Abstract
Signaling molecules are essential for vertebrate embryonic development. Here, two Xenopus homologs of the Drosophila gene fringe, lunatic Fringe (lFng) and radical Fringe (rFng), were identified and the protein product of lFng further characterized. The messenger RNA of lFng is supplied as a maternal message. Its product is a precursor protein consisting of pre-, pro-, and mature regions. The mature lunatic Fringe protein is secreted extracellularly, and it induced mesodermal tissue formation in animal cap assays. These results indicate that secreted lunatic Fringe can induce mesoderm and reveal that the Fringe proteins are a family of vertebrate signaling molecules.
Pattern formation in early vertebrate embryos involves multiple molecular signals. In Xenopus, two families of secreted proteins have been implicated in mesoderm induction: the fibroblast growth factor and the transforming growth factor-β (TGF-β) families (1–4). Blockade of fibroblast growth factor signaling in the embryo by a dominant negative form of fibroblast growth factor receptor leads to the loss of posterior mesoderm(2), indicating the significance of fibroblast growth factor signaling in mesoderm induction. Experiments with a dominant negative form of the activin receptor suggest that molecules with activin-like activities function in Xenopus mesoderm formation (4). In the mouse, elimination of genes involved in the activin pathway has not resulted in defects in mesoderm induction (5), lending support to the suggestion that other molecules, either of the TGF-β family or of an unidentified family, may be involved in mesoderm induction.
We report here the mesoderm-inducing activity of a protein encoded by lunatic Fringe (lFng), a Xenopus homolog of the Drosophila fringe (fng) gene (6). Drosophila fng was discovered for its role in wing disc formation (6). Juxtaposition of fng-expressing and nonexpressing cells is required for the formation of the dorsal-ventral (D-V) boundary of Drosophila wing discs (6). The pattern of fng expression is remarkable when compared with those of three other genes encoding signals for wing disc development: fng is in the dorsal half of the wing disc, wingless at the D-V boundary, hedgehog in the posterior half, and decapentaplegic at the anterior-posterior boundary (6, 7). The fact that these three genes have all been found to have vertebrate homologs functioning in cellular communication important for embryonic patterning makes it interesting to identify vertebrate fng homologs and study their functional roles.
To identify Xenopus fng genes, we used the polymerase chain reaction (PCR) and low-stringency hybridization techniques (8). Complementary DNAs encoding two full-length Xenopus Fng proteins, designated lunatic Fringe and radical Fringe (lFng and rFng), were isolated (Fig. 1, A and B). The predicted products of the Xenopus fng genes are similar to the Drosophila FNG protein (Fig. 1D). Both lFng and rFng have signal sequences at their NH2-termini. The sequences in the NH2-terminal portion are divergent among the three proteins, whereas those in the COOH-terminal region are highly conserved (Fig. 1D). There is a tetrabasic site in each of these proteins immediately NH2-terminal to the conserved region (amino acid residues 84 to 87 in lFng and amino acid residues 62 to 65 in rFng) (Fig. 1, A, B, and D). These features suggest a general structure for each Fng protein: a pre region (the signal peptide) suggestive of secretion, a pro region ending with a tetrabasic site for proteolytic processing, and a mature region that will be functionally active after being cleaved from the rest of the precursor protein (Fig. 1C). Two other dibasic sites previously suggested as putative cleavage sites for Drosophila FNG protein (6) are not conserved in the Xenopus Fng proteins and therefore may not be functionally important. A data bank search indicated the existence of two human fng genes in sequence tags obtained from a human brain complementary DNA library (hF1 and hF2 in Fig. 1D).
Fig. 1.
Analyses of the primary sequences of Xenopus Fringe proteins, lFng and rFng.(A and B)Predicted amino acid sequences of lFng and rFng proteins (16). Putative signal sequences were predicted by Kyte-Doolittle hydrophobicity analyses and are underlined. Doubly underlined sequences are putative proteolytic cleavage sites. Position 163 of lFng and position 149 of rFng are putative N-linked glycosylation sites. (C) A schematic representation of our proposal for a general structure of Fng proteins: the signal peptide as a pre region, followed by a pro and a mature region. (D) Alignment of predicted amino acid sequences of Fringe proteins. Overall amino acid identities are 37% between lFng and Drosophila FNG, 33% between rFng and Drosophila FNG, and 46% between lFng and rFng. A dash in a vertebrate sequence indicates an identity to Drosophila FNG. A dot introduces a gap to allow optimal alignment. Letters in the consensus line (Cons.) are residues conserved among all known sequences. A star indicates identities between the Xenopus sequences; rFng may have an alternatively spliced form that includes an additional sequence at the COOH-terminus (FKSVHCLLYSDTDWCPNHKHNPTT) containing similarities to the other Fng proteins [see(8)]. hF1 is the likely product of human EST sequence R56561 and hF2 is that of human EST sequences F13368 and R13807. R13807encodes additional amino acid residues carboxyl to the hF2 sequence shown here (191 FGDTNHPDPGACLGFVPRVWGNQX-PLMGSS 220). There is no significant primary sequence similarity among Xenopus and Drosophila sequences before residue 120 of Drosophila FNG, residue 84 of lFng, and residue 61 of rFng, indicating that the conserved sequences all fall within the predicted mature regions.
