Abstract
In vertebrates, odors are thought to be detected by a multigene family encoding several hundreds of seven-transmembrane-domain G-protein-coupled receptors found in fish, rat, mouse, dog, and human. Recently, the putative odorant receptor (OR) gene family in the chicken has been characterized. Twelve members have been isolated and subdivided into six subfamilies. Herein, we have further characterized the chicken olfactory receptor subfamily 7 (COR7) composed of two highly related genes (named COR7a and COR7b) which are 98.5% identical. By in situ hybridization experiments, both COR7a and COR7b transcripts were detected in the olfactory epithelium from embryonic day 6 (E6) to the new born stage. Within the olfactory epithelium, the spatial distribution of COR7a and COR7b labeled cells was random. We also observed that every individual positive cell did not coexpress the COR7a and COR7b genes. Interestingly, the COR7b gene was found to be transiently expressed in the notochord from E2 to E6, whereas COR7a or any of the other known members of the COR gene family were not detected in this mesodermal tissue. These data suggest that, in addition to its potential role as an OR in the olfactory system, COR7b may also have a function in the notochord that is essential for the dorsoventral organization of the neural tube and of the somitic mesoderm. We also discuss the possible role(s) of a putative OR present in both the notochord and the sensory olfactory epithelium.
Keywords: olfactory placode, olfactory receptor, olfactory epithelium, development
The initial step for odor detection occurs at the surface of cilia of olfactory neurons where odorants seem to interact with olfactory receptors. Genes encoding putative odorant receptors (ORs) have been recently characterized in several vertebrate species including mouse (1, 2), rat (3), dog (4), fish (5, 6), chicken (7, 8), and human (4, 9). The putative OR family belongs to the large multigene superfamily of seven-transmembrane-domain G-protein-coupled receptors. The size of the OR gene family is extremely variable depending on the species studied. Human and rodent gene families seem to contain several hundred genes (for review, see ref. 10). Whereas the chicken OR gene family contains probably less than 20 genes (7), the catfish contains about 100 genes (6) and the zebra fish contains approximately 30 genes (5). In situ hybridization experiments have revealed that a single putative OR is usually expressed in only a small fraction of the olfactory neuron population, i.e., between 0.1% and 2% of the total number of olfactory neurons (for review, see ref. 11). Similarly, each individual olfactory neuron seems to express only a small subset of putative OR genes, indicating a very specific and fine regulation of OR gene expression in the olfactory epithelium. In the chicken, catfish, and zebra fish, neurons expressing a particular OR gene appear to be randomly distributed throughout the entire olfactory epithelium (5, 7, 8, 12). In contrast, studies performed with rodents have revealed that the OR genes are symmetrically distributed within the olfactory epithelium (13) throughout four broad zones, which consist of a series of elongated bands that extend along the anterior–posterior axis of the nasal cavity (2, 14, 15). Zonal segregation does not seem to be essential for coding a correct odor message since chicken and catfish, which efficiently detect odorants, show random distributions of OR genes within their olfactory epithelium.
Birds such as chickens represent a very attractive model to study the morphogenesis of the olfactory epithelium and olfaction. Despite the common belief that birds are feebly equipped to detect odors, several species such as the kiwi (16, 17), the vulture, or the homing pigeon (18) are known to use their sense of smell to detect food or to find home. The olfactory system of chicken embryos responds to volatiles already on day 19 of incubation [embryonic day 19 (E19)] when the beak has just entered the air sac and the nares are clear of surrounding tissue (19). Being a gregarious animal, the chicken surely uses odorants to determine it’s social behavior. For example, some nesting-litter odors may serve to keep the chicken in the proximity of the nest during early post-hatching life (20).
The avian olfactory system consists of a very simple structure and its morphogenesis has been well described (21). Recently, we have studied the molecular basis of olfaction in the chicken (7) and have reported the isolation and characterization of nine putative chicken OR (COR) genes that were all expressed in the olfactory epithelium. Most of the members of the OR multigene family found in several species are detected in the olfactory sensory epithelium. However, some members, indistinguishable from those expressed in the olfactory epithelium, have been found in various tissues unrelated to olfaction. For example, several OR genes are expressed in cells of the male germ line of dogs and humans (4, 22). These “germ cell receptors” may serve as sensors for chemicals, not necessarily odorant molecules, involved in sperm cell maturation, migration, or fertilization. Other OR genes are expressed in the taste buds of rat (23, 24), in a structure composed of several taste receptor cells organized in discrete spherical clusters known to detect tastant molecules. This “gustatory receptor” gene subfamily includes probably more than 60 members. Furthermore, Drutel et al. (25) have reported the cloning of an additional member of the olfactory receptor family in the rat, called OL1. The OL1 gene is not only expressed in the olfactory epithelium, but interestingly, also in the developing heart, suggesting a potential role for this receptor in cardiac development. Finally, in the chicken, members of the COR gene family were found to be expressed not only in the olfactory epithelium but also, at E5, in cells migrating along the olfactory nerve, suggesting that they could play a role in the morphogenesis of the avian olfactory system (7). Similar observations were later reported with three other chicken OR genes (8). All these observations suggest that, in addition to their role as putative ORs, members of the OR gene family may also have a function in tissues unrelated to olfaction and could have a potential role during early development.
