Abstract
DNA upstream of the transcription start site of the mvaAB operon of Pseudomonas mevalonii, which encodes 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase (EC 1.1.1.88) and HMG-CoA lyase (EC 4.1.3.4), contains a cis-acting regulatory element which functions in the response to mevalonate. The regulatory element resides within a 36-bp region located from 48 to 84 bp upstream of the transcription start site of mvaA. This location was inferred from the beta-galactosidase activities of P. mevalonii harboring plasmid-encoded mvaA-lacZ fusions induced by mevalonate and by DNA gel retardation and competition assays. While protein from P. mevalonii grown on mevalonate produced a band shift, protein from cells grown on succinate gave no band shift, even when mevalonate was added. The operator contains three 10-bp direct repeats with the consensus sequence TGGGTACAGT, which may be important for regulation of the mvaAB operon.
Full text
PDF

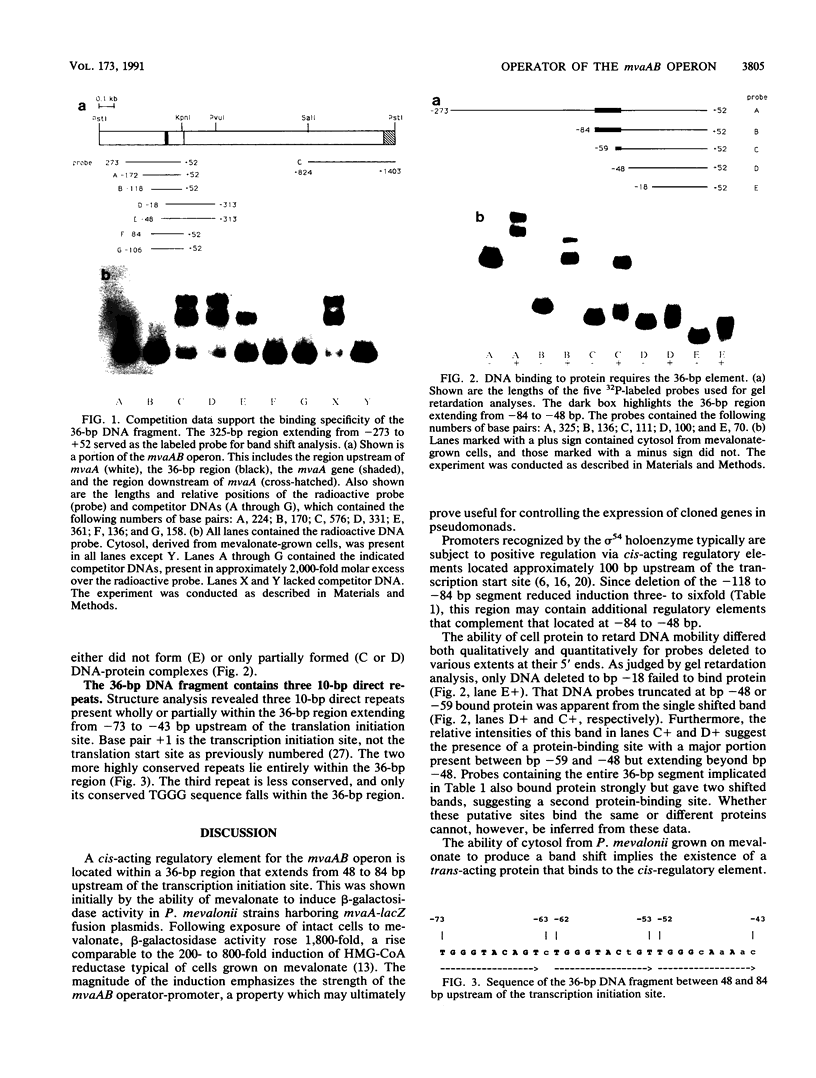

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. H., Rodwell V. W. Nucleotide sequence and expression in Escherichia coli of the 3-hydroxy-3-methylglutaryl coenzyme A lyase gene of Pseudomonas mevalonii. J Bacteriol. 1989 Dec;171(12):6468–6472. doi: 10.1128/jb.171.12.6468-6472.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C., Bagdasarian M. M., Frey J., Timmis K. N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981 Dec;16(1-3):237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- Beach M. J., Rodwell V. W. Cloning, sequencing, and overexpression of mvaA, which encodes Pseudomonas mevalonii 3-hydroxy-3-methylglutaryl coenzyme A reductase. J Bacteriol. 1989 Jun;171(6):2994–3001. doi: 10.1128/jb.171.6.2994-3001.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach M. J., Rodwell V. W. Cloning, sequencing, and overexpression of mvaA, which encodes Pseudomonas mevalonii 3-hydroxy-3-methylglutaryl coenzyme A reductase. J Bacteriol. 1989 Jun;171(6):2994–3001. doi: 10.1128/jb.171.6.2994-3001.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensch W. R., Rodwell V. W. Purification and properties of 3-hydroxy-3-methylglutaryl coenzyme A reductase from Pseudomonas. J Biol Chem. 1970 Aug 10;245(15):3755–3762. [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- CRESSON E. L., FOLKERS K., HOFFMAN C. H., MACRAE G. D., SKEGGS H. R., WOLF D. E., WRIGHT L. D. Discovery of a new acetate-replacing factor. J Bacteriol. 1956 Oct;72(4):519–524. doi: 10.1128/jb.72.4.519-524.