To test whether the lFng product is a secreted protein, we first used the rabbit reticulocyte lysate in vitro translation system (9) (Fig. 2A). The lFng precursor protein entered the secretory pathway (Fig. 2A, lanes 3 to 5)as evidenced by its glycosylation, but was not proteolytically processed in this cell-free system (Fig. 2A). This result suggests that processing of the lFng precursor protein requires specific components and could thus be subject to regulation in the embryo, as was proposed for another Xenopus embryonic inducer, Vg1 (3). Next, we transfected COS cells with a plasmid expressing lFng fused in frame at the COOH-terminus with a stretch of six histidines (His tag) and a T7 antigenic epitope (FrgHispcDNA) (10). Conditioned media were collected from COS cells transfected with either the construct expressing His-tagged lFng or the control vector. A single band was detected in the conditioned medium from lFng-transfected COS cells by protein immunoblotting with the use of a monoclonal antibody against the T7 epitope (Fig. 2B). These results demonstrate that lFng is a secreted protein. The size of the secreted product corresponds to the region carboxyl to the tetrabasic site, although the exact site of cleavage remains to be determined.
Fig. 2.
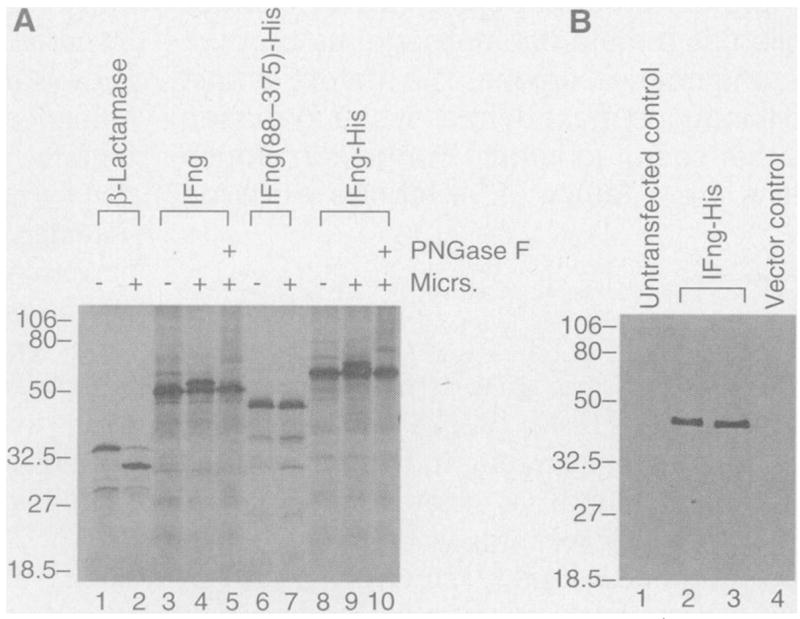
Expression and processing of lFng precursor protein. (A) Expression of lFng in rabbit reticulocyte lysate and its glycosylation by canine pancreatic microsomal membrane preparations. Lane 1: in vitro–translated β-lactamase product. Lane 2: removal of the signal peptide of β-lactamase after addition of the microsomal (Micrs.) preparation. Lane 3: in vitro–translated full-length lFng. Lane 4: same as lane 3, with microsomes added. Note appearance of a larger band. Lane 5: treatment with peptide N-glycosidase F (PN-Gase F) of a reaction similar to that shown in lane 4. Lane 6: in vitro–translated lFng fragment (residues 88 to 375) fused in frame to a His tag and the T7 epitope (FrgHispcDNA). Lane 7: same as lane 6, with microsomes added. Lane 8: in vitro–translated product of FrgHispcDNA. This construct was also used in lanes 2 and 3 of (B). Lane 9: same as lane 8, with microsomes added. Lane 10: same as lane 9: but with further treatment with PNGase F. (B) Secretion of lFng protein by COS cells. Lane 1: medium from control untransfected COS cells. Lanes 2 and 3: conditioned medium from COS cells transfected with FrgHispcDNA (two independent transfections). Lane 4: medium conditioned with COS cells transfected with the vector pcDNAIII alone. Presence of the T7 epitope in lFng fusion protein was revealed on the protein immunoblot by antibodies to T7. The size of this protein is similar to that in lane 7 of (A) and is clearly smaller than that in lane 8 of (A). Molecular size markers in kilodaltons.
