Abstract
Enzyme IIIMtl is part of the mannitol phosphotransferase system of Enterococcus faecalis. It is phosphorylated in a reaction sequence requiring enzyme I and heat-stable phosphocarrier protein (HPr). The phospho group is transferred from enzyme IIIMtl to enzyme IIMtl, which then catalyzes the uptake and concomitant phosphorylation of mannitol. The internalized mannitol-1-phosphate is oxidized to fructose-6-phosphate by mannitol-1-phosphate dehydrogenase. In this report we describe the cloning of the mtlF and mtlD genes, encoding enzyme IIIMtl and mannitol-1-phosphate dehydrogenase of E. faecalis, by a complementation system designed for cloning of gram-positive phosphotransferase system genes. The complete nucleotide sequences of mtlF, mtlD, and flanking regions were determined. From the gene sequences, the primary translation products are deduced to consist of 145 amino acids (enzyme IIIMtl) and 374 amino acids (mannitol-1-phosphate dehydrogenase). Amino acid sequence comparison confirmed a 41% similarity of E. faecalis enzyme IIIMtl to the hydrophilic enzyme IIIMtl-like portion of enzyme IIMtl of Escherichia coli and 45% similarity to enzyme IIIMtl of Staphylococcus carnosus. The putative N-terminal NAD+ binding domain of mannitol-1-phosphate dehydrogenase of E. faecalis shows a high degree of similarity with the N terminus of E. coli mannitol-1-phosphate dehydrogenase (T. Davis, M. Yamada, M. Elgort, and M. H. Saier, Jr., Mol. Microbiol. 2:405-412, 1988) and the N-terminal part of the translation product of S. carnosus mtlD, which was also determined in this study. There is 40% similarity between the dehydrogenases of E. faecalis and E. coli over the whole length of the enzymes. The organization of mannitol-specific genes in E. faecalis seems to be similar to the organization in S. carnosus. The open reading frame for enzyme IIIMtl E. faecalis is followed by a stem-loop structure, analogous to a typical Rho-independent terminator. We conclude that the mannitol-specific genes are organized in an operon and that the gene order is mtlA orfX mtlF mtlD.
