calmodulin-dependent protein kinase IIα promoter. With this promoter, we detected only low expression of Fyn in the neonatal brain. In contrast, there was strong expression of the fyn-transgene in neurons of the adult forebrain. To determine whether the impairment of long-term potentiation (LTP) observed in adult fyn-deficient mice was caused directly by the lack of Fyn in adult hippocampal neurons or indirectly by an impairment in neuronal development, we generated fyn-rescue mice by introducing the wild-type fyn-transgene into mice carrying a targeted deletion in the endogenous fyn gene. In fyn-rescue mice, Schaffer collateral LTP was restored, even though the morphological abnormalities characteristic of fyn-deficient mice were still present. These results suggest that Fyn contributes, at least in part, to the molecular mechanisms of LTP induction.
Fyn, one of the Src-related non-receptor tyrosine kinases, is highly expressed in the central nervous system and the immune system. A physiological role for Fyn in brain function was first suggested by the analysis of fyn-deficient mice generated by homologous recombination. These mice exhibited an impaired long-term potentiation (LTP), a long-lasting enhancement of synaptic transmission that is thought to be the cellular basis for learning and memory, as well as a spatial learning impairment (1). Interestingly, in contrast to Fyn, the disruption of other members of the Src family kinases does not cause any obvious neurological phenotypes (2) or any alteration in LTP (1). These results suggest that Fyn signaling may contribute to neuronal plasticity. However, the analysis of the phenotype in fyn-deficient mice was complicated by the presence of a number of other neurological defects, including uncoordinated hippocampal architecture, reduced neural cell adhesion molecule-dependent neurite outgrowth, and decreased susceptibility to kindling (1, 3, 4). In addition, some other lines of fyn-deficient mice expressing a fyn–β-galactosidease fusion protein showed abnormal suckling behavior, impaired myelination, increased fearfulness, and enhanced seizure susceptibility (5–8). The observed pleiotropic effect of the lack of Fyn suggests that, in the brain, Fyn might be involved in several signal transduction pathways in a variety of cell types and at different developmental stages.
To determine whether Fyn is directly involved in the signaling pathway required for LTP induction or if the phenotype observed is solely due to the malformation of the hippocampus, we attempted to rescue Fyn expression in the fyn-deficient mice by expressing fyn cDNA on a fyn-deficient genetic background. For this purpose, we used the calcium/calmodulin-dependent protein kinase IIα (CaMKIIα) promoter so that the transgene is expressed postnatally only in the forebrain and in neurons and not in glial cells. LTP was normal in the hippocampus expressing transgenic Fyn, even though the morphological abnormalities were still present. These results suggest that impairment of LTP observed in adult fyn-deficient mice was caused directly by the lack of Fyn in adult hippocampal neurons, but not indirectly by an impairment in neuronal development.
MATERIALS AND METHODS
Generation of Fyn-Transgenic Mice.
An EcoRI–DraI fragment of fynB cDNA (pmBF; ref. 9) was inserted into the EcoRV site of the vector, pNN265 (provided by N. Nakanishi, Harvard University), which was constructed by inserting a hybrid intron consisting of an adenovirus splice donor and an IgG splice acceptor (10) in 5′ of the cloning site of the vector pcDNAI/Amp (Invitrogen) to give an efficient transgene expression. To add the CaMKIIα promoter to this construct, fynB cDNA fragment with the 5′ intron and simian virus 40 poly(A) signal sequences was excised using NotI and inserted into the pBluescript-based vector, pNN279 (provided by N. Nakanishi) into which the 8.5-kb fragment including the CaMKIIα promoter (11) was inserted. The transgene was excised from the vector using SalI, and was then gel purified. Transgenic mice were generated by microinjecting the transgene into the pronuclei of fertilized eggs collected from C57BL6/CBA/F1 mice; the injected eggs were then transferred into the oviduct of pseudopregnant females (12). Founders were screened by PCR of tail DNA using the transgene-specific primer pair. Mouse lines were established by crossing founders with the C57BL/6J × 129Sv hybrid strain of hetero- or homozygous fyn-deficient mice (13). Fyn-rescue mice were obtained by breeding animals heterozygote for fyn gene with transgenic F1 individuals.
Northern Blotting, Ribonuclease Protection Assay, and in Situ Hybridization.