Northern (RNA) analysis revealed that lFng messenger RNA is present as a maternal component before zygotic transcription begins (Fig. 3A). Whole-mount in situ hybridization was also used to examine the pattern of lFng expression (11) (Fig. 3, C to K). The apparent concentration of lFng messenger RNA in the animal hemisphere of late blastula and gastrula embryos (Fig. 3C) is likely the result of difficulty in staining the vegetal pole, because lFng messenger RNA could be detected in dissected animal, marginal, and vegetal regions (Fig. 3B). lFng messenger RNA was later expressed in the neural tube (Fig. 3, D and E), in the medial, intermediate, and later neurons (Fig. 3H). Expression of lFng in the eyes was detectable in stage 25 embryos (Fig. 3F) and persisted until stage 28 (Fig. 3G). At stage 35, lFng expression was no longer in the eyes but appeared in the otic vesicles (Fig. 3, I to K). Thus, the pattern of lFng expression suggests that it may have multiple functions during embryogenesis. Expression of lFng in blastula and gastrula suggests a possible role of lFng in mesoderm development, whereas expression in the neural tube suggests that lFng may function in neural development.
Fig. 3.
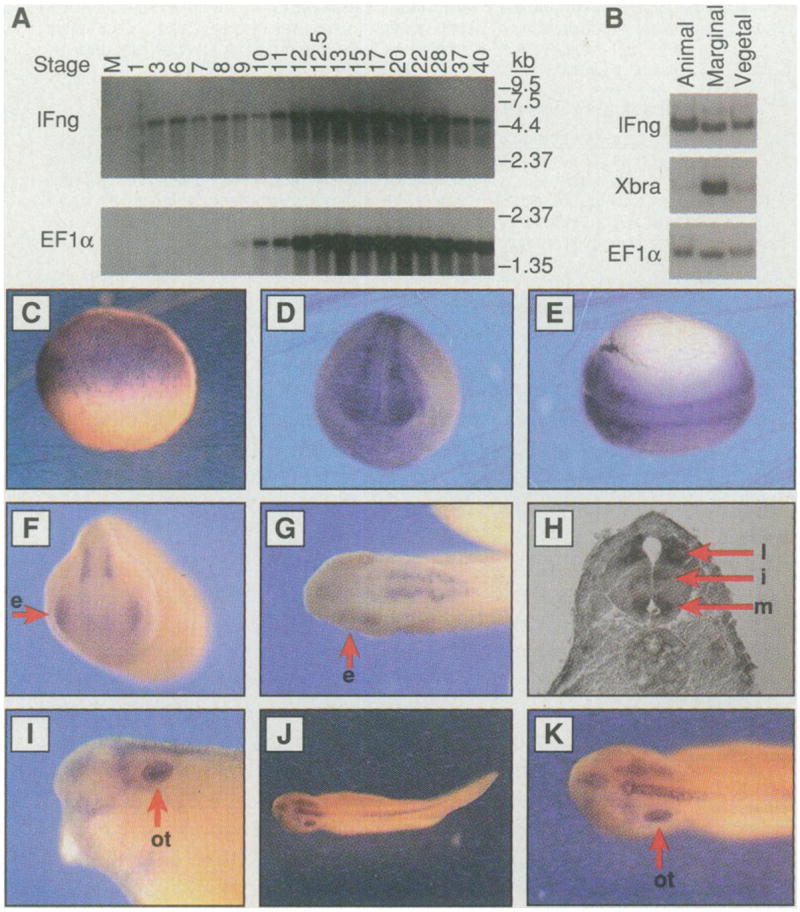
Pattern of lFng expression. (A) Northern (RNA) analyses of lFng and EF1α. Approximately 2 μg of polyadenylated [poly(A)+] RNA was loaded in each lane. Numbering on the top indicates the embryonic stage from which RNA was extracted. M represents RNA from unfertilized eggs. The same blot was used for hybridization with both probes. Longer exposure showed that Ef1α is expressed in unfertilized eggs, although at a lower level than its expression level in later embryonic stages. (B) Detection of lFng messenger RNA in animal, marginal, and vegetal regions of stage 9 embryos by reverse transcriptase–PCR. (C) Lateral view of a late stage 8 embryo; the apparent absence of lFng messenger RNA from the vegetal region was due to difficulty in staining this region. Similar patterns were detected at stages 9 through 11. (D) Anterior and (E) dorsal views of a stage 18 embryo showing lFng expression in the neural tube. (F) Anterior view of a stage 25 embryo. The arrow points to an eye (e). (G) Dorsal view of a stage 28 embryo showing lFng expression in the forebrain, the eyes (e), and the hindbrain. (H) A section at the spinal cord level of a stage 35 embryo, showing lFng expression in lateral, intermediate, and medial neurons (indicated by arrows as l, i, and m, respectively). (I, J, and K) Lateral and dorsal views of a stage 35 embryo. lFng expression persists in the hindbrain and the spinal cord, but not in the eyes. lFng expression appears in the otic vesicles (indicated by the arrow as ot).