Full text
PDF
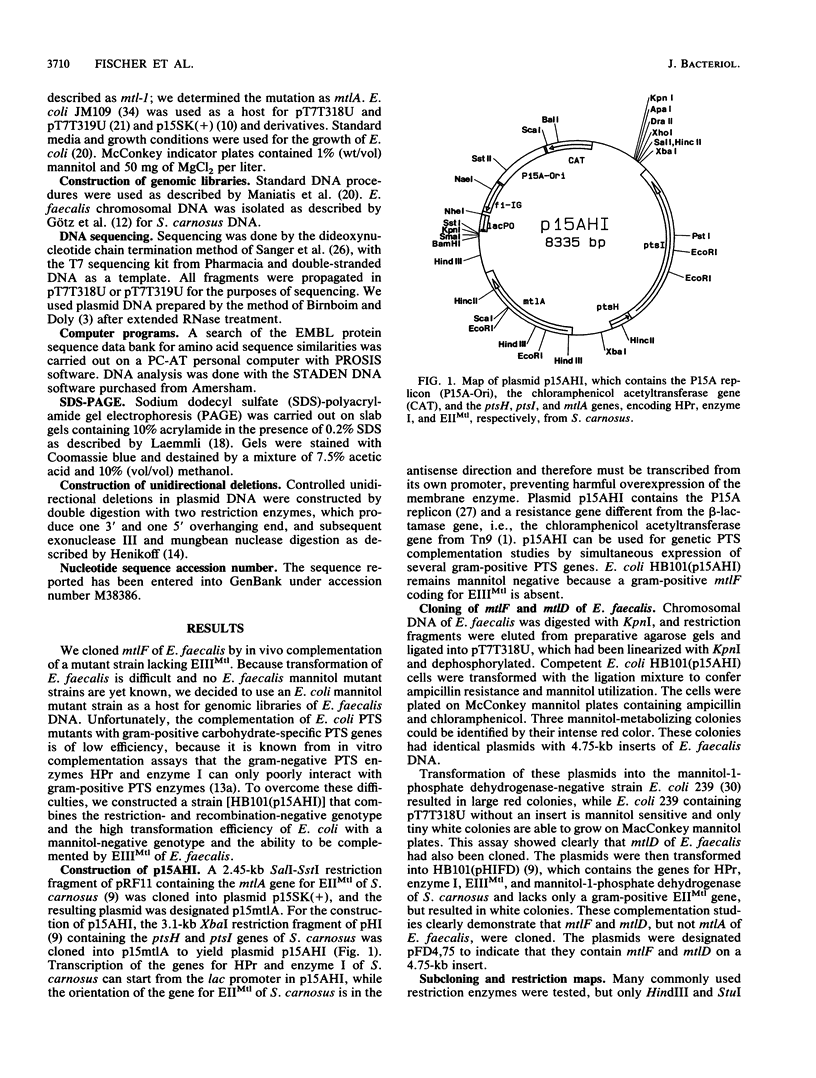
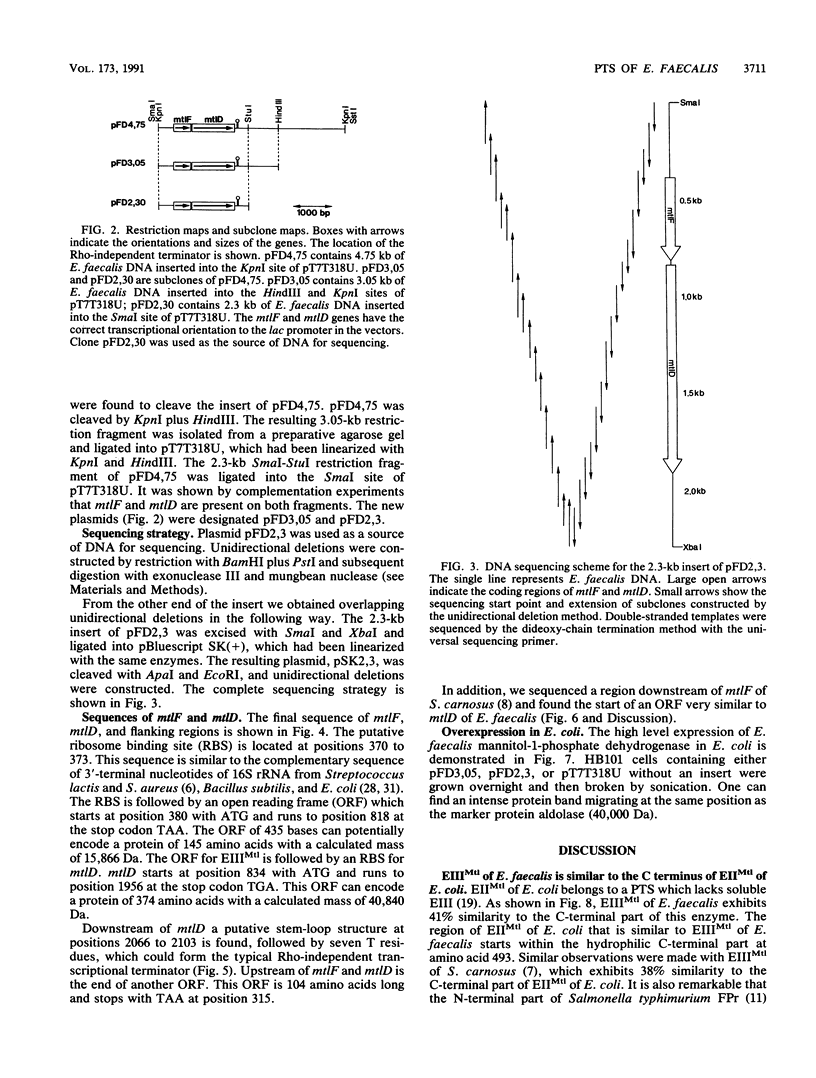

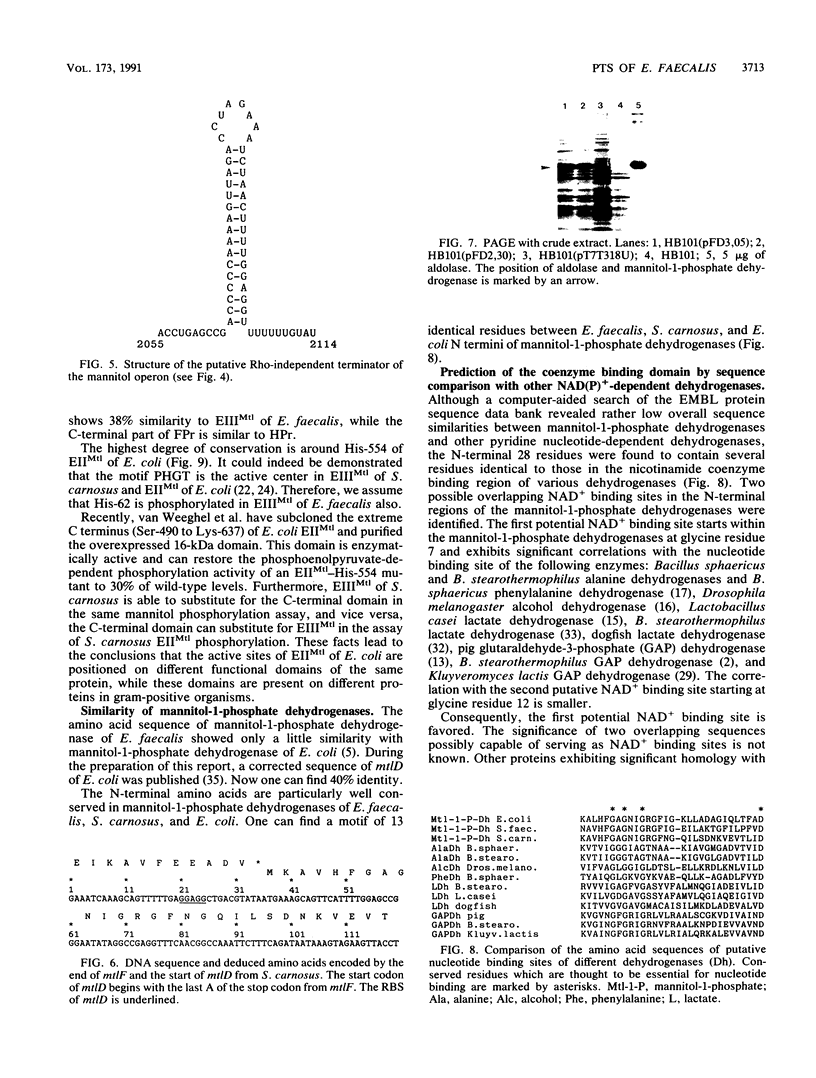


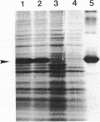
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alton N. K., Vapnek D. Nucleotide sequence analysis of the chloramphenicol resistance transposon Tn9. Nature. 1979 Dec 20;282(5741):864–869. doi: 10.1038/282864a0. [DOI] [PubMed] [Google Scholar]
- Biesecker G., Harris J. I., Thierry J. C., Walker J. E., Wonacott A. J. Sequence and structure of D-glyceraldehyde 3-phosphate dehydrogenase from Bacillus stearothermophilus. Nature. 1977 Mar 24;266(5600):328–333. doi: 10.1038/266328a0. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Davis T., Yamada M., Elgort M., Saier M. H., Jr Nucleotide sequence of the mannitol (mtl) operon in Escherichia coli. Mol Microbiol. 1988 May;2(3):405–412. doi: 10.1111/j.1365-2958.1988.tb00045.x. [DOI] [PubMed] [Google Scholar]
- Fischer R., Eisermann R., Reiche B., Hengstenberg W. Cloning, sequencing and overexpression of the mannitol-specific enzyme-III-encoding gene of Staphylococcus carnosus. Gene. 1989 Oct 30;82(2):249–257. doi: 10.1016/0378-1119(89)90050-4. [DOI] [PubMed] [Google Scholar]
- Geerse R. H., Izzo F., Postma P. W. The PEP: fructose phosphotransferase system in Salmonella typhimurium: FPr combines enzyme IIIFru and pseudo-HPr activities. Mol Gen Genet. 1989 Apr;216(2-3):517–525. doi: 10.1007/BF00334399. [DOI] [PubMed] [Google Scholar]
- Götz F., Zabielski J., Philipson L., Lindberg M. DNA homology between the arsenate resistance plasmid pSX267 from Staphylococcus xylosus and the penicillinase plasmid pI258 from Staphylococcus aureus. Plasmid. 1983 Mar;9(2):126–137. doi: 10.1016/0147-619x(83)90015-x. [DOI] [PubMed] [Google Scholar]