Total RNA was prepared from mouse forebrain using RNA isolation kit (Stratagene). For Northern blotting, 10 μg of total RNA was loaded on a 1% denaturing formaldehyde agarose gel and transferred to a membrane (Duralose, Stratagene). The 252-bp fragment of 3′ non-coding sequence (probe 2, shown in Fig. 1A) and EcoRI–HindIII fragment of fynB cDNA were labeled with 32P and used as probes. For ribonuclease protection assay, a 617-bp fragment of 5′ sequence of the transgene (probe 1, shown in Fig. 1A), including transgene specific 5′ noncoding sequence and the part of fynB cDNA fragment, were subcloned into the vector, pGEM4Z (Promega). From the template DNA linearized at the EcoRI site, the antisense cRNA probe was synthesized using T7 RNA polymerase in the presence of [α-32P]UTP. Ribonuclease protection assay was performed using a commercially available kit (RPAII kit, Ambion, Austin, TX). Five micrograms of total RNA was hybridized with 0.3 ng cRNA probe (specific activity, 0.5 × 108 cpm/μg cRNA) at 45°C for 16–24 hr, and then digested with a mixture of RNase A and T1. The protected bands were separated on a 3.5% denaturing acrylamide gel and detected by autoradiography. The intensity of the bands was quantified by an imaging analyzer (BAS2000, FUJIX, Tokyo). For in situ hybridization, brains were quickly removed and frozen in dry ice. Sagittal sections (14 μm thick) were prepared using a cryostat and mounted on glass slides coated with poly-l-lysine. Sections were dried at 50°C and kept at −80°C until used. The 252-bp fragment of transgene-specific 3′ non-coding sequence (probe 2, shown in Fig. 1A) was inserted into the vector pGEM4Z (Promega). Antisense digoxygenin-labeled RNA probe was transcribed using digoxygenin-11-UTP (Boehringer Mannheim) and T7 RNA polymerase (Promega). Procedures for pretreatment of sections, hybridization, and detection were done according to Hashimoto and Obata (14).
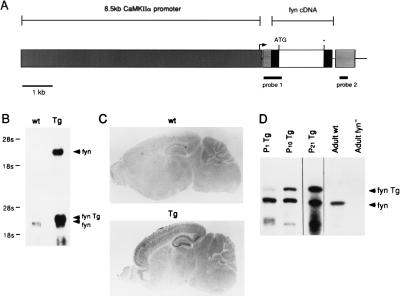
Figure 1.
Generation of fyn-transgenic mice and expression of fyn-transgene. (A) Schematic representation of the transgene construct that is composed of a 8.5-kb fragment, including the CaMKIIα promoter, a hybrid intron consisting of an adenovirus splice donor, and an IgG splice acceptor, an EcoRI–DraI fragment of fynB cDNA, and a simain virus 40 poly(A) signal sequences. Transgenic mice were generated as described. The probes for detection of transgene mRNA and for ribonuclease protection assay are indicated with thick lines (probes 1 and 2, respectively). (B) Northern blot analysis on forebrain. Total RNA prepared from wild-type (wt) and transgenic littermates (Tg) was electrophoresed on formaldehyde agarose gel, transferred onto nylon membrane, and hybridized with probe 1 shown in A Upper and fynB cDNA A Lower. (C) Detection of the transgene mRNA in brain sections. Sagittal brain sections prepared from wild-type (wt) and transgenic (Tg) littermates were hybridized with digoxygenin-labeled cRNA probe 2 shown in A. (D) Developmental change in the level of transgene mRNA. Ribonuclease protection assay was performed on 1-day-old (P1 Tg), 10-day-old (P10 Tg), 3-week-old (P21 Tg) transgenic, adult wild-type (Adult wt), and adult fyn-deficient (Adult fyn−) forebrains. The protected bands corresponding to endogenous fyn mRNA (fyn) and the transgene transcript (fyn Tg) are designated by arrows.
Immunoblotting and in Vitro Immune–Complex Kinase Assay.