Herein we report the characterization and the spatiotemporal expression of two highly related COR genes in the chicken named COR7a and COR7b belonging to the COR7 subfamily. We show that both COR7a and COR7b transcripts were present in the olfactory epithelium from E6 to hatching. Of the total number of COR7ab positive cells detected at each stage of development, ∼40% were COR7a-positive and ∼60% were COR7b-positive. COR7ab-positive cells, presumably olfactory neurons, were randomly distributed throughout the olfactory epithelium. Interestingly, we found that only the COR7b gene was expressed in the notochord between ∼E2 and E6. All the other known members of the COR gene family were not detected in the notochord. These observations suggest a potential role for COR7b as part of an unknow signal transduction pathway perhaps involved in feedback responses originating from the neural tube or the somitic mesoderm.
MATERIALS AND METHODS
Genomic Library Screening.
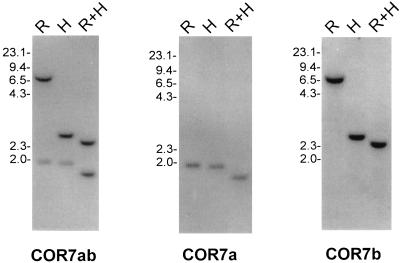
As described in ref. 7. Briefly, 1 × 106 phages from a chicken genomic library constructed in the phage vector λL47 were screened with a 510-bp PCR fragment (named the COR7ab PCR probe) encoding amino acid positions 118–287 of the chicken olfactory receptor COR7 (ref. 7 and Fig. 1). We have isolated four positive phages and have performed Southern blot analysis to determine which of them had a pattern of hybridization identical to a chicken genomic DNA Southern blot analysis (Fig. 2). Two phages, c13 and c24.2, containing the two olfactory receptor genes named COR7a and COR7b, respectively, were further characterized. Inserts were sequenced by using the dideoxynucleoide method of Sanger (26).
Figure 1.
Deduced amino acid sequences of the chicken olfactory receptors COR7a and COR7b. The predicted positions of the seven transmembrane domains are indicated (TM1–TM7). Conserved amino acid positions are indicated with a dot, whereas nonconserved amino acids are indicated with the corresponding one-letter code.
Figure 2.
Chicken genomic DNA blot analysis of the COR7 subfamily. DNA was digested with EcoRI (R), HindIII (H), EcoRI, and HindIII (R+H) and hybridized with the specific COR7a or COR7b probes or with a common probe (COR7ab). The scale (in kb) is indicated.
Genomic Southern Blot Analysis.
Genomic DNA was isolated from chicken embryos at ∼E10 and analyzed as described in ref. 7. 35S-labeled radioactive antisense cRNA encoding COR7ab (510 bp) or 3′ untranslated regions of COR7a (330 bp) or COR7b (480 bp) were used as probes for Southern blot analysis and in situ hybridization (for more details see below).
In situ Hybridization.
As described in ref. 7, 35S-labeled radioactive antisense or sense cRNA were produced for each subfamily by in vitro transcription with T3 or T7 RNA polymerases from an equimolar amount of linearized full-length COR1, COR2, COR4, COR5, and COR6 clones (≈900 bp) for the COR1–6 subfamily probe (7) and from PCR fragments encoding COR7ab (510 bases) (ref. 7 and this paper) for the COR7 subfamily, COR8 (510 bases) for the COR8 subfamily (7), COR9 (425 bases) for the COR9 subfamily (7), COR10 (996 bases) for the COR10 subfamily (original name COR2, GenBank accession no. X94742X94742; ref. 8), and COR11 (936 bases) for the COR11 subfamily (original name COR4, GenBank accession no. X94744X94744; ref. 8). The specific antisense probes for COR7a (330 bp) and COR7b (480 bp) were produced from PCR fragments encoding the 3′ nontranslated regions starting just after the STOP codon of the COR7a gene and of the COR7b gene respectively. The chicken antisense Sonic hedgehog (cSHH) cRNA probe was produced from a cDNA fragment of 1.6 kb encoding the full protein (gift from C. Tabin, Harvard Medical School). The length of cRNA probes were adjusted by limited alkaline hydrolysis to a mass average of approximately 150 bases as described (27).
RESULTS
Cloning of the COR7a and COR7b Genes.
To isolate all the members of the COR7 subfamily, we decided to screen a chicken genomic library at medium stringency with a 500-bp PCR fragment encompassing the TM3 to TM7 region of the partial COR7 gene sequence (7). Four positive phages were isolated and further characterized. By Southern blot analysis using the COR7 PCR fragment as probe, two had patterns of hybridization that were comparable to a chicken genomic DNA blot analysis. One phage (c13) contained an intronless gene encoding a putative OR in the chicken, called COR7a. The other phage (c24.2) contained another intronless gene encoding a putative chicken OR named COR7b. The sequence of the COR7 PCR fragment corresponded to the COR7b gene (100% identical) and was 98.7% identical to the COR7a sequence. The homology between COR7a and COR7b at the nucleic acid level was 98.5% and was 96.6% at the amino acid level (Fig. 1). The two COR7 genes posses less than 44% amino acid identity with all the other known COR genes. Of the 15 nucleotide mutations, only 3 occur in the “third” codon position, 5 were in the second, and 7 were in the first. Interestingly, 73% (11 of 15) of the nucleotide differences between COR7a and COR7b were mutations that resulted in amino acid substitutions. Most of these substitutions changed an aliphatic amino acid into another aliphatic amino acid (6 of 11, 55%) and were dispersed throughout the receptor sequence. There were no hot spots of high diversity as observed in some transmembrane regions of putative olfactory receptors in the rat (3) or the catfish (6).