We studied the function of lFng protein with the animal cap assay, which is a standard test for mesodermal and neural induction. Explants of the animal region, or the animal caps, were isolated at a blastula stage and incubated with a putative inducing factor. These animal caps were then cultured to appropriate stages for histological examination or RNA extraction (13). We first tested for possible inducing activity of conditioned media collected from COS cells transfected either with a lFng-expressing plasmid or with a vector plasmid. lFng-conditioned medium, but not control medium, induced expression of muscle-specific actin, a marker for mesoderm induction (Fig. 4, A to C). We then used the animal cap assays to test lFng protein purified from the conditioned medium (13). It was also active in mesoderm induction as shown by elongation of the animal caps (Fig. 4, D to F) and by histological examinations of sections of lFng-treated animal caps (Fig. 4, G to I). Conditioned medium from rFng-transfected COS cells did not have mesoderm-inducing activity (12).
Fig. 4.
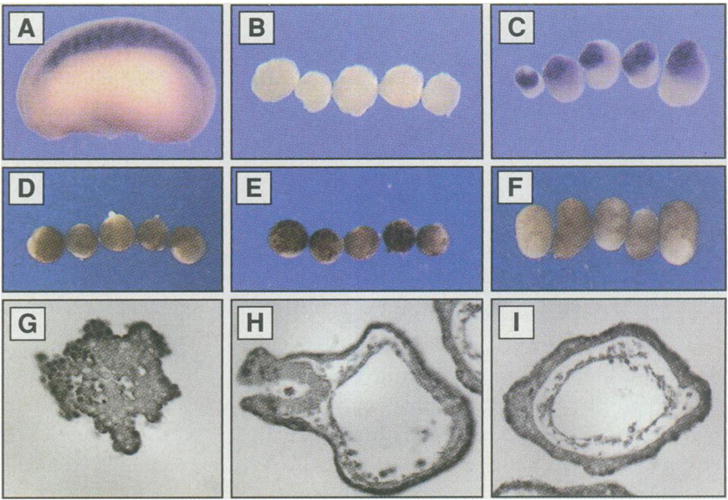
Formation of mesodermal tissues in animal caps treated with mature lFng protein. Medium containing His-tagged lFng protein was used in (C) and purified lFng was used in (F), (H), and (I). Animal caps were treated at stage 8 for 1 hour and cultured to stage 13 in (D) to (F), stage 23 in (B) and (C), or stage 35 in (G) through (I). (A) A stage 23 embryo hybridized in situ with a probe for muscle-specific actin, showing specific expression in the somites. (B) Absence of muscle-specific actin in stage 23 animal caps treated with the control conditioned medium. (C) Expression of muscle-specific actin in stage 23 animal caps treated with conditioned medium from COS cells expressing His-tagged lFng. Conditioned media from COS cells expressing lFng protein lacking the His and T7 tags were also active (12). (D) Animal caps cultured in 0.5× MMR at stage 13. (E) Stage 13 animal caps treated with control elution from nickel agarose beads previously incubated with conditioned medium collected from COS cells transfected with the vector plasmid.(F) Stage 13 animal caps treated with purified lFng protein (6 ng/ml). Note that they are elongated. (G) Section of an animal cap treated with control elution from nickel agarose beads previously incubated with conditioned medium of COS cells transfected with the vector plasmid. (H) Section of an animal cap treated with the purified lFng. (I) Section of another animal cap treated with the purified lFng. The structures formed here are similar to those in bFGF-and bone morphogenetic protein–treated animal caps (1).
