Abstract
Iron- and aluminum-sulfate together, at nanomolar concentrations, trigger the production of reactive oxygen species (ROS) in cultures of human brain cells. Previous studies have shown that following ROS induction, a family of pathogenic brain genes that promote inflammatory signalling, cellular apoptosis and brain cell death is significantly over-expressed. Notably, iron- and aluminum-sulfate induce genes in cultured human brain cells that exhibit expression patterns similar to those observed to be up-regulated in moderate- to late-stage Alzheimer's disease (AD). In this study we have extended our investigations to analyze the expression of micro RNA (miRNA) populations in iron- and aluminum-sulfate treated human neural cells in primary culture. The main finding was that these ROS-generating neurotoxic metal sulfates also up-regulate a specific set of miRNAs that includes miR-9, miR-125b and miR-128. Notably, these same miRNAs are up-regulated in AD brain. These findings further support the idea that iron- and aluminum-sulfates induce genotoxicity via a ROS-mediated up-regulation of specific regulatory elements and pathogenic genes that redirect brain cell fate towards progressive dysfunction and apoptotic cell death.
Keywords: aluminum; human neural cells; inflammation; iron; micro RNA, reactive oxygen species; RNA polymerase II; ROS; sulfates
Metal-induced genotoxicity is an important pathogenic mechanism whereby toxic metals that gain access to nuclear compartments progressively and deleteriously affect the normal structure and function of the genome [1-3]. Iron- plus aluminum-sulfate at relatively low nanomolar concentrations, have been shown to induce genotoxicity in susceptible brain cells through the up-regulation of pro-inflammatory and pro-apoptotic gene families that then redirect cellular fate toward cytoplasmic dysfunction, nuclear DNA fragmentation and cell death [1,3]. These up-regulated genes include those encoding NF-κB subunits, interleukin-1β and β-amyloid precursor proteins, inducible cyclooxygenase-2, cytosolic phospholipase A2, and the pro-apoptotic cell death protein DAXX [1-5]. Interestingly, treatment of human neural (HN) cells in primary culture with 100 nM of iron- plus aluminum-sulfate for one third of their in vitro life-span has been found to emulate many of the up-regulated gene expression changes observed in the neocortex and hippocampus of moderate-to-late stage Alzheimer's disease (AD) [1, 4-7].
To further investigate the nature of these induced genotoxic changes in primary human brain cells, in this study we examined the effects of magnesium-, aluminum-, and/or iron-sulfate on ROS generation and micro RNA (miRNA) speciation in HN cells in primary culture using a 2′,7′-dichlorodihydro-fluorescein diacetate (H2DCFDA) assay, miRNA array and Northern hybridization technologies. Magnesium-sulfate treated HN cells, cultured under identical growth conditions with aluminum- or iron-treated HN cells, were used as controls.
In metal-sulfate treated, and untreated 2 week old HN cells, ROS levels were assayed, after 3.5 days of metal-sulfate exposure, using H2DCFDA at 10 μM in cell culture medium in the dark using protocols provided by the manufacturer (Molecular Probes, Eugene, OR) and as previously described (Fig.1) [1]. H2DCFDA are cell-permeant fluorescent indicators that react with hydroxyl radicals, singlet oxygen or superoxides (collectively termed ROS). Signals were quantified using electronic imaging photography under ultraviolet light (Ex 488 nm; Em 530 nm) and a Zeiss Axioskop/Zeiss MC63 photo control unit/Nikon Optiphot-2 microscope.
Fig 1. Generation of ROS by metal sulfates in HN cell culture.
The evolution of reactive oxygen species (ROS) using 2′,7′-dichlorofluorescein diacetate (H2DCFDA) assay was quantified in 2 week old HN cells 3.5 days after initial treatment with 100 nM magnesium-, iron-, aluminum-, magnesium plus aluminum- or iron- plus aluminum-sulfate [1]. These were compared to age-matched untreated controls containing no metal sulfate (value arbitrarily set to 1.0; dashed horizontal line) [1]. Because aluminum-sulfate alone and iron- plus aluminum-sulfate together were found to generate the most ROS using the H2DCFDA assay (Fig.1), these metal sulfates were next used to study their effects on miRNA expression in HN cells. Data are expressed as relative fluorescence yield, defined as fold-changes over 1.0 (dashed horizontal line). N=5; *p<0.05; **p<0.01 (ANOVA).
