Abstract
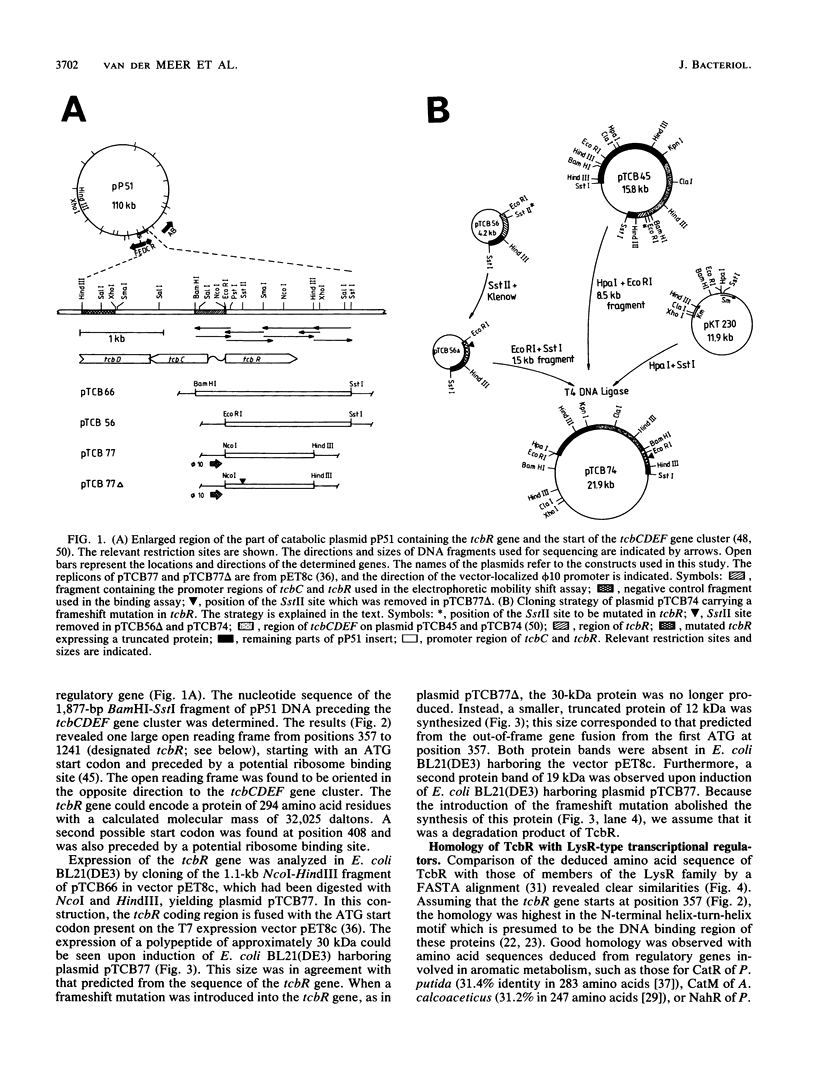
Plasmid pP51 of Pseudomonas sp. strain P51 contains two gene clusters encoding the degradation of chlorinated benzenes, tcbAB and tcbCDEF. A regulatory gene, tcbR, was located upstream and divergently transcribed from the chlorocatechol oxidative gene cluster tcbCDEF. The tcbR gene was characterized by DNA sequencing and expression studies with Escherichia coli and pET8c and appeared to encode a 32-kDa protein. The activity of the tcbR gene product was analyzed in Pseudomonas putida KT2442, in which it appeared to function as a positive regulator of tcbC expression. Protein extracts of both E. coli overproducing TcbR and Pseudomonas sp. strain P51 showed specific DNA binding to the 150-bp region that is located between the tcbR and tcbC genes. Primer extension mapping demonstrated that the transcription start sites of tcbR and tcbC are located in this region and that the divergent promoter sequences of both genes overlap. Amino acid sequence comparisons indicated that TcbR is a member of the LysR family of transcriptional activator proteins and shares a high degree of homology with other activator proteins involved in regulating the metabolism of aromatic compounds.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abril M. A., Michan C., Timmis K. N., Ramos J. L. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol. 1989 Dec;171(12):6782–6790. doi: 10.1128/jb.171.12.6782-6790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiba H., Adhya S., de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981 Nov 25;256(22):11905–11910. [PubMed] [Google Scholar]
- Aldrich T. L., Frantz B., Gill J. F., Kilbane J. J., Chakrabarty A. M. Cloning and complete nucleotide sequence determination of the catB gene encoding cis,cis-muconate lactonizing enzyme. Gene. 1987;52(2-3):185–195. doi: 10.1016/0378-1119(87)90045-x. [DOI] [PubMed] [Google Scholar]
- Aldrich T. L., Rothmel R. K., Chakrabarty A. M. Identification of nucleotides critical for activity of the Pseudomonas putida catBC promoter. Mol Gen Genet. 1989 Aug;218(2):266–271. doi: 10.1007/BF00331277. [DOI] [PubMed] [Google Scholar]
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C., Bagdasarian M. M., Frey J., Timmis K. N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981 Dec;16(1-3):237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chatterjee D. K., Chakrabarty A. M. Restriction mapping of a chlorobenzoate degradative plasmid and molecular cloning of the degradative genes. Gene. 1984 Feb;27(2):173–181. doi: 10.1016/0378-1119(84)90138-0. [DOI] [PubMed] [Google Scholar]
- Chatterjee D. K., Kellogg S. T., Hamada S., Chakrabarty A. M. Plasmid specifying total degradation of 3-chlorobenzoate by a modified ortho pathway. J Bacteriol. 1981 May;146(2):639–646. doi: 10.1128/jb.146.2.639-646.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Don R. H., Weightman A. J., Knackmuss H. J., Timmis K. N. Transposon mutagenesis and cloning analysis of the pathways for degradation of 2,4-dichlorophenoxyacetic acid and 3-chlorobenzoate in Alcaligenes eutrophus JMP134(pJP4). J Bacteriol. 1985 Jan;161(1):85–90. doi: 10.1128/jb.161.1.85-90.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn E., Hellwig M., Reineke W., Knackmuss H. J. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch Microbiol. 1974;99(1):61–70. doi: 10.1007/BF00696222. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. F., Long S. R. DNA footprint analysis of the transcriptional activator proteins NodD1 and NodD3 on inducible nod gene promoters. J Bacteriol. 1989 Oct;171(10):5492–5502. doi: 10.1128/jb.171.10.5492-5502.