To further characterize mesodermal tissues induced by lFng, we assayed for the expression of specific molecular markers in animal caps treated with lFng protein(~6 ng/ml) (14). At stage 12, the general mesoderm marker Xbra and the ventral-lateral mesoderm marker Xwnt-8 were induced (Fig. 5A). The dorsal mesoderm marker noggin was not induced (Fig. 5A). At stage 35, muscle-specific actin, a ventral mesoderm marker globin, and a posterior neural marker XlHbox6 were expressed in lFng-treated caps (Fig. 5B). The midbrain-hindbrain junction marker En-2 was not induced by lFng at this concentration (Fig. 5B). Expression of the general neural marker NCAM was also detected in animal caps treated with lFng (Fig. 5B).
Fig. 5.
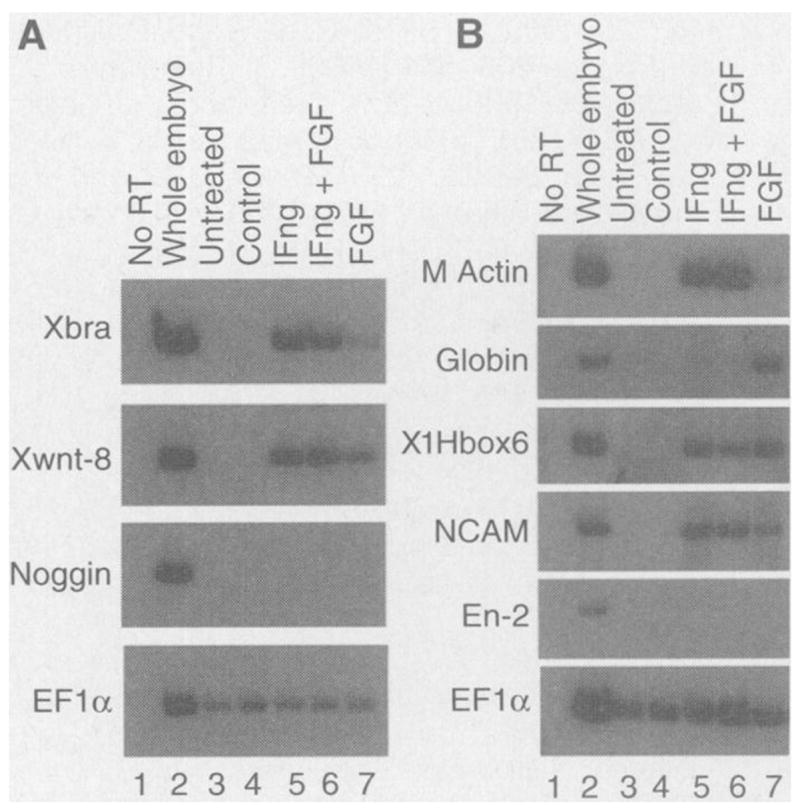
Expression of molecular markers in lFng-treated animal caps. (A) Expression of markers at stage 12 in response to lFng (6 ng/ml) protein. Xbra is a general mesoderm marker, Xwnt-8 a ventral-lateral mesoderm marker, and noggin a dorsal mesoderm marker. (B) Expression of markers at stage 35 in response to lFng protein. Muscle-specific actin (M actin) isa general mesoderm marker, NCAM a general neural marker, En-2 a marker for the midbrain-hindbrain junction, XlHBox6 a posterior neural marker, and globin a ventral mesoderm marker. Animal caps were treated at stage 8 with the purified lFng protein(~6 ng/ml), bFGF (40 ng/ml), or both, for 1 hour and cultured to either stage 12 [in (A)] or stage 35 [in (B)] before RNA extraction. For both (A) and (B): lane 1, control reactions with no reverse transcriptase (RT) added in the reverse transcription step; lane 2, expression of markers in whole embryos; lane 3, expression of markers in caps not treated with any inducer; lane 4, caps treated with control elution from nickel agarose beads previously incubated with conditioned medium of vector-transfected COS cells; lane 5, expression of markers in caps treated with lFng protein (6 ng/ml) purified from conditioned medium of COS cells expressing His-tagged lFng; lane 6, caps treated with lFng (6 ng/ml) and bFGF (40 ng/ml); lane 7, expression in caps treated with bFGF (40 ng/ml). EF1α serves as a control for the level of input RNA used in RT-PCR.
