Abstract
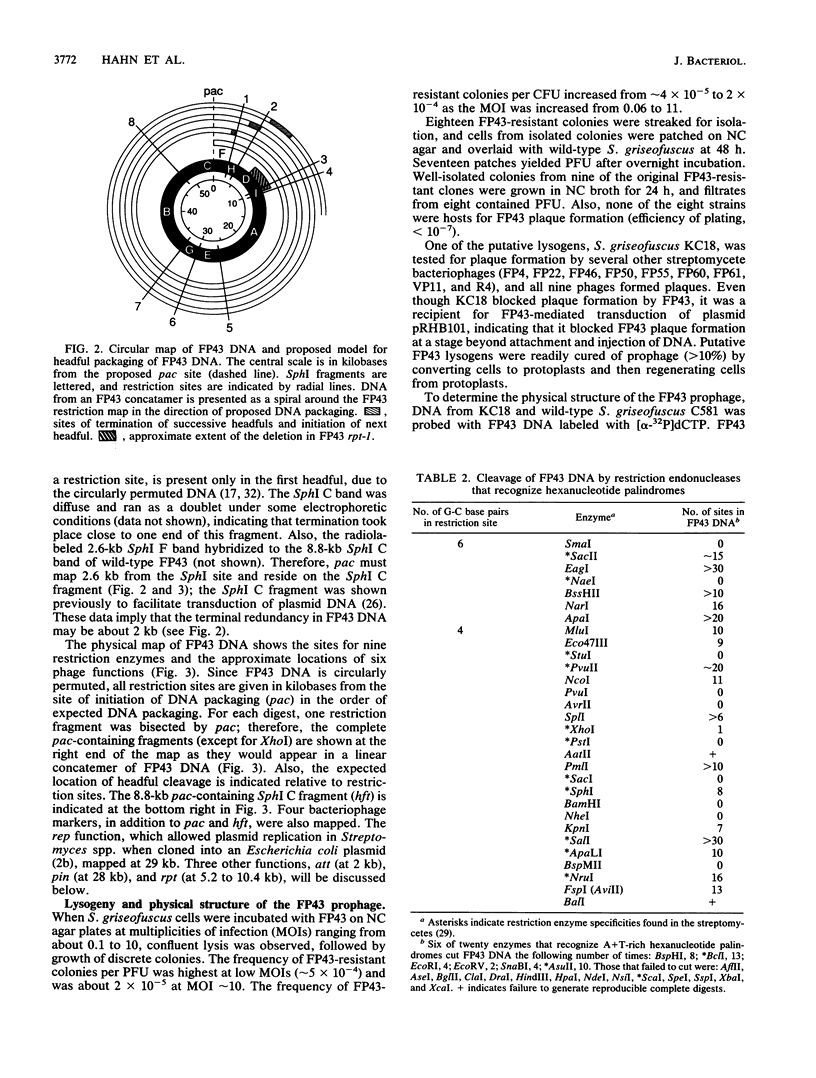
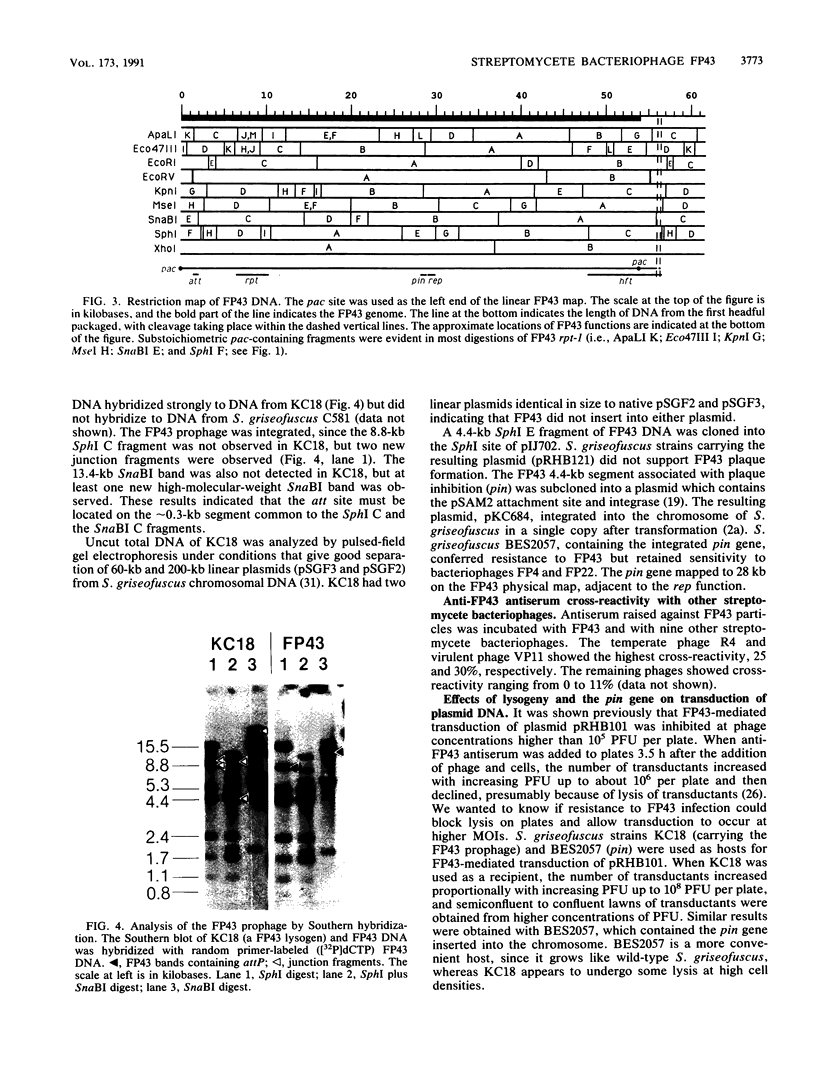
FP43 is a temperate bacteriophage for Streptomyces griseofuscus that forms plaques on many Streptomyces species. FP43 virions contain 56 kb of double-strand DNA that is circularly permuted and terminally redundant, and contains 65% G + C. A physical map of the FP43 genome was constructed, and the origin for headful packaging (pac) was localized to an 8.8-kb region of the genome (hft) that mediates high-frequency transduction by FP43 of plasmid pRHB101. The phage attachment site (attP), a replication origin (rep), a region that inhibits plaque formation (pin), and a 3-kb deletion (rpt) that caused a 100-fold reduction in plasmid transduction were mapped.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackermann H. W., Eisenstark A. The present state of phage taxonomy. Intervirology. 1974;3(4):201–219. doi: 10.1159/000149758. [DOI] [PubMed] [Google Scholar]
- Anné J., Wohlleben W., Burkardt H. J., Springer R., Pühler A. Morphological and molecular characterization of several actinophages isolated from soil which lyse Streptomyces cattleya or S. venezuelae. J Gen Microbiol. 1984 Oct;130(10):2639–2649. doi: 10.1099/00221287-130-10-2639. [DOI] [PubMed] [Google Scholar]
- Baltz R. H. Genetic recombination in Streptomyces fradiae by protoplast fusion and cell regeneration. J Gen Microbiol. 1978 Jul;107(1):93–102. doi: 10.1099/00221287-107-1-93. [DOI] [PubMed] [Google Scholar]
- Birch A. W., Cullum J. Temperature-sensitive mutants of the Streptomyces plasmid pIJ702. J Gen Microbiol. 1985 Jun;131(6):1299–1303. doi: 10.1099/00221287-131-6-1299. [DOI] [PubMed] [Google Scholar]
- Cox K. L., Baltz R. H. Restriction of bacteriophage plaque formation in Streptomyces spp. J Bacteriol. 1984 Aug;159(2):499–504. doi: 10.1128/jb.159.2.499-504.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowding J. E. Characterization of a bacteriophage virulent for Streptomyces coelicolor A3(2). J Gen Microbiol. 1973 May;76(1):163–176. doi: 10.1099/00221287-76-1-163. [DOI] [PubMed] [Google Scholar]