Micro RNAs constitute an RNA-polymerase II (RNA Pol II)-transcribed family of evolutionarily conserved, non-coding RNAs that recognize the 3′ untranslated regions (UTRs) of specific messenger RNA (mRNA) populations. Through their complementary base-pairing with the 3′ UTRs of target mRNA, miRNAs function in transcriptional processing or provide guidance for further mRNA modification [7-17]. The variable expression of miRNAs during cell growth and aging has suggested a role for miRNA in the regulation of gene expression during development and differentiation, and while human genomes encode several hundred different miRNAs, a significantly smaller sub-set is either enriched in the brain or specific to that organ [7,10-12]. Although up-regulated miRNAs are generally regarded as negative regulators of these multiple processes, down-regulation of miRNA might be expected to enhance mRNA translation and thereby up-regulate the expression of specific genes. Moreover, multiple miRNAs may regulate the expression of a single mRNA, or one miRNA may have multiple mRNA targets and thereby have multiple effects on the expression of families of genes [7-12]. For example, base-sequence analysis of synapsin mRNA, encoding a neurotransmitter release-regulating phosphoprotein that is significantly down-regulated in AD, shows that this mRNA contains 14 potential binding sites for different miRNAs in its 3′ UTR [13,14]. Synapsin mRNA has been further identified as a specific target for miR-125b, and it has been suggested that up-regulation of miR-125b may have some bearing on the down-regulation of synapsin mRNA-mediated gene expression [7,15,18].
Experimental details used in this study are described in footnote 1.
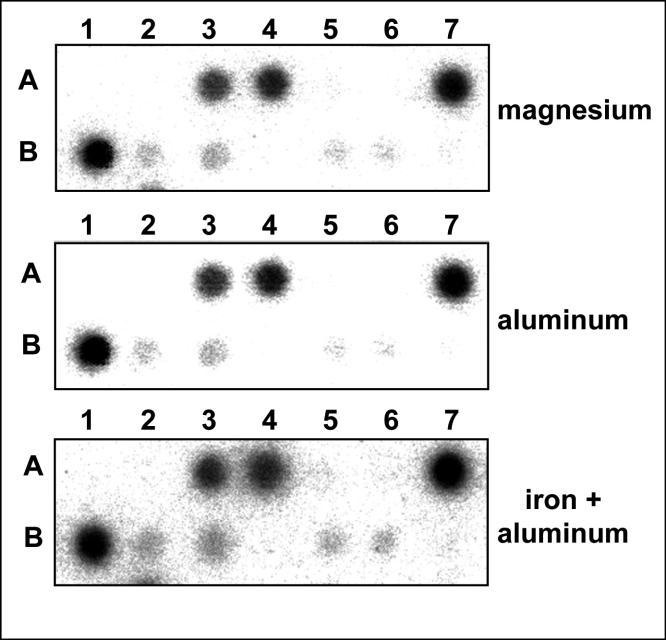
Aluminum-sulfate and iron- and aluminum-sulfate together were found to induce in HN cells a significant up-regulation in ROS 3.5 days after treatment as measured by H2DCFDA assay (Fig. 1). This increased ROS, in turn, is known to up-regulate specific mRNAs encoding a family of pro-inflammatory, pathogenic genes [1,3-5]. Typical miRNA array data, analyzing miRNA expression in HN cells treated for 7 days with 100 nM magnesium-sulfate (control), aluminum-sulfate or iron- plus aluminum-sulfate, is shown in Fig. 2. The miRNAs for miR-9, miR124a, miR-125b, miR-128, miR-132 and 5S rRNA were found to be relatively abundant, and all other miRNAs listed in Table 1 were much less abundant (7; Fig. 2; data not shown). Similar to the hippocampal region of control human brain tissue, the relative order of miRNA abundance in control HN cells was 5S miRNA>miR-128>>miR-125b>>miR-9>miR-124a>>miR-132>>>>all other miRNA levels studied (Table 1) [7]. While miR-9 is brain-specific [2], miR-125b and miR-128 are brain-enriched; both miR-9 and miR-128 RNA are also highly enriched in adenine-plus-uridine nucleotide content. Like miR-124a and miR-125b, miR-9 and miR-128 are retinoic acid inducible in cultured human NTera-2 clone D1 cells [16] and in HN cells in primary culture [7; unpublished observations]. Unlike miR-9, miR125b and miR-128 levels, relative miR-124a, miR-132 or 5S rRNA abundance did not significantly change in aluminum sulfate or iron- plus aluminum-sulfate treated HN cells when compared to magnesium-sulfate treated controls. Levels for miR-9, miR124a, miR-125b, miR-128, and miR-132 in HN cells in the presence of magnesium-, aluminum- or aluminum-plus iron -sulfate were further studied using Northern analysis and these results are shown in Fig. 3. In agreement with the miRNA array data (Fig. 2), miR-9, miR-125b and miR-128 were found to be the most significantly up-regulated miRNAs in this study. As HN cell cultures are composed of both neuronal and glial cells, the contributions of either brain cell type to miRNA production and speciation remain to be investigated. Systematic, computational nucleic acid sequence analysis of the interaction of brain-specific and brain-enriched miRNAs with the 3′ UTRs of target neural mRNAs should provide further insight into the role of neurotoxic metal in selective miRNA speciation and their regulation of mRNA complexity [7,19].