The relation of lFng with previously known mesoderm inducers is not clear, although functional interactions have been observed between lFng and fibroblast growth factor-basic (bFGF) (Fig. 5B). Considering the biochemical and functional properties of lFng, an attractive possibility is that other molecules may control the generation of active mature lFng protein by regulating the proteolytic processing of lFng precursor protein. Determination of the distribution of the mature lFng protein in Xenopus embryos will provide information about the function and regulation of the endogenous lFng protein.
REFERENCES AND NOTES
- 1.Klein PS, Melton DA. Endocrinol Rev. 1994;15:326. doi: 10.1210/edrv-15-3-326. [DOI] [PubMed] [Google Scholar]; Dawid IB, Taira M. Bioessays. 1994;16:385. doi: 10.1002/bies.950160603. [DOI] [PubMed] [Google Scholar]; Slack JM. Curr Biol. 1994;4:116. doi: 10.1016/s0960-9822(94)00027-8. [DOI] [PubMed] [Google Scholar]; Jones CM, Smith JC. ibid. 1995;5:574. [Google Scholar]; Slack JMW, Darlington BG, Heath JK, Godsave SF. Nature. 1987;326:197. doi: 10.1038/326197a0. [DOI] [PubMed] [Google Scholar]; Smith JC. Development. 1987;99:3. doi: 10.1242/dev.99.1.3. [DOI] [PubMed] [Google Scholar]; Weeks DL, Melton DA. Cell. 1987;51:861. doi: 10.1016/0092-8674(87)90109-7. [DOI] [PubMed] [Google Scholar]; Kimelman D, Kirschner M. :869. ibid. [Google Scholar]; Rosa FM, et al. Science. 1988;239:783. doi: 10.1126/science.3422517. [DOI] [PubMed] [Google Scholar]; Slack JMW, Darlington BG, Gillespie LL, Godsave SF, Paterno GD. Development. 1989;107:141. doi: 10.1242/dev.107.Supplement.141. [DOI] [PubMed] [Google Scholar]; Asashima M, et al. Roux’s Arch Dev Biol. 1990;198:330. doi: 10.1007/BF00383771. [DOI] [PubMed] [Google Scholar]; Smith JC, Price BMJ, Van Nimmen K, Huylebroeck D. Nature. 1990;345:729. doi: 10.1038/345729a0. [DOI] [PubMed] [Google Scholar]; Sokol S, Wong GG, Melton DA. Science. 1990;249:561. doi: 10.1126/science.2382134. [DOI] [PubMed] [Google Scholar]; Thomsen G, et al. Cell. 1990;63:485. doi: 10.1016/0092-8674(90)90445-k. [DOI] [PubMed] [Google Scholar]; Green JBA, Smith JC. Nature. 1990;347:391. doi: 10.1038/347391a0. [DOI] [PubMed] [Google Scholar]; Green JBA, New HV, Smith JC. Cell. 1992;71:731. doi: 10.1016/0092-8674(92)90550-v. [DOI] [PubMed] [Google Scholar]; van den Eijnden-Van Raaij AJM, et al. Nature. 1990;345:732. doi: 10.1038/345732a0. [DOI] [PubMed] [Google Scholar]; Jones CM, Kuehn MR, Hogan BLM, Smith JC, Wright CVE. Development. 1995;121:3651. doi: 10.1242/dev.121.11.3651. [DOI] [PubMed] [Google Scholar]; Koster M, et al. Mech Dev. 1991;33:191. doi: 10.1016/0925-4773(91)90027-4. [DOI] [PubMed] [Google Scholar]; Dale L, Howes G, Price BMJ, Smith JC. Development. 1992;115:573. doi: 10.1242/dev.115.2.573. [DOI] [PubMed] [Google Scholar]; Jones CM, Lyons KM, Lapan PM, Wright CVE, Hogan BJM. :639. ibid. [Google Scholar]
- 2.Amaya E, Musci TJ, Kirschner MW. Cell. 1991;66:257. doi: 10.1016/0092-8674(91)90616-7. [DOI] [PubMed] [Google Scholar]; Cornell R, Kimelman D. Development. 1994;120:453. doi: 10.1242/dev.120.2.453. [DOI] [PubMed] [Google Scholar]; LaBonne C, Whitman M. :463. ibid. [Google Scholar]