Two Genes in the COR7 Subfamily.
Genomic DNA blot analysis was performed at high or low stringency (data not shown) with the COR7 PCR probe (named COR7ab because it can hybridize to both COR7a and COR7b genes) to estimate the size of the COR7 gene subfamily (Fig. 2). In each lane, only two distinct bands were present, suggesting that the COR7 gene subfamily is composed of two genes. Sequence analysis of the 3′ and 5′ untranslated regions of the COR7a and COR7b genes revealed no obvious homology, and therefore, we used the first 330 bp or 480 bp after the stop codon of COR7a or COR7b, respectively, to generate specific probes. COR7a and COR7b blot analysis with the specific probes revealed, as expected, only one band per restriction enzyme digestion (Fig. 2). The complementary patterns of positive bands with the two specific probes corresponded to the pattern of the COR7ab PCR probe, indicating that only COR7a and COR7b composed the COR7 gene subfamily.
COR7a and COR7b Expression in the Olfactory Epithelium During Development.
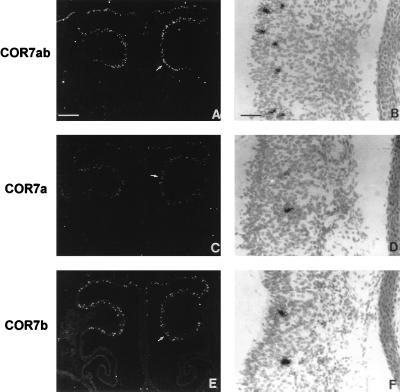
The chicken olfactory epithelium is located in the back of the beak covering the roof of the nasal cavity and the superior conchae. This epithelium is a pseudostratified columnar epithelium composed of three major cell types: olfactory neurons, supporting cells, and olfactory stem cells. We analyzed the expression of COR7a and COR7b in the olfactory epithelium by in situ hybridization experiments with serial coronal sections of the nasal cavity (≈14 μm) hybridized with single-stranded antisense RNA probes derived from the COR7a- and COR7b-specific regions described above. At E6, E8, E10, E12, E14, E16, and E18 and at birth, we detected positive signals corresponding to cellular bodies as clusters of silver grains within the olfactory epithelium (Fig. 3). With the exception of the notochord, which was positive for COR7b (see below), no positive signals were observed elsewhere in other regions of the embryo. The Fig. 3 shows the expression at E16 in the nasal cavity of COR7a, COR7b, or both (COR7ab). Bright-field enlargments of the olfactory epithelium labeled with the COR7ab probe (Fig. 3B), the COR7a probe (Fig. 3D) or the COR7b probe (Fig. 3F) are shown. The labeled cells are likely to correspond to olfactory neurons since earlier observations reported by Leibovici et al. (8) indicate that olfactory nerve axotomy, which induce the death of mature olfactory neurons, abolished the expression of COR genes. At E16, but also from E6 to E18, positive COR7a and COR7b olfactory neurons were distributed randomly within the olfactory epithelium. No zonal expressions neither in the dorso-ventral and in the anterior–posterior dimensions were observed. The cell bodies of the mature olfactory neurons are located in the middle layer of the olfactory epithelium, while those of the supporting cells are found at the outer surface, and those of the basal cells, which are the precursors of the olfactory neurons, are found at the base of the epithelium. At E7 and E8, most of the labeled cells were localized at the base of the olfactory epithelium. Later, from E14 to E18, most of the COR7a- and COR7b-positive cells were found in the middle cell body layer corresponding to the olfactory neurons. Sometimes, labeled cells were found at the base or at the surface of the epithelium. At all stages, the number of silver grains was always smaller for a COR7a-positive cell than for a COR7b-labeled cell, indicating that the COR7a gene expression was apparently smaller. It is possible that the COR7a probe hybridized to a smaller portion of mRNA transcripts, either because it had a smaller size (300 bases in comparison with 480 bases for the COR7b probe) or because a small intron is present in the COR7a probe. In situ experiments with E4 and E5 olfactory epithelium sections were not performed since we have already shown that signals with the COR7ab probe are hardly, if at all, detectable at these early stages (0–1 positive cell per section at E5, see table 2 in ref. 7). However, from E6 to hatching, the relative levels of gene expression for COR7a and COR7b remained stable during development with ≈40% of COR7a-positive cells and ≈60% of COR7b cells. The total number of COR7ab-labeled cells increased regularly from E6 to E16 (data not shown). There were ≈2 positive cells per section at E6, ≈12 cells per section at E7, ≈70 cells per section at E8 and E10, ≈84 cells per section at E12, 112 positive cells per section at E14, and finally ≈204 positive cells per section at E16. Later in development, the number of labeled cells decreased for an unknown reason to ≈60 cells per section at E18 and increased to ≈173 cells per section at hatching. The decreased of transcription levels for COR7a and 7b at E18 may represent apoptosis of distinct sensory neurons while specific connections are being established between the olfactory epithelium and the bulb, or since the number of sections (≈30) and animals (≈10) tested by in situ is rather limited, it may not be statistically significant. Interestingly, the sum of individual COR7a- and COR7b-positive cells corresponded to the number of positive cells detected with the COR7ab PCR probe, indicating that a single olfactory neuron does not coexpress the two COR7 genes.