The forebrains were homogenized in 10 vol of RIPA buffer (Tris⋅HCl, pH 7.5/1% Nonidet P-40/0.1% sodium deoxycholate/0.1% SDS/0.15 M NaCl/1 mM EDTA/1 mM sodium orthovanadate/10 μg/ml aprotinin/10 μg/ml leupeptin/1 mM phenylmethylsulfony fluoride), then centrifuged at 10,000 × g for 10 min. Supernatants were rapidly frozen in liquid nitrogen and stored at −80°C until used. Protein concentration was determined using BCA protein assay kit (Pierce). For immunoblotting, the RIPA lysates were separated on 10% SDS/PAGE gel, transferred to nitrocellulose (Schleicher & Schuell), and probed with anti-Fyn (provided by M. Yasuda, National Institute of Physiological Science, Japan), anti-Src (mAB327, Oncogene Science), or anti-phosphotyrosine antibodies (PT66, Sigma). After incubation with horseradish peroxidase-conjugated anti-mouse IgG, signals were detected by chemiluminescence (ECL, Amersham). Quantification of signals was performed using an imaging analyzer (Quanty One, PDI Imageware Systems, Huntington Station, NY). For kinase assay, the RIPA lysate (200 μg of protein) was immunoprecipitated with an anti-Fyn antibody conjugated with protein G–Sepharose (Pharmacia). Kinase reaction of the immunoprecipitants was started by adding 1 μM [γ-32P]ATP (specific activity, 100 μCi/nmol; 1 Ci = 37 GBq) with 2.5 mg of acid-treated enolase (Sigma). After incubation at 25°C for 5 min, the reaction was stopped by boiling in the presence of SDS, and then separated on a SDS/10% polyacrylamide gel. The intensity of phosphorylated bands was quantified by an imaging analyzer (BAS2000, FUJIX).
Histological Staining.
Anesthetized animals were perfused with 4% paraformaldehyde/0.5% picric acid in 0.1 M phosphate buffer (pH 7.4), and then their brains were removed and incubated in the same fixative at 4°C until used. Brains were sliced with a vibratome (Oxford) in 70-μm-thick sections. Some sections were used for Nissl staining (cresyl violet) and some for immunohistochemistry. To visualize the fine dendritic structures, immunohistochemical staining was performed using anti-MAP2 monoclonal antibody (AP20, Leinco Technologies, St. Louis). After incubation with biotinylated anti-mouse IgG, the signal was detected using vectastain ABC kit (Vector Laboratories).
Electrophysiological Procedures.
Brains were quickly removed and placed in an ice-cold (0°C to 4°C) artificial cerebrospinal fluid (ACSF) containing 124 mM NaCl/4.4 mM KCl/25 mM NaHCO3/1.0 mM Na2HPO4/2.0 mM CaCl2/2.0 mM MgSO4/10 mM glucose. Hippocampal slices (400 mm thick) were transferred to an interface recording chamber containing cool ACSF (15–20°C). ACSF was perfused through the chamber (1–3 ml per min) as the temperature in the recording chamber was slowly increased to 30°C. Electrophysiological experiments were started after waiting at least 1 hr for the slices to recover. A fine bipolar stimulating electrode was used to stimulate Schaffer collateral fibers in the stratum radiatum of the CA1 region of the hippocampus. The stimulation strength was set to elicit responses equivalent to 25% of the maximum field excitatory postsynaptic potential (EPSP). Synaptic responses were elicited at 0.02 Hz and responses were recorded with low resistance (5–10 MΩ) glass microelectrodes filled with ACSF. Tetanus-induced LTP was elicited with trains (1 sec) of 100 Hz stimulation delivered twice with an interval of 20 sec.
RESULTS AND DISCUSSION
Transgenic Fyn Is Expressed at High Levels in the Neurons of Postnatal Forebrain Postnatally.
A founder mouse carrying the fyn-transgene (Fig. 1A) was obtained. Northern blot analysis and in situ hybridization were performed to evaluate the level and pattern of transgene expression in the brain of F1 offspring. Transgenic fyn mRNA of the expected size (≈4 kb) was detected in the forebrain of 1-month-old transgenic mice (Fig. 1B). This band was larger than the endogenous fyn mRNA (2.8 kb) due to an additional non-coding sequence derived from the transgene construct. Quantitative analysis showed that the level of transgenic fyn mRNA is approximately four times higher than the endogenous one. In situ hybridization on adult brain sections revealed that the regional distribution of transgenic fyn mRNA was similar to CaMKIIα mRNA (15). Whereas fyn mRNA is broadly distributed in the brain (16, 17), the transgenic fyn transcript was localized to neurons and restricted to the forebrain, including the neocortex, hippocampus, striatum, and the amygdaloid nucleus (Fig. 1C). To quantify transgenic fyn mRNA during postnatal development, a ribonuclease protection assay was performed on fyn-transgenic mouse forebrains (Fig. 1D). The time course of the fyn-transgene expression was similar to that of CaMKIIα (18, 19). A very weak signal was detected in newborn brain and the levels of transgene expression increased over a 3-week postnatal period.
