Abstract
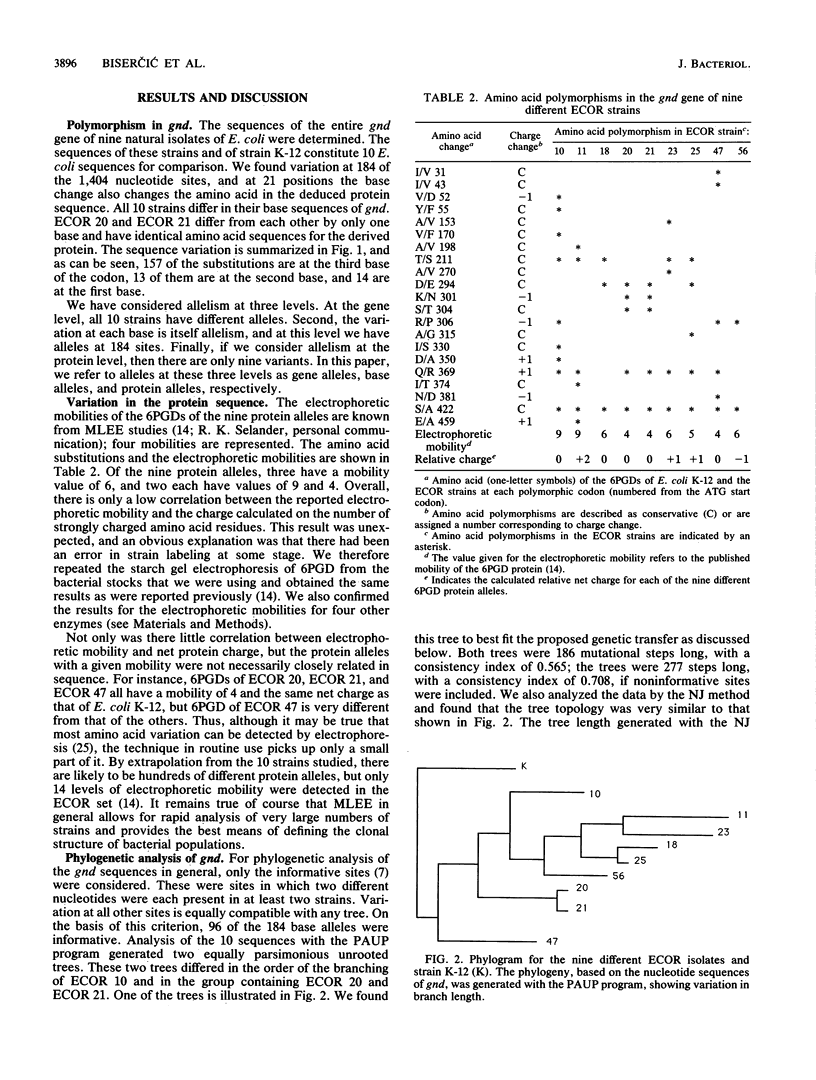
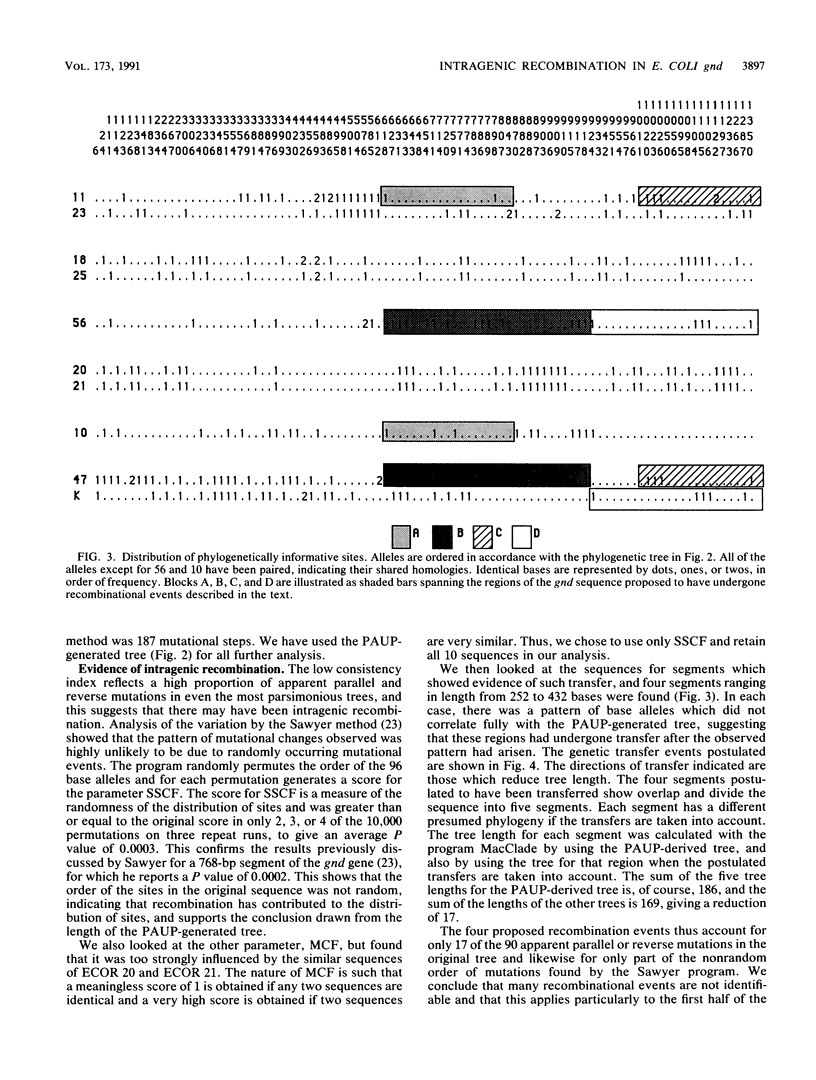
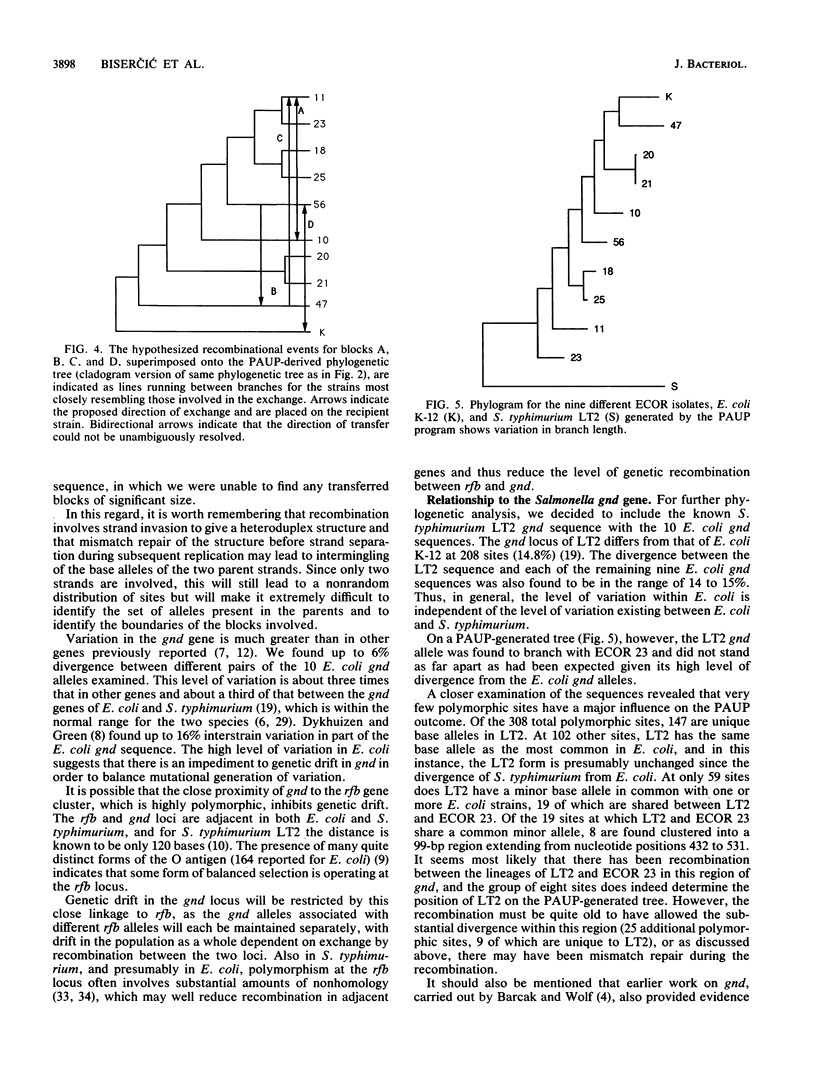
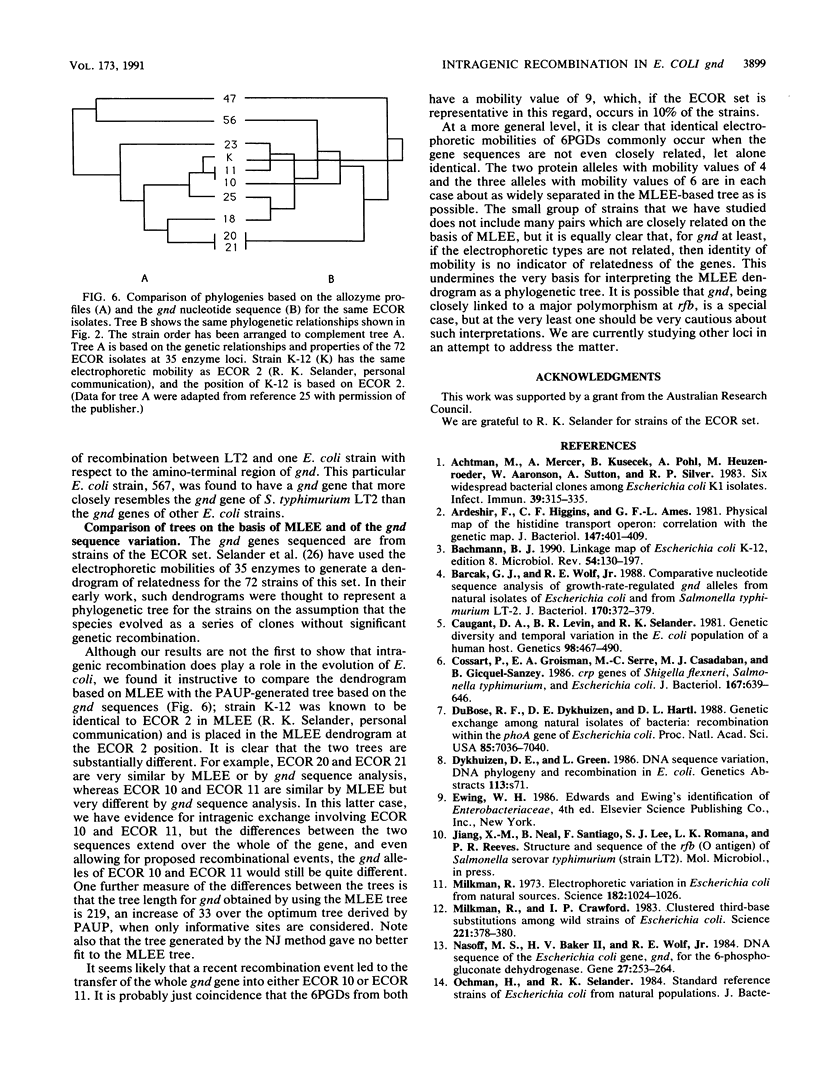
Nine natural isolates of Escherichia coli were examined, and the sequence of the entire 1,404 bases of the gnd gene (6-phosphogluconate dehydrogenase, EC 1.1.1.44) was determined. These isolates, along with E. coli K-12, constitute 10 strains for analysis. (The sequence of the E. coli K-12 gnd gene is known.) A total of 184 sites were polymorphic, and up to 6% sequence divergence was observed between pairs of strains. The deduced amino acid sequences showed much more variation than had been shown by multilocus enzyme electrophoresis, and in addition the net charge calculated did not correlate strongly with electrophoretic mobility. A phylogenetic tree for the sequences that was based on maximum parsimony differed significantly from a tree for the same strains that was based on multilocus enzyme electrophoresis for 35 enzymes (R. K. Selander, D. A. Caugant, and T. S. Whittam, p. 1625-1648, in F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger, ed., Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, 1987). These data, together with analysis of sequence variation between the strains, indicated that intragenic recombination and transfer of the whole of gnd have occurred in the evolution of these strains. There is evidence of one recombination event between E. coli and Salmonella typhimurium.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achtman M., Mercer A., Kusecek B., Pohl A., Heuzenroeder M., Aaronson W., Sutton A., Silver R. P. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect Immun. 1983 Jan;39(1):315–335. doi: 10.1128/iai.39.1.315-335.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardeshir F., Higgins C. F., Ames G. F. Physical map of the Salmonella typhimurium histidine transport operon: correlation with the genetic map. J Bacteriol. 1981 Aug;147(2):401–409. doi: 10.1128/jb.147.2.401-409.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcak G. J., Wolf R. E., Jr Comparative nucleotide sequence analysis of growth-rate-regulated gnd alleles from natural isolates of Escherichia coli and from Salmonella typhimurium LT-2. J Bacteriol. 1988 Jan;170(1):372–379. doi: 10.1128/jb.170.1.372-379.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caugant D. A., Levin B. R., Selander R. K. Genetic diversity and temporal variation in the E. coli population of a human host. Genetics. 1981 Jul;98(3):467–490. doi: 10.1093/genetics/98.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P., Groisman E. A., Serre M. C., Casadaban M. J., Gicquel-Sanzey B. crp genes of Shigella flexneri, Salmonella typhimurium, and Escherichia coli. J Bacteriol. 1986 Aug;167(2):639–646. doi: 10.1128/jb.167.2.639-646.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBose R. F., Dykhuizen D. E., Hartl D. L. Genetic exchange among natural isolates of bacteria: recombination within the phoA gene of Escherichia coli. Proc Natl Acad Sci U S A. 1988 Sep;85(18):7036–7040. doi: 10.1073/pnas.85.18.7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkman R., Crawford I. P. Clustered third-base substitutions among wild strains of Escherichia coli. Science. 1983 Jul 22;221(4608):378–380. doi: 10.1126/science.6346486. [DOI] [PubMed] [Google Scholar]