Fig. 2. miRNA array analysis.
Nylon membrane-bound DNA-equivalents (15 μM) of 13 miRNAs (known to be enriched in brain) and 5S rRNA were probed with total 32P-radiolabelled miRNA fractions (<25 nucleotides) isolated from magnesium-sulfate, aluminum-sulfate or iron- plus aluminum-sulfate treated HN cells (Table 1) [1,7]. For each panel row A1-A7; miR-219, miR-183, miR-9, miR-125b, miR-124b, miR-125a, miR-128. Row B1−7; 5S rRNA, miR-132, miR-124a, miR-137, miR-139, miR-153, miR-135. Each individual miRNA signal was quantified against the 5S rRNA signal within the same sample. miR-9, miR-125b and miR-128, but not 5S rRNA gave consistently the highest hybridization signals in iron-plus aluminum-sulfate treated cells when compared to magnesium-sulfate treated controls (N=5). We hypothesize that these induced changes in miRNA expression may be ROS-specific in nature since (a) aluminum-sulfate alone gave qualitatively similar but quantitatively smaller signals for miR-9, miR-125b and miR-128 patterns than did iron- plus aluminum-sulfate together (Fig. 1), and (b) incubation of HN cells with 1 uM of hydrogen peroxide (a potent ROS-generating reagent) also emulated these specific miRNA expression profiles (unpublished observations).
Table 1.
DNA equivalent sequence (5′ to 3′) of miRNA probes used in this study and Genbank accession numbers for that miRNA sequence [7]. A 21 nucleotide 5S ribosomal RNA (5S rRNA) probe was derived from the immediate 5′ end of the 107 nucleotide human 5S rRNA.
| RNA | DNA sequence | Genbank |
|---|---|---|
| miR-9 | TCTTTGGTTATCTAGCTGTATGA | AJ459704 |
| miR-124a | TAAGGCACGCGGTGAATGCCA | AJ459733 |
| miR-125b | TCCCTGAGACCCTAACTTGTGA | AY865852 |
| miR-128 | TCACAGTGAACCGGTCTCTTTT | AJ459739 |
| miR-132 | TAACAGTCTACAGCCATGGTCGT | AJ459743 |
| miR-219 | TGATTGTCCAAACGCAATTC | AJ550423 |
| miR-124b | TTAAGGCACGCGGGTGAATGC | AJ459734 |
| miR-125a | TCCCTGAGACCCTTTAACCTGTG | AJ459735 |
| miR-135 | TATGGCTTTTTATTCCTATGTGAA | AJ459746 |
| miR-137 | TATTGCTTAAGAATACGCGTAG | AJ459748 |
| miR-139 | TCTACAGTGCACGTGTCT | AJ459750 |
| miR-153 | TTGCATAGTC ACAAAAGTGA | AJ459765 |
| miR-183 | ACTACGAATGATAACATCCGTGG | AY194163 |
| 5S rRNA | ATACTCTGGTTTCTCTTCAGA | NR002758 |
Fig 3. Northern analysis.
From miRNA relative abundance data derived from an initial DNA array screening and electronic autoradiography (Fig. 2), specific miRNA signals in magnesium-sulfate, aluminum-sulfate or iron- plus aluminum-sulfate treated HN cells were assayed using Northern gel analysis. Again, signals were quantified against 5S rRNA levels within the same sample. N=5; *p<0.05; **p<0.01 (ANOVA).