Figure 3.
mRNA localization of COR7a and COR7b in the olfactory epithelium. Serial adjacent sections (14 μm) of the nasal cavity at E16 were hybridized with 35S-labeled antisense RNA probes prepared from COR7a (C and D), COR7b (E and F), and COR7ab (A and B) genes. B, D, and F represent enlargements of the olfactory epithelium covering the olfactory conchae (arrows in A, C, and E). With dark-field illumination, positive cells appear as white spots on a black background. At bright-field and higher magnifications, positive cells are covered with dense silver grains. Note the random distributions of COR7a and COR7b transcripts within the olfactory epithelium and their localization in the middle layer of the epithelium that correspond to the nuclei of olfactory neurons. The sum of labeled cells with COR7a (≈110 cells) and COR7b (≈135 cells) corresponds approximately to the number of positive cells detected with the nonspecific probe COR7ab (≈255 cells), indicating that there is no coexpression of the two genes in the same cell. [Bars = 270 μm (A, C, and E) and 8 μm (B, D, and F).]
Expression of COR7b in the Notochord.
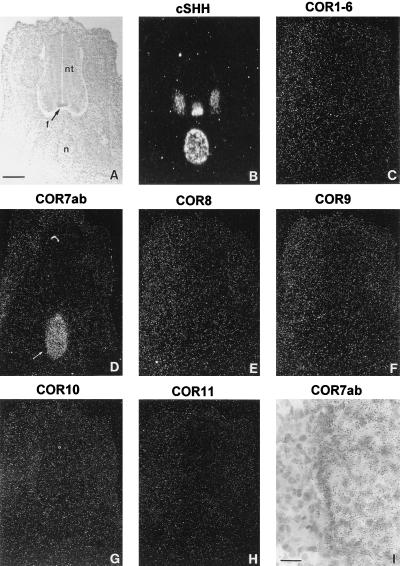
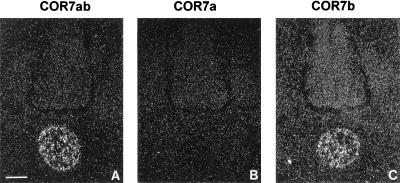
The notochord is a mesodermal tissue located just ventral to the neural tube (Fig. 4A) and plays a key role during the dorso-ventral development of the neural tube. It induces the differentiation of the floor plate by contact dependent signal(s) and of motor neurons by diffusion signal(s) (28–30). Removal of the notochord leads to the formation of spinal chord that is devoid of floor plate, motor-neurons, and ventral neurons (31), whereas grafted notochord gives rise to additional ectopic floor plate and motor neurons (31, 32). In addition, the notochord has also been implicated in other developmental processes such as the formation of the vertebrate cartilage (33, 34) and the modulation of neural crest cell migration and differentiation (35). Surprisingly, in situ hybridization analysis of serial coronal sections of whole chicken embryos at E5 with the COR7ab PCR probe revealed a positive signal in the notochord (Fig. 4D). No signal was observed in any other region (excepted in the olfactory system, see above). Since the COR gene family is composed of 12 members subdivided into six subfamilies, we decided to analyze adjacent sections by in situ hybridization with probes derived from each subfamily. The six probes were prepared as described in the experimental procedures. No signal was detected in the notochord with COR1–6 (Fig. 4C), nor with COR8 (Fig. 4E), COR9 (Fig. 4F), COR10 (Fig. 4G), or COR11 (Fig. 4H). As a positive control, we hybridized an adjacent section with an antisense RNA probe from the chicken Sonic Hedgehog gene (cSHH) (36), which can be used as marker for the notochord and the floor plate (Fig. 4B). We then tested whether COR7a, COR7b, or both were expressed in the notochord. As shown in Fig. 5C, only COR7b transcripts were detected with the corresponding specific probe. No signal was detected with the antisense probe specific for COR7a (Fig. 5B). The positive signals obtained with the COR7ab probe (Fig. 5A) or with the COR7b probe were quantitatively comparable, indicating that, indeed, the signal corresponded to COR7b only. In addition, in situ hybridization experiments performed with parasagittal sections and hybridized with the COR7ab probe revealed that the COR7b expression was constant along the rostro-caudal axis and that no gradient was observed (data not shown). We concluded that the majority of the cells in the notochord are positive for COR7b. Although with 35S probes (diffuse signals) and the highest possible magnification (to visualize individual cells), we could not exclude that a small cell subpopulation may not express COR7b, control experiments done with the sense probe encoding COR7ab failed to produce any signals.
Figure 4.
Expression of the chicken olfactory receptor gene family in the notochord. (A) Coronal sections (14 μm) of the neural tube (nt) and the notochord (n) at E5 were stained with toluidine blue. (B) Adjacent sections were hybridized with an antisense RNA probe from the chicken Sonic Hedgehog (cSHH) used as a marker of the notochord and of the floor plate (f). (C–H) Serial adjacent sections were hybridized with antisense RNA probes prepared from all the known members of the COR gene family: COR1–6 (C), COR7ab (D), COR8 (E), COR9 (F), COR10 (G), and COR11 (H). Note that positive signals were detected only with the COR7ab probe. (I) Higher magnification of a region (arrow in D) of the notochord labeled with the COR7ab antisense RNA probe. [Bars = 34 μm (A–G) and 5 μm (I).]