- Harris J. I., Perham R. N. Glyceraldehyde 3-phosphate dehydrogenase from pig muscle. Nature. 1968 Sep 7;219(5158):1025–1028. doi: 10.1038/2191025a0. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hensel R., Mayr U., Yang C. Y. The complete primary structure of the allosteric L-lactate dehydrogenase from Lactobacillus casei. Eur J Biochem. 1983 Aug 15;134(3):503–511. doi: 10.1111/j.1432-1033.1983.tb07595.x. [DOI] [PubMed] [Google Scholar]
- Kreitman M. Nucleotide polymorphism at the alcohol dehydrogenase locus of Drosophila melanogaster. Nature. 1983 Aug 4;304(5925):412–417. doi: 10.1038/304412a0. [DOI] [PubMed] [Google Scholar]
- Kuroda S., Tanizawa K., Sakamoto Y., Tanaka H., Soda K. Alanine dehydrogenases from two Bacillus species with distinct thermostabilities: molecular cloning, DNA and protein sequence determination, and structural comparison with other NAD(P)(+)-dependent dehydrogenases. Biochemistry. 1990 Jan 30;29(4):1009–1015. doi: 10.1021/bi00456a025. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee C. A., Saier M. H., Jr Mannitol-specific enzyme II of the bacterial phosphotransferase system. III. The nucleotide sequence of the permease gene. J Biol Chem. 1983 Sep 10;258(17):10761–10767. [PubMed] [Google Scholar]
- Mead D. A., Szczesna-Skorupa E., Kemper B. Single-stranded DNA 'blue' T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1986 Oct-Nov;1(1):67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- Pas H. H., Robillard G. T. S-phosphocysteine and phosphohistidine are intermediates in the phosphoenolpyruvate-dependent mannitol transport catalyzed by Escherichia coli EIIMtl. Biochemistry. 1988 Aug 9;27(16):5835–5839. doi: 10.1021/bi00416a002. [DOI] [PubMed] [Google Scholar]
- Pas H. H., ten Hoeve-Duurkens R. H., Robillard G. T. Bacterial phosphoenolpyruvate-dependent phosphotransferase system: mannitol-specific EII contains two phosphoryl binding sites per monomer and one high-affinity mannitol binding site per dimer. Biochemistry. 1988 Jul 26;27(15):5520–5525. doi: 10.1021/bi00415a020. [DOI] [PubMed] [Google Scholar]