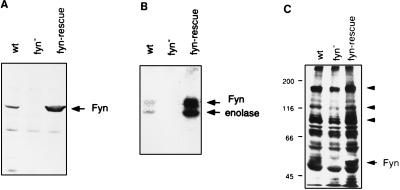
We next generated mice carrying the fyn-transgene on the fyn-deficient genetic background by mating fyn-transgenic mice with fyn-deficient mice. We refer to these mice as fyn-rescue mice. We then determined in brain extracts of fyn-rescue mice the level of the Fyn protein, derived from the transgene, as well as the level of Fyn kinase activity. We detected by immunoblot analysis (Fig. 2A) a protein band identical to the endogenous Fyn in the forebrain of 3-month-old rescue mice. Quantitative immunoblotting revealed that the level of transgenic Fyn protein was approximately four times higher than in wild-type littermates. An in vitro immune complex kinase assay showed that the fyn-transgene product was catalytically active; it could phosphorylate itself as well as the substrate enolase (Fig. 2B). The specific activity of transgenic Fyn kinase, estimated by the intensity of the phosphorylated bands compared with the relative amount of Fyn protein, was equivalent to the activity of the endogenous Fyn. We next examined the effects of fyn-transgene expression on the level of tyrosine-phosphorylated proteins in the forebrain by immunoblotting using anti-phosphotyrosine antibody. In fyn-deficient mice, multiple proteins (approximately 180, 120, and 100 kDa) were significantly less phosphorylated, suggesting that they are preferentially phosphorylated by Fyn in vivo (Fig. 2C). In fyn-rescue mice, the phosphorylation of these proteins was restored to the levels in wild-type animals. In addition, we detected a 60-kDa protein identified as phosphorylated transgenic Fyn.
Figure 2.
Fyn protein, kinase activity, and tyrosine phosphorylation in the fyn-rescue mice. (A) The extracts prepared from forebrains of 2- to 3-month-old wild-type (wt), fyn-deficient (fyn−) and fyn-rescue (fyn-rescue) mice were separated on a SDS/10% polyacrylamide gel and blotted with an anti-Fyn antibody. The band corresponding to Fyn protein is designated by an arrow. (B) Proteins from the same extracts were immunoprecipitated with an anti-Fyn antibody conjugated with protein G–Sepharose. The precipitates were incubated with [γ-32P]ATP in the presence of enolase, and then resolved on a SDS/10% polyacrylamide gel. Fyn and enolase are indicated by arrows. (C) Proteins from the same brain extracts were blotted with an anti-phosphotyrosine antibody, PT66. The arrowheads indicate the bands that show less phosphorylation in fyn-deficient mice.
Morphological Abnormalities Are Present in fyn-Rescue Hippocampus.
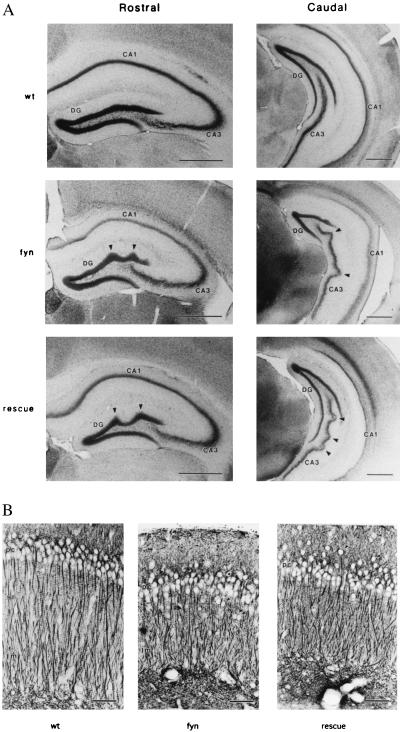
We then examined the anatomy of the hippocampus in fyn-rescue mice. As shown in Fig. 3A, we observed the abnormal cell arrangement characteristic of the fyn-deficient mice. The caudal part of CA3 pyramidal cell layer was undulated (Fig. 3A, arrowheads) and the dorsal part of the dentate gyrus granular cell layer was occasionally perturbed in the rescue mice. To visualize the apical dendritic arborizations in CA1 pyramidal cells, immunohistochemical staining was performed using antibody to microtubule-associated protein MAP2 (Fig. 3B). As previously reported in the fyn-deficient mice, CA1 pyramidal cells were less tightly packed and ectopic pyramidal cells (Fig. 3B, arrowheads) were frequently observed in fyn-rescue mice. The dendritic regions were relatively narrow and the length of the apical dendrites were shortened in both fyn-deficient and fyn-rescue mice in comparison to wild-type animals, although the dendritic arborization running across the stratum radiatum and lacunosum-moleculare was basically conserved in both mutant mice. These histological observations indicated that expression of the fyn-transgene, controlled by the CaMKIIα promoter, did not rescue the morphological defects in the hippocampus of fyn-deficient mice. The undulation of hippocampal cell layers, commonly observed in the independent lines of fyn-deficient mice (1, 17) may reflect a failure in the regulation of cell number or in neuronal migration due to the lack of Fyn in neurons during development before the onset of the CaMKIIα promoter driven expression of the fyn-transgene. Alternatively, the developmental phenotype may reflect the lack of Fyn in non-neuronal cells.