Figure 5.
Specific expression of the COR7b gene in the notochord. Serial coronal sections (14 μm) at E5 were hybridized with specific antisense RNA probes of the COR7 gene subfamily that can distinguish between COR7a (B) and COR7b (C) transcripts. Quantitatively, the signal detected with the specific COR7b probe is comparable to the signal obtained with the nonspecific COR7ab probe shown in A. Note the absence of labeled cells with the COR7a probe, indicating that only the COR7b transcripts are present in the notochord. (Bar = 34 μm.)
Transient Expression of COR7b in the Notochord.
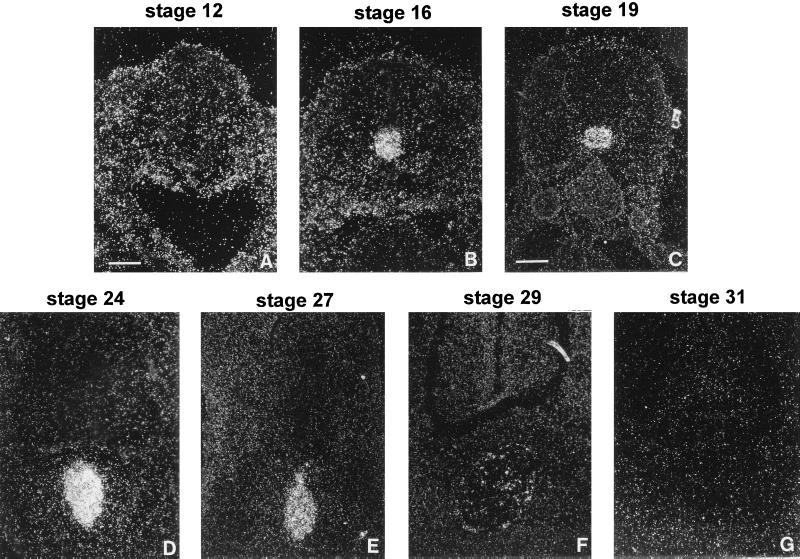
To characterize the level of COR7b gene expression in the notochord during embryonic development, we performed in situ hybridization experiments at different stages with the COR7a- or COR7b-specific probes, and as a control with the nonspecific COR7ab probe. No signals were detected in the notochord or in any tissue at embryonic stage 9 (29–33 hr of incubation) or stage 12 (45–50 hr) according to the Hamburger and Hamilton’s classification (37). However, positive cells for COR7b and COR7ab were first detected at stage 16 (51–56 hr) and then at stage 17 (52–64 hr), 19 (E3), 24 (E4), 27 (E5), and 29 (E6) but not later in development (stage 31 or E7, E8, E9, E10, E12, E14, E16, and E18). No signal was observed with the COR7a-specific probe at all the tested stages. Coronal sections of the notochord at different stages of the development and hybridized with the COR7ab probe are shown in Fig. 6. The number of COR7b-positive cells increased gradually until stage 24 where it reached a maximum, then the signal decreased rapidly (Fig. 6, stage 29), and completely disappeared at stage 31. No signals were detected in the notochord at any stage with antisense probes derived from the other known subfamily members, such as COR1–6, COR7a, COR8, COR9, COR10, or COR11, indicating that among the COR gene family, COR7b is uniquely expressed in the notochord. Note that the level of COR7b gene expression (transcripts per cell) was difficult to estimate because it was very small (COR7b-positive signals in the notochord were only detectable after ∼20 days of exposure), when compared with the high level of expression for the control cSHH gene (only 3–5 days of exposure were required).
Figure 6.
Transient expression of COR7b in the notochord. Coronal sections of the notochord at stage 12 (A), 16 (B), 19 (C), 24 (D), 27 (E), 29 (F), and 31 (G) were hybridized with antisense RNA probes specific for COR7a or COR7b and with the nonspecific probe COR7ab. Only the signals corresponding to COR7b transcripts obtained with the COR7ab probe are shown. Chicken embryos reach stage 12 after 45–50 hr of incubation, stage 16 after 51–56 hr, stage 19 after 3 days (E3), stage 24 at E4, stage 27 at E5, stage 29 at E6, and stage 31 at E7. COR7b transcripts are detected from stage 16 to stage 29. Note that the COR7b signal intensity reaches a peak at E4. [Bars = 15 μm (A, B, and D) and 34 μm (C and E–G).]
DISCUSSION
COR7b cDNA in the Notochord.
To ensure that COR7b mRNA transcripts were present in the notochord, we performed reverse transcription-coupled PCR (RT–PCR) with primers containing the start codon (ATG) and the stop codon (TGA) of the COR7b gene. A single product of ∼1000 nucleotides was amplified by RT–PCR with cDNA from the notochord but not from the retina (data not shown). The product was sequenced, and we observed that the insert encoded a complete open reading frame (from ATG to TGA) identical to the genomic DNA sequence derived from the COR7b gene.