- Milkman R. Electrophoretic variation in Escherichia coli from natural sources. Science. 1973 Dec 7;182(4116):1024–1026. doi: 10.1126/science.182.4116.1024. [DOI] [PubMed] [Google Scholar]
- Nasoff M. S., Baker H. V., 2nd, Wolf R. E., Jr DNA sequence of the Escherichia coli gene, gnd, for 6-phosphogluconate dehydrogenase. Gene. 1984 Mar;27(3):253–264. doi: 10.1016/0378-1119(84)90070-2. [DOI] [PubMed] [Google Scholar]
- Ochman H., Selander R. K. Evidence for clonal population structure in Escherichia coli. Proc Natl Acad Sci U S A. 1984 Jan;81(1):198–201. doi: 10.1073/pnas.81.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Whittam T. S., Caugant D. A., Selander R. K. Enzyme polymorphism and genetic population structure in Escherichia coli and Shigella. J Gen Microbiol. 1983 Sep;129(9):2715–2726. doi: 10.1099/00221287-129-9-2715. [DOI] [PubMed] [Google Scholar]
- Orskov F., Orskov I., Evans D. J., Jr, Sack R. B., Sack D. A., Wadström T. Special Escherichia coli serotypes among enterotoxigenic strains from diarrhoea in adults and children. Med Microbiol Immunol. 1976 Jun 1;162(2):73–80. doi: 10.1007/BF02121318. [DOI] [PubMed] [Google Scholar]
- Plos K., Hull S. I., Hull R. A., Levin B. R., Orskov I., Orskov F., Svanborg-Edén C. Distribution of the P-associated-pilus (pap) region among Escherichia coli from natural sources: evidence for horizontal gene transfer. Infect Immun. 1989 May;57(5):1604–1611. doi: 10.1128/iai.57.5.1604-1611.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves P., Stevenson G. Cloning and nucleotide sequence of the Salmonella typhimurium LT2 gnd gene and its homology with the corresponding sequence of Escherichia coli K12. Mol Gen Genet. 