Figure 3.
Hippocampal morphology in fyn-deficient and fyn-rescue mice. (A) Nissl staining of rostral and caudal coronal hippocampal sections of wild-type (wt), fyn-deficient (fyn), and fyn-rescue (rescue) mice. Characteristic abnormal arrangements (arrowheads) of rostral dentate gyrus (DG) and caudal CA3 region in fyn-deficient mice were also observed in the rescue mice. (Bars = 500 μm.) (B) Dendritic organization of the CA1 field in rescue mice. Apical dendrites of the pyramidal cells were revealed by immunohistochemical staining using anti-MAP2 antibody. CA1 pyramidal cell layer (pc) was less tightly packed and ectopic pyramidal cells (arrowheads) were frequently observed in fyn-deficient (fyn) and fyn-rescue (rescue) mice. Dendrites ran across the stratum radiatum (sr) and lacunosum-moleculare (lm), but their lengths were shorter than in wild-type litter mates. (Bar = 50 μm.)
LTP Is Normal in fyn-Rescue Mice.
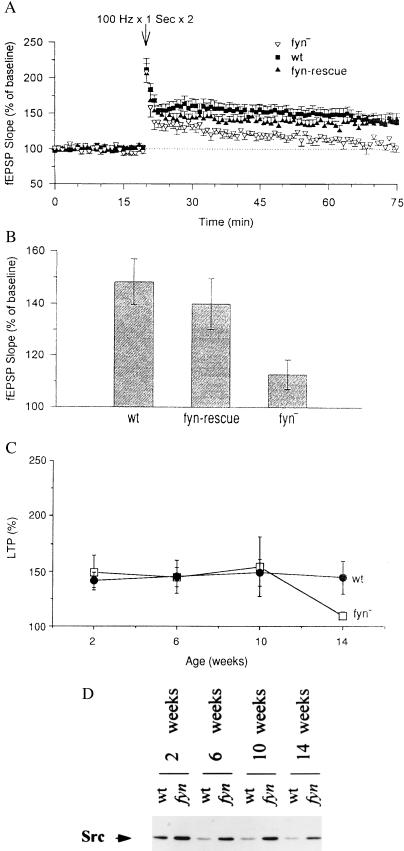
To determine if synaptic plasticity was restored in the fyn-rescue mice despite their anatomical abnormality, we tested LTP in adult hippocampal slices comparable to those used in the initial studies of the fyn-deficient mice. As described previously (1), a tetanic stimulation evoking a postsynaptic response equivalent to 25% of the maximal EPSP was applied to hippocampal slices prepared from three groups of mice: wild-type, fyn-deficient, and fyn-rescue mice. We found that LTP was induced in wild-type and fyn-rescue slices, but not in fyn-deficient slices (Fig. 4A). The average value of relative EPSP slope 60 min after tetanic stimulation in fyn-rescue slices was significantly larger than in fyn-deficient slices (135 ± 7.2% and 112.7 ± 5.7%, respectively; t = 2.39, P < 0.05). In fact, the LTP in fyn-rescue slices was similar to that in wild-type slices (148.3 ± 8.7%; Fig. 4B). Thus, transgenic Fyn introduced into postnatal hippocampal neurons allowed us to distinguish between the morphological and the physiological defects in fyn-deficient mice. These results indicate that the impairment of LTP in fyn-deficient hippocampus is not caused by the developmental abnormality in the hippocampal architecture, but by the lack of Fyn in hippocampal neurons.
Figure 4.