- 3.Thomsen GH, Melton DA. Cell. 1993;74:433. doi: 10.1016/0092-8674(93)80045-g. [DOI] [PubMed] [Google Scholar]; Dale L, Matthews G, Colman A. EMBO J. 1993;12:4471. doi: 10.1002/j.1460-2075.1993.tb06136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hemmati-Brivanlou A, Melton DA. Nature. 1992;359:609. doi: 10.1038/359609a0. [DOI] [PubMed] [Google Scholar]; Kessler DS, Melton DA. Science. 1994;266:596. doi: 10.1126/science.7939714. [DOI] [PubMed] [Google Scholar]; Schulte-Merker S, Smith JC, Dale L. EMBO J. 1994;13:3533. doi: 10.1002/j.1460-2075.1994.tb06660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vassalli A, Matzuk MM, Gardner HA, Lee KF, Jaenisch R. Genes Dev. 1994;8:414. doi: 10.1101/gad.8.4.414. [DOI] [PubMed] [Google Scholar]; Matzuk MM, et al. Nature. 1995;374:354. doi: 10.1038/374354a0. [DOI] [PubMed] [Google Scholar]; Matzuk MM, Kumar TR, Bradley A. :356. ibid. [Google Scholar]
- 6.Irvine KD, Wieschaus E. Cell. 1994;79:595. doi: 10.1016/0092-8674(94)90545-2. [DOI] [PubMed] [Google Scholar]
- 7.Kim J, Irvine KD, Carroll SB. ibid. 1995;82:795. doi: 10.1016/0092-8674(95)90476-x. [DOI] [PubMed] [Google Scholar]; Campbell G, Weaver T, Tomlinson A. ibid. 1993;74:1113. doi: 10.1016/0092-8674(93)90732-6. [DOI] [PubMed] [Google Scholar]; Tabata T, Kornberg TB. ibid. 1994;76:89. doi: 10.1016/0092-8674(94)90175-9. [DOI] [PubMed] [Google Scholar]
- 8.Several pairs of degenerate PCR primers for fringe were designed and the following pair worked: the upstream sequence CGI YTI TGY TGY AAR ATG coding for ALCCKM, and the downstream sequence CAR AAI CCI GCI CCI CCI GT coding for GGAGFC (16). Complementary DNAs (cDNAs) from stages 10.5, 13, and 28 Xenopus embryos were used as templates. The PCR conditions were as follows: 94°C for 3 min, 53°C for 1 min, and 72°C for 2 min for 1 cycle, followed by 36 cycles of 94°C for 45 s, 53°C for 1 min, and 72°C for 2 min. A band of about 280 base pairs was obtained and subcloned into pBluescrpt SK. About 100 colonies were obtained, and 24 individual cDNAs have been isolated. Nine of these clones were sequenced and found to encode two distinct Xenopus Fng proteins: five clones encoding lFng and four clones encoding rFng. The fragments were used as probes to screen stage 17 and stage 28 Xenopus embryonic cDNA libraries. Positive clones were isolated (10 for lFng and 7 for rFng), and the longest cDNAs (1.4 kb for lFng and 2.5 kb for rFng) were sequenced. Both of the sequences contained stop codons in all three frames upstream of the first AUG. The longest open reading frames encoded full-length proteins for lFng and rFng. In the case of rFng, the amino acid sequence shown in Fig. 1, B and D, is likely to be the product of an alternatively spliced form. At the COOH-terminus of the sequence shown, a stop codon is introduced by a sequence of 67 nucleotides that fits the consensus of an intron. If this intron is spliced out, a sequence encoding FKSVHCLLYSDTDWCPNHKHNPTT (16) will be added to the COOH-terminus. Low-stringency hybridization was carried out with 30%formamide, 8× standard saline citrate (SSC), 5× Denhardt’s solution, 0.5% SDS, and salmon sperm DNA (100 μg/ml). Fifteen positive plaques were isolated for further screening, and a single plaque went through secondary screening. A cDNA of 2.3 kb was isolated after tertiary screening. It was sequenced and found to encode rFng. In the 3′ end of the coding region, this cDNA also contains the intron described above so that it predicts the same product as shown in Fig. 1B.