1989 May;217(1):182–184. doi: 10.1007/BF00330960. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S. A., Dykhuizen D. E., Hartl D. L. Confidence interval for the number of selectively neutral amino acid polymorphisms. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6225–6228. doi: 10.1073/pnas.84.17.6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S. Statistical tests for detecting gene conversion. Mol Biol Evol. 1989 Sep;6(5):526–538. doi: 10.1093/oxfordjournals.molbev.a040567. [DOI] [PubMed] [Google Scholar]
- Selander R. K., Caugant D. A., Ochman H., Musser J. M., Gilmour M. N., Whittam T. S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986 May;51(5):873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander R. K., Levin B. R. Genetic diversity and structure in Escherichia coli populations. Science. 1980 Oct 31;210(4469):545–547. doi: 10.1126/science.6999623. [DOI] [PubMed] [Google Scholar]
- Selander R. K., Musser J. M., Caugant D. A., Gilmour M. N., Whittam T. S. Population genetics of pathogenic bacteria. Microb Pathog. 1987 Jul;3(1):1–7. doi: 10.1016/0882-4010(87)90032-5. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Shields D. C., Wolfe K. H., Li W. H. Chromosomal location and evolutionary rate variation in enterobacterial genes. Science. 1989 Nov 10;246(4931):808–810. doi: 10.1126/science.2683084. [DOI] [PubMed] [Google Scholar]