LTP in fyn-rescue and young fyn-deficient mice and Src compensation in fyn-deficient mice. (A) Field EPSP recorded before and after tetanic stimulation in slices from adult wild-type (wt, ▪), fyn-deficient (fyn−, ▵), and fyn-rescue mice (fyn-rescue, ▴). Each point is the mean ± SEM of the field EPSP slope. Wild-type mice: n = 8 animals, 13 slices; fyn-deficient mice: n = 9 animals, 16 slices; fyn-rescue mice: n = 11 animals, 32 slices. (B) LTP measured 1 hr after tetanic stimulation. Wild-type (wt) mice: 148.3 ± 8.7% (n = 8); fyn-rescue (fyn-rescue) mice: 135.0 ± 7.2% (n = 9); fyn-deficient (fyn−) mice: 112.7 ± 5.7% (n = 11). Wild-type mice versus fyn-rescue mice: t = 1.14, not significant; fyn-rescue mice versus fyn-deficient mice: t = 2.39, P < 0.05. (C) Time course of LTP in wild-type (wt, •) and fyn-deficient (fyn−, □) mice. Each point is the percentage of LTP obtained at different age. Wild-type mice: 2 weeks, n = 5; 6 weeks, n = 6; 10 weeks, n = 6; 14 weeks, n = 9. fyn-deficient mice: 2 weeks, n = 12; 6 weeks, n = 7; 10 weeks, n = 4, 14 weeks, n = 6. (D) Src levels in wild-type (wt) and fyn-deficient (fyn) mice of different postnatal ages. Protein extract from mice aged 2, 6, 10, 12, and 14 weeks. The levels of Src are indicated by the arrow.
Age-Dependent Impairment of LTP in fyn-Deficient Mice.
The fyn-deficient mice examined here and in the original studies were between 14 weeks and 6 months of age and showed a clear blunting of LTP (1). However, in the course of these experiments, we also examined younger fyn-deficient mice and found, surprisingly, that LTP was similar to that seen in wild-type animals (Fig. 4C). Thus, the LTP phenotype in the fyn-deficient mice is age dependent and appears only when the mice are over 10 weeks of age. This result further supports the idea that the LTP phenotype is not related to the developmental abnormality since young fyn-deficient mice show normal LTP, but also have a clearly disturbed hippocampal structure.
Compensatory Response of Src Is Also Age-Dependent.
The mechanism of this age-dependent rescue of the adult Fyn phenotype is not known, but it could be mediated by developmentally regulated compensation by other kinases at younger ages (20, 21). Since we had earlier found that the Src kinase is elevated in the fyn-deficient mice (21), we now examined the developmental time course of this compensation. As shown in Fig. 4D, at all ages Src is expressed at higher levels in the fyn-deficient mice than in wild-type mice. However, at 14 weeks of age where the LTP deficit begins to be observed in the fyn-deficient mice, the level of compensatory Src expression is reduced. Thus, although src-deficient mice do not show any obvious neurological phenotype (2) and exhibited normal LTP (1), Src may be involved in the mechanisms of synaptic plasticity at least in the context of a Fyn deficiency. Also, LTP induced by pairing the synaptic input with strong postsynaptic depolarization of CA1 pyramidal cells in fyn-deficient mice is blocked by the tyrosine kinase inhibitor genistein, suggesting that other tyrosine kinases may contribute to LTP induction (T. J. O’Dell, unpublished results).
How Does Fyn Contribute to the Mechanisms of LTP Induction?
The findings that Fyn contributes directly to LTP raises the question: How does it produce its effects? As we have shown previously (1), the fact that strong stimulation could induce LTP in fyn-deficient mice suggests that Fyn is not essential for LTP induction, but may act as a modulatory molecule that influences the threshold for the induction of LTP. This effect might be mediated through tyrosine phosphorylation of other proteins required for LTP. What might these proteins be? One candidate is protein kinase C. In hematopoietic cells, Fyn is involved in phosphatidylinositol signal transduction pathway (22, 23). If Fyn also interacts with phospholipase C in neurons, the activity of protein kinase C required for LTP induction (24–26) might be altered in fyn-deficient mice. A second candidate is the N-methyl-d-aspartate (NMDA) receptor. Thus, tyrosine phosphorylation has also been shown to modulate the function of certain neurotransmitter-gated ion channels (27–29). In particular, the 2A and 2B subunits of the NMDA receptor are the targets of tyrosine phosphorylation (29–31), and indeed tyrosine phosphorylation of NMDA receptor 2B is increased after LTP induction (32, 33). The NMDA receptor may not be the only target for Fyn as NMDA receptor associated tyrosine phosphoproteins are also regulated by Fyn (S.G.N.G., unpublished results). The NMDA receptors 2A and 2B can actually be phosphorylated by Fyn in vitro (34) and these were shown to be hypophosphorylated in fyn-deficient mice (unpublished results). Finally, a third possibility is that Fyn might be involved in neural cell adhesion molecule signaling which is also required for normal LTP and spatial learning (35–37). In fact, neural cell adhesion molecule-dependent neurite outgrowth of fyn-deficient neurons is impaired in culture (3).