In summary, this is the first study describing the neurotoxic effects of aluminum-sulfate and aluminum- plus iron-sulfate, at 100 nM concentrations, on miRNA expression patterns in untransformed human brain cells in primary culture using a combined miRNA-DNA array and Northern analysis approach. These experiments employed 3 week old HN cells that are primary co-cultures of neurons and glia, however, the complex nature of neuronal and glial diversity, and the contribution of age-dependency to neuronal degeneration and apoptotic cell death warrant further examination in future studies. We note that the amount of bioavailable metal in the HN cell culture medium is likely to be considerably lower than the applied amount. We envision that it is the repeatedly added, relatively high flux of exogenous metal sulfate into the system, perhaps via ROS-mediated effects, that rapidly perturbs specific gene expression transcription processes that operate on a time scale of seconds to minutes [1-3,20]. The mis-regulation of specific miRNAs may be due, in part, to a change in brain cell redox balance driven by metal-ion mediated ROS generation, ensuing effects on redox-sensitive transcription factors such as NF-κB, and the induction of specific pathogenic gene programs [1,7]. The molecular biology, transport and in vivo speciation of aluminum- and iron-sulfates in genetic systems are not well understood, however, as little as 50 nM ambient aluminum has been shown to dramatically perturb RNA Pol II-directed gene transcription in isolated human brain cell nuclei [2,3]. These studies further suggest the involvement of soluble aluminum- and iron-sulfate in several different aspects of human brain gene expression, and in particular, with the molecular-genetic processes associated with transcriptional and post-transcriptional control. For example, synapsin mRNA has been found to be down-regulated in both AD brain and in iron- plus aluminum-sulfate treated HN cells in primary culture [1,7,18]. The down-regulation of synaptic proteins in AD, such as those encoded by synapsin mRNA, may occur, in part, because synapsin mRNA is a recognized target for the repressive effects engendered by up-regulation of miR-125b binding [7,15,18]. Therefore, similar observed alterations in miRNA speciation in AD brain [7] and in iron- plus aluminum-sulfate-treated human brain cells may be expected to contribute, in both cases, to the molecular-genetic changes that accompany pathogenic gene expression, synaptic deficits, progressive brain cell dysfunction and apoptotic cell death.
ACKNOWLEDGEMENTS
Thanks are extended to Drs. Jian-Guo Cui, Yuan Yuan Li, Yuhai Zhao, Hilary Thompson and Darlene Guillot for expert technical assistance. This work was interrupted and delayed in publication due to the extensive devastation caused by hurricane Katrina (29 August 2005) to the Louisiana State University Heath Sciences Center, New Orleans, LA, USA. Part of this study was presented in abstract form at the 7th Keele meeting on Aluminum 24−28 February 2007 in Merida, Yucatan, Mexico. These studies were supported in part by NIH NIA AG18031.
Glossary
ABBREVIATIONS
- AD
Alzheimer's disease
- ANOVA
analysis of variance
- HN cells
human neural cells
- mRNA
messenger RNA
- miRNA
micro RNA
- NF-κB
nuclear factor for kappa B
- ROS
reactive oxygen species
- UTR
un-translated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
FOOTNOTE 1 - Experimental details
In the construction of brain cell-specific miRNA array panels, 13 individual brain-enriched miRNAs and human 5S ribosomal RNA (5S rRNA) were spotted onto GeneScreen Plus nylon membranes using a Biomek® 2000 laboratory automation workstation (Beckmann, Fullerton, CA) and were crosslinked, baked, hybridized and probed according to the manufacturer's protocol (NEN® Research Products, Boston MA) [7, 10-12]. A guanidine isothiocyanate- and silica gel-based membrane total RNA purification system and miRNA isolation kit (PureLink™ Invitrogen, Carlsbad, CA) were used to isolate total small RNAs (5S rRNA, tRNA and miRNA) from magnesium-, aluminum- and iron- plus aluminum-sulfate treated HN cells. Total RNA concentrations and spectral purity were quantified using RNA 6000 Nano LabChip analysis and a 2100 Spectral Analyzer (Caliper Technologies, Mountainview, CA; Agilent Technologies, Palo Alto, CA) [1,4,5,7]. 