- Smith N. H., Beltran P., Selander R. K. Recombination of Salmonella phase 1 flagellin genes generates new serovars. J Bacteriol. 1990 May;172(5):2209–2216. doi: 10.1128/jb.172.5.2209-2216.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N. H., Selander R. K. Sequence invariance of the antigen-coding central region of the phase 1 flagellar filament gene (fliC) among strains of Salmonella typhimurium. J Bacteriol. 1990 Feb;172(2):603–609. doi: 10.1128/jb.172.2.603-609.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus A., Leslie J. F., Milkman R. Molecular evolution of the Escherichia coli chromosome. I. Analysis of structure and natural variation in a previously uncharacterized region between trp and tonB. Genetics. 1988 Oct;120(2):345–358. doi: 10.1093/genetics/120.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma N. K., Quigley N. B., Reeves P. R. O-antigen variation in Salmonella spp.: rfb gene clusters of three strains. J Bacteriol. 1988 Jan;170(1):103–107. doi: 10.1128/jb.170.1.103-107.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma N., Reeves P. Identification and sequence of rfbS and rfbE, which determine antigenic specificity of group A and group D salmonellae. J Bacteriol. 1989 Oct;171(10):5694–5701. doi: 10.1128/jb.171.10.5694-5701.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittam T. S., Ochman H., Selander R. K. Geographic components of linkage disequilibrium in natural populations of Escherichia coli. Mol Biol Evol. 1983 Dec;1(1):67–83. doi: 10.1093/oxfordjournals.molbev.a040302. [DOI] [PubMed] [Google Scholar]
- Whittam T. S., Ochman H., Selander R. K. Multilocus genetic structure in natural populations of Escherichia coli. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1751–1755. doi: 10.1073/pnas.80.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]