Taken together our data suggest that the Fyn signaling pathway is involved in the regulation of synaptic plasticity in a manner that is independent of its role in development. Consistent with an important role for Fyn in synaptic physiology, transgenic mice carrying a constitutively active form of Fyn in which the C-terminal negative regulatory tyrosine residue (Y531) was replaced by phenylalanine show enhanced seizure susceptibility and spontaneous death (N.K., unpublished results). In a larger sense, our experiments provide direct evidence that using the CaMKIIα promoter and the fyn-transgene, it is possible to rescue LTP in animals whose gene has been knocked out by homologous recombination. Combined with region-specific and regulated (38) expression of transgenes, transgene rescue provides an additional method for evaluating the contribution of individual genes to synaptic plasticity and behavior.
Acknowledgments
We thank Harriet Ayers and Irma Trumpet for typing, Monica Mendelsohn for the generation of the transgenic mice, Dr. M. Osman for animal care, S. Yuasa and S. Furuya for technical advice for histological part of this study, R. M. Perlmutter for providing fynB cDNA pmBF, N. Nakanishi for pNN265 and pNN279, and M. Yasuda for anti-Fyn monoclonal antibody. This research was supported by the Howard Hughes Medical Institute and in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science and Culture of Japan. N.K. was supported by a long-term fellowship from the Ministry of Education, Science and Culture of Japan.
ABBREVIATIONS
- CaMKIIα
calcium/calmodulin-dependent protein kinase IIα
- LTP
long-term potentiation
- EPSP
excitatory postsynaptic potential
- NMDA
N-methyl-d-aspartate
- ACSF
artificial cerebrospinal fluid
References
- 1.Grant S G N, O’Dell T J, Karl K A, Stein P L, Soriano P, Kandel E R. Science. 1992;258:1903–1910. doi: 10.1126/science.1361685. [DOI] [PubMed] [Google Scholar]
- 2.Soriano P, Montgomery C, Gesk R, Bradley A. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- 3.Beggs H E, Soriano P, Maness P F. J Cell Biol. 1994;127:825–833. doi: 10.1083/jcb.127.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cain D P, Grant S G N, Saucier D, Hargreaves E L, Kandel E R. Epilepsy Res. 1995;22:107–114. doi: 10.1016/0920-1211(95)00029-1. [DOI] [PubMed] [Google Scholar]
- 5.Yagi T, Aizawa S, Tokunaga T, Shigetani Y, Takeda N, Ikawa Y. Nature (London) 1993;366:742–745. doi: 10.1038/366742a0. [DOI] [PubMed] [Google Scholar]
- 6.Umemori H, Sato S, Yagi T, Aizawa S, Yamamoto T. Nature (London) 1994;367:572–576. doi: 10.1038/367572a0. [DOI] [PubMed] [Google Scholar]
- 7.Miyakawa T, Yagi T, Watanabe S, Niki H. Mol Brain Res. 1994;27:179–182. doi: 10.1016/0169-328x(94)90201-1. [DOI] [PubMed] [Google Scholar]
- 8.Miyakawa T, Yagi T, Taniguchi M, Matsuura H, Tateishi K, Niki H. Mol Brain Res. 1995;28:349–352. doi: 10.1016/0169-328x(94)00251-9. [DOI] [PubMed] [Google Scholar]
- 9.Cooke M P, Perlmutter R M. New Biol. 1989;1:66–74. [PubMed] [Google Scholar]
- 10.Choi T, Huang M, Gorman C, Jaenisch R. Mol Cell Biol. 1991;11:3070–3074. doi: 10.1128/mcb.11.6.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayford M, Wang J, Kandel E R, O’Dell T J. Cell. 1995;81:891–904. doi: 10.1016/0092-8674(95)90009-8. [DOI] [PubMed] [Google Scholar]
- 12.Hogan B, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1986. pp. 90–182. [Google Scholar]
- 13.Stein P L, Lee H-M, Rich S, Soriano P. Cell. 1992;70:741–750. doi: 10.1016/0092-8674(92)90308-y. [DOI] [PubMed] [Google Scholar]
- 14.Hashimoto T, Obata K. Neurosci Res. 1991;12:514–527. doi: 10.1016/s0168-0102(09)80004-7. [DOI] [PubMed] [Google Scholar]