The chicken olfactory system represents a very attractive model to study olfaction not only because of its simple structure but because of the small size of the chicken olfactory receptor gene repertoire that reduces the complexity of the system dramatically. Herein, we have characterized two highly related putative OR genes, named COR7a and COR7b. These receptor genes are expressed randomly in the avian olfactory epithelium and with a percentage of expression of ∼40% for COR7a and ∼60% for COR7b. Interestingly, COR7b alone is expressed in the notochord, indicating that some member(s) of the olfactory receptor gene family are expressed outside the olfactory system and may have additional function(s), unrelated to olfaction (see below).
Expression of Putative ORs Outside the Olfactory System.
Out of the several hundred olfactory receptor genes estimated in the rodent genome, only a small fraction have been sequenced and analyzed. The majority of these olfactory receptor genes are expressed in the olfactory epithelium, and despite the fact that it has not yet been definitely proven, it is likely that these receptors are involved in the detection of odorants. However, recent data indicate that several members of the olfactory receptor gene family are expressed in tissues unrelated to olfaction. Some of these “putative ORs” have been found to be expressed in taste buds (23, 24), in germ cells (4, 22), in the developing heart (25), and along the olfactory nerve (7, 8). Herein, we report the transient expression of such a receptor in the notochord. Thus, these data suggest strongly that some members of the olfactory receptor family are implicated in other functions.
A “non-olfactory receptor” cannot be distinguished from a “putative OR” by comparison of their molecular sequences but by the analysis of their patterns of expression. For example, in dog, it has been shown that among a single subfamily of related OR genes, some are expressed in the olfactory epithelium whereas others are expressed in germ cells (22). Moreover, herein we show that the COR7a and COR7b genes, which posses 96% of amino acid identity, are both expressed in the olfactory epithelium, but only one (COR7b) is present in the notochord.
Just recently, indirect functional evidence for an interaction between a G-protein-coupled receptor (ODR-10) and the odorant diacetyl has been reported in Caenorhabditis elegans (38). The odr-10 gene encodes a seven-transmembrane-domain OR that has no sequence homology (less than 10%) with any members of the putative OR family characterized in lower and higher vertebrates. This gene is only expressed by AWA neurons, two of the 32 chemosensory neurons in C. elegans. More than 40 highly divergent members of this family of G-protein-coupled receptor in the C. elegans have been identified (39). Eleven of 14 tested receptor genes appear to be expressed in chemosensory neurons, but as observed in vertebrates, three of them are expressed in nonsensory cells. For example, the gene srg-12 is expressed in excretory cells and in the gut. The linked sra-10 and sra-11 genes are expressed in other sensory neurons, in interneurons, in pharyngeal neurons, and in muscles. Similarly, herein we show that both the COR7a and COR7b genes, which posses 96% of amino acid identity, are expressed in the olfactory epithelium, but only one is detected in the notochord. Below we discuss the potential role(s) of a chicken putative OR both in the olfactory epithelium and in the notochord.
COR7a and COR7b Are Putative ORs.
The reported expression of COR7a and COR7b in the olfactory epithelium, presumably in the sensory olfactory neurons, strongly suggests that they could function as ORs. However, until proved otherwise, they should be regarded as orphan receptors. As reported with members of two other COR subfamily (7, 8), we observe that the COR7a and COR7b genes are expressed at early stages (∼E5) of the development of the olfactory system. Are the COR functional at those early stages? What is (are) the ligand(s) present during development that could activate these receptors? Is the ligand an odor molecule? Unfortunately, we are unable to provide any answers to those important questions. However, our data suggest that the COR genes are expressed prior to olfactory synaptogenesis, which first takes place at E8–E10 (21), indicating that the choice of receptor expression by an individual olfactory neuron is predetermined and not influenced by a retrograde effect of the olfactory bulb. We also observe a dramatic increase in the number of COR-positive cells between E7 and E8, which coincides with the synaptogenesis of olfactory axons in the bulb. It is possible that appropriate synaptic connections in the olfactory bulb promote the maturation and survival of olfactory neurons. This may also explain why, between E6 and E8, we observe positive cells throughout the entire thickness of the olfactory epithelium, and then gradually during the course of development, a greater portion of the COR-positive cells are observed in the middle nuclear layer corresponding to the position of mature olfactory neuron nuclei. The finding that even at E16 or E18, we still detect COR-positive cells at the base of the olfactory epithelium suggests either that the olfactory epithelium is continuously regenerating or indicates the presence of newly formed immature olfactory neurons.
Although the COR7a and COR7b genes are very similar at the nucleic acid level, we do not observe coexpression of these genes in the same neuron. Indeed, the sum of the positive cells for each individual specific probe (COR7a plus COR7b) is always equal to the number of positive cells detected with a common (nonspecific) probe (COR7ab). This is true for every tested stage. These data are corroborated with expression studies of other members of the COR gene family, indicating that a single olfactory neuron is committed to express only one receptor or a small subset of receptors from its limited COR gene repertoire.
Potential Role(s) for COR7b in the Notochord.