28S/18S ratios consistently exceeded 1.4 and the A260/280 of total RNA (based on peak area) was typically ≥1.8. Poly A+ messenger RNA was found to range in size from 0.5−8 kilobases. In a preliminary screen, miRNA arrays were probed with total labeled miRNAs isolated from 3 week old HN cells that had been exposed to 100 nM magnesium-sulfate (control), or aluminum-sulfate, or aluminum- plus iron-sulfate, for 7 days, or about one-third of their in vitro life-span [1]. Derived from human fetal brain tissue, HN cells are primary, untransformed co-cultures of human neurons and glia (Clonetics CC-2599; Cambrex-Lonza Biosciences, Walkersville MD) that are an imperfect but highly informative in vitro model for the study of molecular, genetic, inflammatory and neurotoxicological aspects of AD [1,3,5,7,18]. When maintained in culture for 3 weeks HN cells contain approximately equal populations of differentiated human neurons and glia [1]. These cells survive in culture for about 4.5 weeks at which point they begin to die through the process of nuclear DNA fragmentation or apoptosis [1,3,4]. HN cells containing 100 nM aluminum-sulfate or iron- plus aluminum-sulfate (but not 100 nM magnesium-sulfate treated cells) in their growth medium generally die via apoptotic processes about 10 days after toxic metal-ion treatment [unpublished observations]. All details on HN cell cultures, magnesium-, aluminum-, and iron-sulfate reagent preparation, incubation details, miRNA extraction using Trizol reagent (Invitrogen), enrichment and labeling methods have been previously described in detail [1,7]. After hybridization and washing, miRNA arrays were scanned, pixel intensities and expression signals were quantified, and features were extracted using Genespring ver 7.2 algorithms (Silicon Genetics, Redwood City, CA) [1,3-5,7]. Statistical significance of toxic metal ion-induced miRNA levels over (magnesium-sulfate) controls was analyzed using a two-way factorial analysis of variance (p, ANOVA; Statistical Analysis System; SAS Institute, Cary, NC). Specific miRNAs showing strong hybridization signals on the miRNA panels were further studied using Northern blot analysis. Magnesium-, aluminum-, or iron- plus aluminum-sulfate treated HN cell total RNA extracts containing miRNA or 5S rRNA (25 μg) were alternately run out on 15% TBE-urea denaturing gels, transferred to GeneScreen membranes, crosslinked, baked, hybridized and probed with DNA oligomers corresponding to specific miRNAs. These miRNAs had been radiolabeled using [γ-32P]-δATP (6000 Ci/mmol) and a T4 polynucleotide kinase labeling system (Invitrogen) (Table 1) as previously described [7].
REFERENCES
- 1.Alexandrov PN, Zhao Y, Pogue AI, Tarr MA, Kruck TPA, Percy ME, Cui JG, Lukiw WJ. J. Alzheimer's Dis. 2005;8:117–127. doi: 10.3233/jad-2005-8204. [DOI] [PubMed] [Google Scholar]
- 2.Sarkander HI, Balb G, Schlosser H, Stoltenburg G, Lux RM. In: Brain Aging: Neuropathology and Neuropharmacology. Cervos-Navarro J, Sarkander HI, editors. Raven Press; New York: 1983. pp. 259–274. [Google Scholar]
- 3.Lukiw WJ. In: Aluminum and Alzheimer's Disease, the Science that Describes the Link. Exley C, editor. Elsevier Publishers; London: 2001. pp. 147–168. [Google Scholar]
- 4.Colangelo V, Schurr J, Ball MJ, Pelaez RP, Bazan NG, Lukiw WJ. J. Neurosci. Res. 2002;70:462–473. doi: 10.1002/jnr.10351. [DOI] [PubMed] [Google Scholar]
- 5.Lukiw WJ. Neurochem. Res. 2004;29:1287–1297. doi: 10.1023/b:nere.0000023615.89699.63. [DOI] [PubMed] [Google Scholar]
- 6.Nakamichi N, Yoneda Y. Japan J. Pharmacol. 2002;89:337–348. doi: 10.1254/jjp.89.337. [DOI] [PubMed] [Google Scholar]
- 7.Lukiw WJ. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 8.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Eur. J. Neurosci. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- 10.Kim VN, Nam JW. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krichevsky AM, King KS, Donahue CP, Kosik KS. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du T, Zamore PD. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 14.Brown JR, Sanseau P P. Drug Discovery Today. 2005;10:595–601. doi: 10.1016/S1359-6446(05)03399-4. [DOI] [PubMed] [Google Scholar]
- 15.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Nature Genetics. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 16.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.John B, Sander C, Marks DS. Methods Mol. Biol. 2006;342:101–113. doi: 10.1385/1-59745-123-1:101. [DOI] [PubMed] [Google Scholar]
- 18.Lukiw WJ, Rogaev EI, Bazan NG. Alzheimer's Research. 1996;2:221–227. [Google Scholar]
- 19.Carninci P. Trends Genet. 2006;22:501–510. doi: 10.1016/j.tig.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Lukiw WJ, Percy ME, Kruck TP. J. Inorg. Biochem. 2005;99:1895–1898. doi: 10.1016/j.jinorgbio.2005.04.021. [DOI] [PubMed] [Google Scholar]