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Bagdasarian M., Bagdasarian M. M., Timmis K. N. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz B., Chakrabarty A. M. Organization and nucleotide sequence determination of a gene cluster involved in 3-chlorocatechol degradation. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4460–4464. doi: 10.1073/pnas.84.13.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz B., O'Halloran T. V. DNA distortion accompanies transcriptional activation by the metal-responsive gene-regulatory protein MerR. Biochemistry. 1990 May 22;29(20):4747–4751. doi: 10.1021/bi00472a001. [DOI] [PubMed] [Google Scholar]
- Ghosal D., You I. S. Nucleotide homology and organization of chlorocatechol oxidation genes of plasmids pJP4 and pAC27. Mol Gen Genet. 1988 Jan;211(1):113–120. doi: 10.1007/BF00338401. [DOI] [PubMed] [Google Scholar]
- Ghosal D., You I. S. Operon structure and nucleotide homology of the chlorocatechol oxidation genes of plasmids pJP4 and pAC27. Gene. 1989 Nov 30;83(2):225–232. doi: 10.1016/0378-1119(89)90108-x. [DOI] [PubMed] [Google Scholar]
- Harker A. R., Olsen R. H., Seidler R. J. Phenoxyacetic acid degradation by the 2,4-dichlorophenoxyacetic acid (TFD) pathway of plasmid pJP4: mapping and characterization of the TFD regulatory gene, tfdR. J Bacteriol. 1989 Jan;171(1):314–320. doi: 10.1128/jb.171.1.314-320.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartnett C., Neidle E. L., Ngai K. L., Ornston L. N. DNA sequences of genes encoding Acinetobacter calcoaceticus protocatechuate 3,4-dioxygenase: evidence indicating shuffling of genes and of DNA sequences within genes during their evolutionary divergence. J Bacteriol. 1990 Feb;172(2):956–966. doi: 10.1128/jb.172.2.956-966.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Haughn G. W., Calvo J. M., Wallace J. C. A large family of bacterial activator proteins. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6602–6606. doi: 10.1073/pnas.85.18.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Wallace J. C., Brown J. P. Finding protein similarities with nucleotide sequence databases. Methods Enzymol. 1990;183:111–132. doi: 10.1016/0076-6879(90)83009-x. [DOI] [PubMed] [Google Scholar]
- Kaphammer B., Kukor J. J., Olsen R. H. Regulation of tfdCDEF by tfdR of the 2,4-dichlorophenoxyacetic acid degradation plasmid pJP4. J Bacteriol. 1990 May;172(5):2280–2286. doi: 10.1128/jb.172.5.2280-2286.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaphammer B., Olsen R. H. Cloning and characterization of tfdS, the repressor-activator gene of tfdB, from the 2,4-dichlorophenoxyacetic acid catabolic plasmid pJP4. J Bacteriol. 1990 Oct;172(10):5856–5862. doi: 10.1128/jb.172.10.5856-5862.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhm A. E., Schlömann M., Knackmuss H. J., Pieper D. H. Purification and characterization of dichloromuconate cycloisomerase from Alcaligenes eutrophus JMP 134. Biochem J. 1990 Mar 15;266(3):877–883. [PMC free article] [PubMed] [Google Scholar]
- Neidle E. L., Hartnett C., Bonitz S., Ornston L. N. DNA sequence of the Acinetobacter calcoaceticus catechol 1,2-dioxygenase I structural gene catA: evidence for evolutionary divergence of intradiol dioxygenases by acquisition of DNA sequence repetitions. J Bacteriol. 1988 Oct;170(10):4874–4880. doi: 10.1128/jb.170.10.4874-4880.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidle E. L., Hartnett C., Ornston L. N. Characterization of Acinetobacter calcoaceticus catM, a repressor gene homologous in sequence to transcriptional activator genes. J Bacteriol. 