- Fishman S. E., Hershberger C. L. Amplified DNA in Streptomyces fradiae. J Bacteriol. 1983 Aug;155(2):459–466. doi: 10.1128/jb.155.2.459-466.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn D. R., McHenney M. A., Baltz R. H. Characterization of FP22, a large streptomycete bacteriophage with DNA insensitive to cleavage by many restriction enzymes. J Gen Microbiol. 1990 Dec;136(12):2395–2404. doi: 10.1099/00221287-136-12-2395. [DOI] [PubMed] [Google Scholar]
- Jackson E. N., Jackson D. A., Deans R. J. EcoRI analysis of bacteriophage P22 DNA packaging. J Mol Biol. 1978 Jan 25;118(3):365–388. doi: 10.1016/0022-2836(78)90234-6. [DOI] [PubMed] [Google Scholar]
- Katz E., Thompson C. J., Hopwood D. A. Cloning and expression of the tyrosinase gene from Streptomyces antibioticus in Streptomyces lividans. J Gen Microbiol. 1983 Sep;129(9):2703–2714. doi: 10.1099/00221287-129-9-2703. [DOI] [PubMed] [Google Scholar]
- Kuhstoss S., Richardson M. A., Rao R. N. Site-specific integration in Streptomyces ambofaciens: localization of integration functions in S. ambofaciens plasmid pSAM2. J Bacteriol. 1989 Jan;171(1):16–23. doi: 10.1128/jb.171.1.16-23.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962 Jul;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- Matsushima P., Baltz R. H. Genetic transformation of Micromonospora rosaria by the Streptomyces plasmid pIJ702. J Antibiot (Tokyo) 1988 Apr;41(4):583–585. doi: 10.7164/antibiotics.41.583. [DOI] [PubMed] [Google Scholar]
- Matsushima P., Baltz R. H. Streptomyces lipmanii expresses two restriction systems that inhibit plasmid transformation and bacteriophage plaque formation. J Bacteriol. 1989 Jun;171(6):3128–3132. doi: 10.1128/jb.171.6.3128-3132.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima P., Cox K. L., Baltz R. H. Highly transformable mutants of Streptomyces fradiae defective in several restriction systems. Mol Gen Genet. 1987 Mar;206(3):393–400. doi: 10.1007/BF00428877. [DOI] [PubMed] [Google Scholar]