The most surprising feature reported herein is the expression of a putative OR in the notochord between E2 and E6. Several hypotheses can be envisaged to account for this unexpected expression in a nonneuronal tissue. Since our data are based on in situ hybridization experiments, we cannot be sure whether COR7b transcripts are translated into functional receptors, therefore it may be possible, although unlikely, that COR7b in the notochord has no relevant function. Alternatively, the notochord expression of COR7b may reflect a specific leakage in the tightly controlled transcriptional regulatory machinery of this particular gene, since the highly related COR7a gene is completely silent in this tissue. Since COR7b transcripts are detected, it is likely that the receptor protein is produced by the positive cells, but it may be incorrectly folded or just present at the cell surface with no function in the notochord. Another possibility is that the COR7b indeed plays a role as a receptor in the notochord. What this role would be remains highly speculative at present. The notochord is a mesodermal tissue that is found just ventral of the neural tube but is present only transiently during the embryonic development. Like other organizing structures such as the Hensen’s node or the zone of polarizing activity (ZPA) of the limb bud that express common molecular markers (for example, SHH), the notochord has inductive properties. For instance, it plays a key role in the establishment of the dorso-ventral polarity of the neural tube (40). The differentiation of floor plate cells and motor neurons is dependent on inductive signals emanating from the notochord (31, 40, 41). Removal of the notochord at stage 10 prevents later floor plate and ventral neuronal differentiation (31), while conversely, notochord grafts (stages 10–12) placed adjacent to the neural tube induce ectopic floor plate and ventral neuron differentiation 2 days later (32). In addition, it has been shown that, in vivo, the notochord and the floor plate possess also a dorso-ventral polarizing activity on the paraxial mesoderm (somites) (42). The dorsal part of the somites will segregate into the dermomyotome (which will yield the striated muscles and dermis) and the ventral part will segregate in the sclerotome (which will yield the vertebrae, the intervertebral disks, and ribs). Removal of the notochord promotes the differentiation of somites into dermomyotome, and conversely implantation of notochord grafts induces a ventralizing effect promoting the differentiation into sclerotome. Finally, it has been shown that the presence of the notochord is inhibitory to migrating neural crest cells (35). It has also been found that the notochord and its matrix attract motor axons. However, once these axons reach the immediate vicinity of the notochord, they circumnavigate its matrix as if they were inhibited by it (35). At present it is not clear what signal/ligand molecule(s) could be detected by the COR7b receptor in the notochord. We can postulate that the COR7b may detect an unknown signal or factor received by the notochord. Perhaps this signal is a feedback response originating from the neural tube, the somites, or the migrating neural crest cells in response to the inductive signals emanating from the notochord.
If the COR7b receptor is functional in mesodermal cells of the notochord, these cells may possess, at least transiently, proteins involved in a signal transduction pathways similar or related to olfactory pathways. Clearly, the potential role of COR7b in the notochord during the early phases of development remains to be determined. However these data suggest strongly that some “putative ORs” may be implicated in the detection of biological molecules not directly involved in olfaction.
Acknowledgments
This research was supported by a grant from the Swiss National Science Foundation (31.32623.91).
ABBREVIATIONS
- E
embryonic day
- OR
odorant receptor
- COR
chicken OR
Footnotes
References
- 1.Nef P, Hermans-Borgmeyer I, Artieres-Pin H, Beasley L, Dionne V E, Heinemann S F. Proc Natl Acad Sci USA. 1992;89:8948–8952. doi: 10.1073/pnas.89.19.8948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ressler K J, Sullivan S L, Buck L B. Cell. 1993;73:597–609. doi: 10.1016/0092-8674(93)90145-g. [DOI] [PubMed] [Google Scholar]
- 3.Buck L, Axel R. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 4.Parmentier M, Libert F, Schurmans S, Schiffmann S, Lefort A, Eggerickx D, Ledent C, Mollereau C, Gerard C, Perret J, Grootegoed A, Vassart G. Nature (London) 1992;355:453–455. doi: 10.1038/355453a0. [DOI] [PubMed] [Google Scholar]
- 5.Barth A L, Justice N J, Ngai J. Neuron. 1996;16:23–34. doi: 10.1016/s0896-6273(00)80020-3. [DOI] [PubMed] [Google Scholar]