- Reiche B., Frank R., Deutscher J., Meyer N., Hengstenberg W. Staphylococcal phosphoenolpyruvate-dependent phosphotransferase system: purification and characterization of the mannitol-specific enzyme IIImtl of Staphylococcus aureus and Staphylococcus carnosus and homology with the enzyme IImtl of Escherichia coli. Biochemistry. 1988 Aug 23;27(17):6512–6516. doi: 10.1021/bi00417a047. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selzer G., Som T., Itoh T., Tomizawa J. The origin of replication of plasmid p15A and comparative studies on the nucleotide sequences around the origin of related plasmids. Cell. 1983 Jan;32(1):119–129. doi: 10.1016/0092-8674(83)90502-0. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster J. R. Kluyveromyces lactis glyceraldehyde-3-phosphate dehydrogenase and alcohol dehydrogenase-1 genes are linked and divergently transcribed. Nucleic Acids Res. 1990 Jul 25;18(14):4271–4271. doi: 10.1093/nar/18.14.4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon E., Lin E. C. Mutations affecting the dissimilation of mannitol by Escherichia coli K-12. J Bacteriol. 1972 Aug;111(2):566–574. doi: 10.1128/jb.111.2.566-574.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Charldorp R., Van Knippenberg P. H. Sequence, modified nucleotides and secondary structure at the 3'-end of small ribosomal subunit RNA. Nucleic Acids Res. 1982 Feb 25;10(4):1149–1158. doi: 10.1093/nar/10.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga R. K., Terpstra P., Hol W. G. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986 Jan 5;187(1):101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- Wilks H. M., Hart K. W., Feeney R., Dunn C. R., Muirhead H., Chia W. N., Barstow D. A., Atkinson T., Clarke A. R., Holbrook J. J. A specific, highly active malate dehydrogenase by redesign of a lactate dehydrogenase framework. Science. 1988 Dec 16;242(4885):1541–1544. doi: 10.1126/science.3201242. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]