1989 Oct;171(10):5410–5421. doi: 10.1128/jb.171.10.5410-5421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins E. J., Gordon M. P., Caceres O., Lurquin P. F. Organization and sequence analysis of the 2,4-dichlorophenol hydroxylase and dichlorocatechol oxidative operons of plasmid pJP4. J Bacteriol. 1990 May;172(5):2351–2359. doi: 10.1128/jb.172.5.2351-2359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J. L., Michan C., Rojo F., Dwyer D., Timmis K. Signal-regulator interactions. Genetic analysis of the effector binding site of xylS, the benzoate-activated positive regulator of Pseudomonas TOL plasmid meta-cleavage pathway operon. J Mol Biol. 1990 Jan 20;211(2):373–382. doi: 10.1016/0022-2836(90)90358-S. [DOI] [PubMed] [Google Scholar]
- Reineke W., Knackmuss H. J. Microbial degradation of haloaromatics. Annu Rev Microbiol. 1988;42:263–287. doi: 10.1146/annurev.mi.42.100188.001403. [DOI] [PubMed] [Google Scholar]
- Rosenberg A. H., Lade B. N., Chui D. S., Lin S. W., Dunn J. J., Studier F. W. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56(1):125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- Rothmel R. K., Aldrich T. L., Houghton J. E., Coco W. M., Ornston L. N., Chakrabarty A. M. Nucleotide sequencing and characterization of Pseudomonas putida catR: a positive regulator of the catBC operon is a member of the LysR family. J Bacteriol. 1990 Feb;172(2):922–931. doi: 10.1128/jb.172.2.922-931.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell M. A. Homology between nucleotide sequences of promoter regions of nah and sal operons of NAH7 plasmid of Pseudomonas putida. Proc Natl Acad Sci U S A. 1986 Jan;83(2):369–373. doi: 10.1073/pnas.83.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell M. A., Sukordhaman M. Evidence that the transcription activator encoded by the Pseudomonas putida nahR gene is evolutionarily related to the transcription activators encoded by the Rhizobium nodD genes. J Bacteriol. 1989 Apr;171(4):1952–1959. doi: 10.1128/jb.171.4.1952-1959.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell M. A., Wender P. E. Identification of the nahR gene product and nucleotide sequences required for its activation of the sal operon. J Bacteriol. 1986 Apr;166(1):9–14. doi: 10.1128/jb.166.1.9-14.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlömann M., Schmidt E., Knackmuss H. J. Different types of dienelactone hydrolase in 4-fluorobenzoate-utilizing bacteria. J Bacteriol. 1990 Sep;172(9):5112–5118. doi: 10.1128/jb.172.9.5112-5118.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Streber W. R., Timmis K. N., Zenk M. H. Analysis, cloning, and high-level expression of 2,4-dichlorophenoxyacetate monooxygenase gene tfdA of Alcaligenes eutrophus JMP134. J Bacteriol. 1987 Jul;169(7):2950–2955. doi: 10.1128/jb.169.7.2950-2955.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- You I. S., Ghosal D., Gunsalus I. C. Nucleotide sequence of plasmid NAH7 gene nahR and DNA binding of the nahR product. J Bacteriol. 1988 Dec;170(12):5409–5415. doi: 10.1128/jb.170.12.5409-5415.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer J. R., Eggen R. I., Zehnder A. J., de Vos W. M. Sequence analysis of the Pseudomonas sp. strain P51 tcb gene cluster, which encodes metabolism of chlorinated catechols: evidence for specialization of catechol 1,2-dioxygenases for chlorinated substrates. J Bacteriol. 1991 Apr;173(8):2425–2434. doi: 10.1128/jb.173.8.2425-2434.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer J. R., van Neerven A. R., de Vries E. J., de Vos W. M., Zehnder A. J. Cloning and characterization of plasmid-encoded genes for the degradation of 1,2-dichloro-, 1,4-dichloro-, and 1,2,4-trichlorobenzene of Pseudomonas sp. strain P51. J Bacteriol. 1991 Jan;173(1):6–15. doi: 10.1128/jb.173.1.6-15.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]