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Chou J., Cohen S. N. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980 Aug;143(2):971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R. The xylABC promoter from the Pseudomonas putida TOL plasmid is activated by nitrogen regulatory genes in Escherichia coli. Mol Gen Genet. 1986 Apr;203(1):129–136. doi: 10.1007/BF00330393. [DOI] [PubMed] [Google Scholar]
- Gill J. F., Jr, Beach M. J., Rodwell V. W. Mevalonate utilization in Pseudomonas sp. M. Purification and characterization of an inducible 3-hydroxy-3-methylglutaryl coenzyme A reductase. J Biol Chem. 1985 Aug 5;260(16):9393–9398. [PubMed] [Google Scholar]
- Gill J. F., Jr, Beach M. J., Rodwell V. W. Transport of mevalonate by Pseudomonas sp. strain M. J Bacteriol. 1984 Oct;160(1):294–298. doi: 10.1128/jb.160.1.294-298.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan-Starck T. C., Rodwell V. W. Pseudomonas mevalonii 3-hydroxy-3-methylglutaryl-CoA reductase. Characterization and chemical modification. J Biol Chem. 1989 Oct 25;264(30):17913–17918. [PubMed] [Google Scholar]
- Jordan-Starck T. C., Rodwell V. W. Role of cysteine residues in Pseudomonas mevalonii 3-hydroxy-3-methylglutaryl-CoA reductase. Site-directed mutagenesis and characterization of the mutant enzymes. J Biol Chem. 1989 Oct 25;264(30):17919–17923. [PubMed] [Google Scholar]
- Kustu S., Santero E., Keener J., Popham D., Weiss D. Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989 Sep;53(3):367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Scher D. S., Rodwell V. W. 3-Hydroxy-3-methylglutaryl coenzyme A lyase from Pseudomonas mevalonii. Biochim Biophys Acta. 1989 Jun 28;1003(3):321–326. doi: 10.1016/0005-2760(89)90239-7. [DOI] [PubMed] [Google Scholar]
- Staudt L. M., Singh H., Sen R., Wirth T., Sharp P. A., Baltimore D. A lymphoid-specific protein binding to the octamer motif of immunoglobulin genes. Nature. 1986 Oct 16;323(6089):640–643. doi: 10.1038/323640a0. [DOI] [PubMed] [Google Scholar]
- Strauss F., Varshavsky A. A protein binds to a satellite DNA repeat at three specific sites that would be brought into mutual proximity by DNA folding in the nucleosome. Cell. 1984 Jul;37(3):889–901. doi: 10.1016/0092-8674(84)90424-0. [DOI] [PubMed] [Google Scholar]
- Tsung K., Brissette R. E., Inouye M. Identification of the DNA-binding domain of the OmpR protein required for transcriptional activation of the ompF and ompC genes of Escherichia coli by in vivo DNA footprinting. J Biol Chem. 1989 Jun 15;264(17):10104–10109. [PubMed] [Google Scholar]