- 9.Full-length lFng, His-tagged lFng, and the His-tagged COOH-terminal fragment of lFng (with amino acid residues 88 to 375) were each generated by in vitro translation in the presence of 35S-labeled methionine with a coupled transcription-translation rabbit reticulocyte lysate system (Promega). To examine glycosylation and signal peptide cleavage, we used a canine pancreatic microsomal membrane preparation (Promega)at a concentration of 2 μl of microsomal membrane preparation in a 25-μl reaction. β-Lactamase was used as a positive control for signal peptide cleavage by the microsomal membrane preparation. To confirm that the appearance of higher molecular weight bands was the result of glycosylation, we used the peptide N-glycosidase F (PNGase F) (New England Biolabs, Beverly, MA). PNGase F cleaves between the innermost N-acetylglucosamine and asparagine residues from N-linked glycoproteins. PNGase F was added to the in vitro translation products after the proteins were denatured and incubated for 3 hours at 37°C. The products were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) followed by autoradiography.
- 10.To analyze the expression of lFng protein in mammalian cells, we cultured COS-7 cells in Dulbecco’s modified essential medium (DMEM) (Gibco-BRL) supplemented with 5% fetal bovine serum (Hyclone, Logan, UT) and grown to 50% confluence in 100-mm dishes. Next, 20 μg of FrgpcDNA, FrgHis-pcDNA, or control vector was transfected into COS-7 cells with 25 μg of Lipofectin (Gibco-BRL). Twenty-four hours after transfection, the medium was removed and fresh medium was added. The cells were further cultured for 96 hours, and conditioned media were collected and stored in small samples at −70°C. For detection of the protein secreted by transfected COS-7 cells, nickel agarose was used to purify the His-tagged lFng protein from 10 ml of conditioned medium of FrgHis-pcDNA-transfected cells. His-tagged lFng protein from a 5-ml equivalent of the conditioned medium was analyzed on SDS-PAGE, transferred to a nitrocellulose filter, and detected by a T7 antibody that recognized the T7 epitope present in the fusion protein. The concentration of lFng protein was estimated by comparison with a T7-tagged recombinant protein of known concentration.
- 11.Whole-mount in situ hybridization was performed essentially as described [ Harland RM. In: Methods in Cell Biology. Kay BK, Peng HB, editors. Vol. 36. Academic Press; San Diego, CA: 1991. pp. 685–695.].
- 12.L. Wen and Y. Rao, data not shown.
- 13.Animal cap assays were previously described (15). Blastula animal cap explants were isolated and treated for 1 hour with lFng-containing medium or purified His-tagged lFng protein. His-tagged lFng was purified with nickel agarose (Qiagen) from conditioned medium from COS cells transfected with pcDNAIII expressing His-tagged lFng. Nickel agarose was equilibrated in DMEM and then incubated with either lFng-conditioned medium or control medium (from COS cells transfected with the vector plasmid) at 4°C for 3 hours. After binding, the beads were washed three times with DMEM and resuspended in 0.5× MMR (modified Ringer’s) solution containing bovine serum albumin (100 μg/ml). Pictures of animal caps were taken at stage 13 for overall morphology. Some caps were fixed at stage 23 and hybridized in situ with a muscle-specific actin probe. Other caps were fixed at stage 35 for examination of histology. Sections (8 μm thick) were made from paraffin-embedded animal caps.
- 14.Sequences of most of the PCR primers have been described in (15). Additional primers are as follows: Xwnt-8 (upstream: AGA TGA CGG CAT TCC AGA; downstream: TCT CCC GAT ATC TCA GGA), noggin (upstream: TGC TGA GAC TCT TGG ACT; downstream: AAT GCT TCG CCA AGC GAA), globin (upstream: GCC TAC AAC CTG AGA GTG G; downstream: CAG GCT GGT GAG CTG CCC), and lFng (upstream: GAG AGC AAT ATG CGT CCT G; downstream: GTA AGC AGC TTT CCA CGG) (DNAgency).
- 15.Rao Y. Genes Dev. 1994;8:939. doi: 10.1101/gad.8.8.939. [DOI] [PubMed] [Google Scholar]
- 16.Abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, lle; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
- 17.We would like to thank K. Irvine and E. Wieschaus for providing us with the Drosophila fng probe; C. Tabin, E. Laufer, S. Sokol, Y. Wang, R. Johnson, and T. Vogt for sharing information and nomenclature of the vertebrate fng genes; and R. Cagan, J. Gordon, R. Kopan, M. Nonet, D. Ornitz, J. Sanes, and A. Strauss for helpful comments on the manuscript.