- 15.Burgin K E, Waxham M N, Rickling S, Westgate S A, Mobley W C, Kelly P T. J Neurosci. 1990;10:1788–1798. doi: 10.1523/JNEUROSCI.10-06-01788.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Umemori H, Wanaka A, Kato H, Takeuchi M, Tohyama M, Yamamoto T. Mol Brain Res. 1992;16:303–310. doi: 10.1016/0169-328x(92)90239-8. [DOI] [PubMed] [Google Scholar]
- 17.Yagi T, Shigetani Y, Okado N, Tokunaga T, Ikawa Y, Aizawa S. Oncogene. 1993;8:3343–3351. [PubMed] [Google Scholar]
- 18.Beaman-Hall C M, Hozza M J, Vallano M L. J Neurochem. 1992;58:1259–1267. doi: 10.1111/j.1471-4159.1992.tb11337.x. [DOI] [PubMed] [Google Scholar]
- 19.Sugiura H, Yamauchi T. Brain Res. 1992;593:97–104. doi: 10.1016/0006-8993(92)91269-k. [DOI] [PubMed] [Google Scholar]
- 20.Stein P L, Vogel H, Soriano P. Genes Dev. 1994;8:1999–2007. doi: 10.1101/gad.8.17.1999. [DOI] [PubMed] [Google Scholar]
- 21.Grant S G N, Karl K A, Kiebler M A, Kandel E R. Genes Dev. 1995;9:1909–1921. doi: 10.1101/gad.9.15.1909. [DOI] [PubMed] [Google Scholar]
- 22.Shiroo M, Goff L, Biffen M, Shivnan E, Alexander D. EMBO J. 1992;11:4887–4897. doi: 10.1002/j.1460-2075.1992.tb05595.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karnitz L M, Sutor S L, Abraham R T. J Exp Med. 1994;179:1799–1808. doi: 10.1084/jem.179.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malinow R, Schulman H, Tsien R W. Science. 1989;245:862–866. doi: 10.1126/science.2549638. [DOI] [PubMed] [Google Scholar]
- 25.Wang J H, Feng D P. Proc Natl Acad Sci USA. 1992;89:2576–2580. doi: 10.1073/pnas.89.7.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abeliovich A, Chen C, Goda Y, Silva A J, Stevens C F, Tonegawa S. Cell. 1993;75:1253–1262. doi: 10.1016/0092-8674(93)90613-u. [DOI] [PubMed] [Google Scholar]
- 27.Moss S J, Gorrie G H, Amato A, Smart T G. Nature (London) 1995;377:344–348. doi: 10.1038/377344a0. [DOI] [PubMed] [Google Scholar]
- 28.Valenzuela C F, Machu T K, McKernan R M, Whiting P, VanRenterghem B B, McManaman J L, Brozowski S J, Smith G B, Olsen R W, Harris R A. Mol Brain Res. 1995;31:165–172. doi: 10.1016/0169-328x(95)00048-w. [DOI] [PubMed] [Google Scholar]
- 29.Moon I S, Apperson M L, Kennedy M B. Proc Natl Acad Sci USA. 1994;91:3954–3958. doi: 10.1073/pnas.91.9.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y T, Yu X M, Salter M W. Proc Natl Acad Sci USA. 1996;93:1721–1725. doi: 10.1073/pnas.93.4.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen C, Leonard J P. J Neurochem. 1996;67:194–200. doi: 10.1046/j.1471-4159.1996.67010194.x. [DOI] [PubMed] [Google Scholar]
- 32.Rastas J A P, Brent V A, Voss K, Errington M L, Bliss T V P, Gard J W. Proc Natl Acad Sci USA. 1996;93:10452–10456. doi: 10.1073/pnas.93.19.10452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenblum K, Dudai Y, Richter-Levin G. Proc Natl Acad Sci USA. 1996;93:10457–10460. doi: 10.1073/pnas.93.19.10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki T, Okumura-Noji K. Biochem Biophys Res Commun. 1995;216:582–588. doi: 10.1006/bbrc.1995.2662. [DOI] [PubMed] [Google Scholar]
- 35.Lüthi A L, Laurent J-P, Figurov A, Muller D, Schachner M. Nature (London) 1994;372:777–779. doi: 10.1038/372777a0. [DOI] [PubMed] [Google Scholar]
- 36.Muller D, Wang C, Skibo G, Toni N, Cremer H, Calaora V, Rougon G, Kiss J Z. Neuron. 1996;17:413–422. doi: 10.1016/s0896-6273(00)80174-9. [DOI] [PubMed] [Google Scholar]
- 37.Cremer H, Lange R, Christoph A, Plomanm M, Vopper G, Roes J, Brown R, Baldwin S, Kraemer P, Scheff S, Barthels D, Rajewsky K, Wille W. Nature (London) 1994;367:455–457. doi: 10.1038/367455a0. [DOI] [PubMed] [Google Scholar]
- 38.Mayford M, Bach M E, Huang Y-Y, Wang L, Hawkins R D, Kandel E R. Science. 1996;274:1678–1683. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]