Abstract
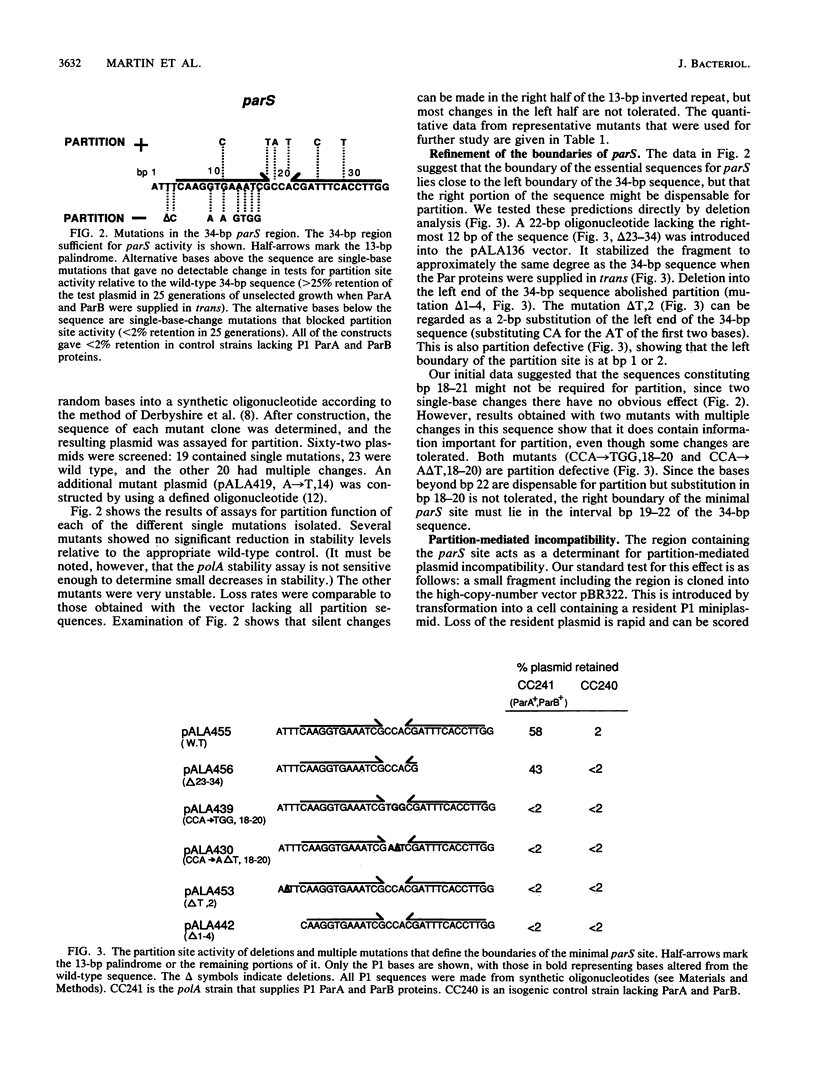
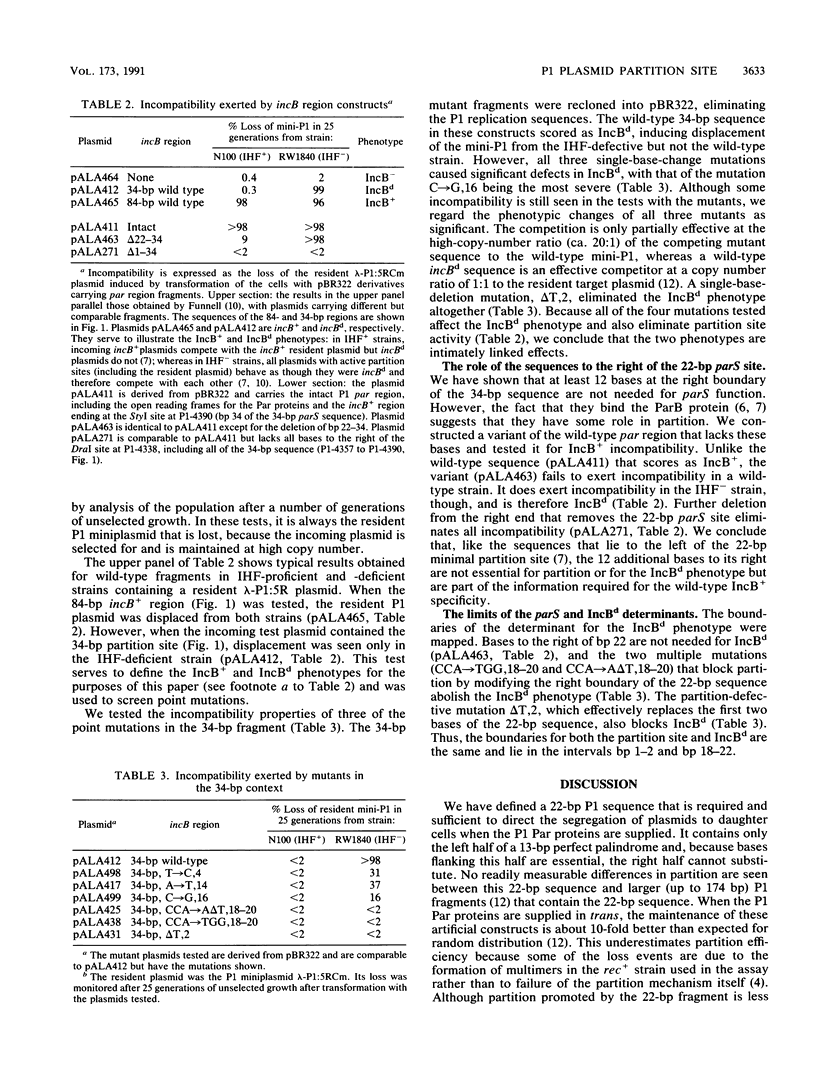
P1 plasmid partition requires two plasmid-encoded Par proteins and a cis-acting site. The site, parS, lies in a region consisting of a 13-bp palindrome and an adjacent AT-rich sequence. A series of point mutations were analyzed for their effects on partition site activity. The results indicated that only the left arm of the palindrome and some adjacent bases were needed. The limits of the functional site were further refined to a maximum of 22 bp, which includes binding sites for the P1 ParB protein. Mutations in the 22-bp site cause concomitant defects in partition and the ability to exert partition-mediated incompatibility. Like the region immediately to the left of the 22-bp region, the right arm of the palindrome is not essential for partition but does contain information that affects the specificity of incompatibility.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abeles A. L., Friedman S. A., Austin S. J. Partition of unit-copy miniplasmids to daughter cells. III. The DNA sequence and functional organization of the P1 partition region. J Mol Biol. 1985 Sep 20;185(2):261–272. doi: 10.1016/0022-2836(85)90402-4. [DOI] [PubMed] [Google Scholar]
- Austin S., Abeles A. Partition of unit-copy miniplasmids to daughter cells. I. P1 and F miniplasmids contain discrete, interchangeable sequences sufficient to promote equipartition. J Mol Biol. 1983 Sep 15;169(2):353–372. doi: 10.1016/s0022-2836(83)80055-2. [DOI] [PubMed] [Google Scholar]
- Austin S., Abeles A. Partition of unit-copy miniplasmids to daughter cells. II. The partition region of miniplasmid P1 encodes an essential protein and a centromere-like site at which it acts. J Mol Biol. 1983 Sep 15;169(2):373–387. doi: 10.1016/s0022-2836(83)80056-4. [DOI] [PubMed] [Google Scholar]
- Austin S., Ziese M., Sternberg N. A novel role for site-specific recombination in maintenance of bacterial replicons. Cell. 1981 Sep;25(3):729–736. doi: 10.1016/0092-8674(81)90180-x. [DOI] [PubMed] [Google Scholar]