- Matsushima P., McHenney M. A., Baltz R. H. Efficient transformation of Amycolatopsis orientalis (Nocardia orientalis) protoplasts by Streptomyces plasmids. J Bacteriol. 1987 May;169(5):2298–2300. doi: 10.1128/jb.169.5.2298-2300.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima P., McHenney M. A., Baltz R. H. Transduction and transformation of plasmid DNA in Streptomyces fradiae strains that express different levels of restriction. J Bacteriol. 1989 Jun;171(6):3080–3084. doi: 10.1128/jb.171.6.3080-3084.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHenney M. A., Baltz R. H. Transduction of plasmid DNA in Streptomyces spp. and related genera by bacteriophage FP43. J Bacteriol. 1988 May;170(5):2276–2282. doi: 10.1128/jb.170.5.2276-2282.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHenney M. A., Baltz R. H. Transduction of plasmid DNA in macrolide producing streptomycetes. J Antibiot (Tokyo) 1989 Nov;42(11):1725–1727. doi: 10.7164/antibiotics.42.1725. [DOI] [PubMed] [Google Scholar]
- Roberts R. J. Restriction enzymes and their isoschizomers. Nucleic Acids Res. 1990 Apr 25;18 (Suppl):2331–2365. doi: 10.1093/nar/18.suppl.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solenberg P. J., Baltz R. H. Transposition of Tn5096 and other IS493 derivatives in Streptomyces griseofuscus. J Bacteriol. 1991 Feb;173(3):1096–1104. doi: 10.1128/jb.173.3.1096-1104.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susskind M. M., Botstein D. Molecular genetics of bacteriophage P22. Microbiol Rev. 1978 Jun;42(2):385–413. doi: 10.1128/mr.42.2.385-413.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youderian P., Sugiono P., Brewer K. L., Higgins N. P., Elliott T. Packaging specific segments of the Salmonella chromosome with locked-in Mud-P22 prophages. Genetics. 1988 Apr;118(4):581–592. doi: 10.1093/genetics/118.4.581. [DOI] [PMC free article] [PubMed] [Google Scholar]