- 6.Ngai J, Dowling M M, Buck L, Axel R, Chess A. Cell. 1993;72:657–666. doi: 10.1016/0092-8674(93)90395-7. [DOI] [PubMed] [Google Scholar]
- 7.Nef S, Allaman I, Fiumelli H, de Castro E, Nef P. Mech Dev. 1996;55:65–77. doi: 10.1016/0925-4773(95)00491-2. [DOI] [PubMed] [Google Scholar]
- 8.Leibovici M, Lapointe F, Aletta P, Ayer-Le Lievre C. Dev Biol. 1996;175:118–131. doi: 10.1006/dbio.1996.0100. [DOI] [PubMed] [Google Scholar]
- 9.Ben Arie N, Lancet D, Taylor C, Khen M, Walker N, Ledbetter D H, Carrozzo R, Patel K, Sheer D, Lehrach H, North M A. Hum Mol Genet. 1994;3:229–235. doi: 10.1093/hmg/3.2.229. [DOI] [PubMed] [Google Scholar]
- 10.Nef P. Receptors Channels. 1993;1:259–266. [PubMed] [Google Scholar]
- 11.Buck L B. Annu Rev Neurosci. 1996;19:517–544. doi: 10.1146/annurev.ne.19.030196.002505. [DOI] [PubMed] [Google Scholar]
- 12.Ngai J, Chess A, Dowling M M, Necles N, Macagno E R, Axel R. Cell. 1993;72:667–680. doi: 10.1016/0092-8674(93)90396-8. [DOI] [PubMed] [Google Scholar]
- 13.Nef P, Heinemann S F, Dionne V E. Chem Senses. 1991;16:562–563. [Google Scholar]
- 14.Vassar R, Ngai J, Axel R. Cell. 1993;74:309–318. doi: 10.1016/0092-8674(93)90422-m. [DOI] [PubMed] [Google Scholar]
- 15.Strotmann J, Wanner I, Helfrich T, Beck A, Breer H. Cell Tissue Res. 1994;278:11–20. doi: 10.1007/BF00305773. [DOI] [PubMed] [Google Scholar]
- 16.Wenzel B M. Nature (London) 1968;220:1133–1134. doi: 10.1038/2201133a0. [DOI] [PubMed] [Google Scholar]
- 17.Wenzel B M. Beidler Handbook of Sensory Physiology. Vol. 4. Heidelberg: Springer; 1969. [Google Scholar]
- 18.Papi F. Experimentia. 1990;46:352–363. [Google Scholar]
- 19.Tolhurst B E, Vince M A. Anim Behav. 1976;24:772–779. doi: 10.1016/s0003-3472(76)80007-3. [DOI] [PubMed] [Google Scholar]
- 20.Burne T H J, Rogers L J. Int J Comp Psychol. 1995;8:98–113. [Google Scholar]
- 21.Ayer-Le Lievre C, Lapointe F, Leibovici M. Biol Cell. 1995;84:25–34. [Google Scholar]
- 22.Vanderhaeghen P, Schurmans S, Vassart G, Parmentier M. J Cell Biol. 1993;123:1441–1452. doi: 10.1083/jcb.123.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abe K, Kusakabe Y, Tanemura K, Emori Y, Arai S. FEBS Lett. 1993;316:253–256. doi: 10.1016/0014-5793(93)81302-g. [DOI] [PubMed] [Google Scholar]
- 24.Abe K, Kusakabe Y, Tanemura K, Emori Y, Arai S. J Biol Chem. 1993;268:12033–12039. [PubMed] [Google Scholar]
- 25.Drutel G, Arrang J M, Diaz J, Wisnewsky C, Schwartz K, Schwartz J C. Receptors Channels. 1995;3:33–40. [PubMed] [Google Scholar]
- 26.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5466. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox K H, Deleon D V, Angerer L M, Angerer R C. Dev Biol. 1984;101:485–502. doi: 10.1016/0012-1606(84)90162-3. [DOI] [PubMed] [Google Scholar]
- 28.Watterson R L, Goodheart C R, Lindberg G. Anat Rec. 1955;122:539–559. doi: 10.1002/ar.1091220405. [DOI] [PubMed] [Google Scholar]
- 29.Yamada T, Placzek M, Tanaka H, Dodd J, Jessell T M. Cell. 1991;64:635–647. doi: 10.1016/0092-8674(91)90247-v. [DOI] [PubMed] [Google Scholar]
- 30.Yamada T, Pfaff S L, Edlund T, Jessell T M. Cell. 1993;73:673–686. doi: 10.1016/0092-8674(93)90248-o. [DOI] [PubMed] [Google Scholar]
- 31.Placzek M, Tessier-Lavigne M, Yamada T, Jessell T, Dodd J. Science. 1990;250:985–988. doi: 10.1126/science.2237443. [DOI] [PubMed] [Google Scholar]
- 32.van Straaten H W, Hekking J W, Wiertz-Hoessels E J, Thors F, Drukker J. Anat Embryol. 1988;177:317–324. doi: 10.1007/BF00315839. [DOI] [PubMed] [Google Scholar]
- 33.Hall B K. Adv Anat Embryol Cell Biol. 1977;53:1–49. [PubMed] [Google Scholar]
- 34.Kosher R A, Lash J W. Dev Biol. 1975;42:362–378. doi: 10.1016/0012-1606(75)90340-1. [DOI] [PubMed] [Google Scholar]
- 35.Stern C D, Artinger K B, Bronner-Fraser M. Development (Cambridge, UK) 1991;113:207–216. doi: 10.1242/dev.113.1.207. [DOI] [PubMed] [Google Scholar]
- 36.Riddle R D, Johnson R L, Laufer E, Tabin C. Cell. 1993;75:1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- 37.Hamburger V, Hamilton H L. J Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
- 38.Sengupta P, Chou J H, Bargmann C I. Cell. 1996;84:899–909. doi: 10.1016/s0092-8674(00)81068-5. [DOI] [PubMed] [Google Scholar]
- 39.Troemel E R, Chou J H, Dwyer N D, Colbert H A, Bargmann C I. Cell. 1995;83:207–218. doi: 10.1016/0092-8674(95)90162-0. [DOI] [PubMed] [Google Scholar]
- 40.Yamada T, Pfaff S L, Edlund T, Jessell T M. Cell. 1993;73:673–86. doi: 10.1016/0092-8674(93)90248-o. [DOI] [PubMed] [Google Scholar]
- 41.Placzek M, Jessell T M, Dodd J. Development (Cambridge, UK) 1993;117:205–218. doi: 10.1242/dev.117.1.205. [DOI] [PubMed] [Google Scholar]
- 42.Pourquie O, Coltey M, Teillet M A, Ordahl C, Le Douarin N M. Proc Natl Acad Sci USA. 1993;90:5242–5246. doi: 10.1073/pnas.90.11.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]