- Das A., Court D., Adhya S. Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor rho. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1959–1963. doi: 10.1073/pnas.73.6.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. A., Austin S. J. Recognition of the P1 plasmid centromere analog involves binding of the ParB protein and is modified by a specific host factor. EMBO J. 1988 Jun;7(6):1881–1888. doi: 10.1002/j.1460-2075.1988.tb03021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. A., Martin K. A., Austin S. J. Specificity switching of the P1 plasmid centromere-like site. EMBO J. 1990 Apr;9(4):991–998. doi: 10.1002/j.1460-2075.1990.tb08201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire K. M., Salvo J. J., Grindley N. D. A simple and efficient procedure for saturation mutagenesis using mixed oligodeoxynucleotides. Gene. 1986;46(2-3):145–152. doi: 10.1016/0378-1119(86)90398-7. [DOI] [PubMed] [Google Scholar]
- Friedman S. A., Austin S. J. The P1 plasmid-partition system synthesizes two essential proteins from an autoregulated operon. Plasmid. 1988 Mar;19(2):103–112. doi: 10.1016/0147-619x(88)90049-2. [DOI] [PubMed] [Google Scholar]
- Funnell B. E. Participation of Escherichia coli integration host factor in the P1 plasmid partition system. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6657–6661. doi: 10.1073/pnas.85.18.6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley W. S. Mapping of the polA locus of Escherichia coli K12: genetic fine structure of the cistron. Genetics. 1980 May;95(1):15–38. doi: 10.1093/genetics/95.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K. A., Friedman S. A., Austin S. J. Partition site of the P1 plasmid. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8544–8547. doi: 10.1073/pnas.84.23.8544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Hoppensteadt F. C. On plasmid incompatibility. Plasmid. 1978 Sep;1(4):421–434. doi: 10.1016/0147-619x(78)90001-x. [DOI] [PubMed] [Google Scholar]



