Abstract
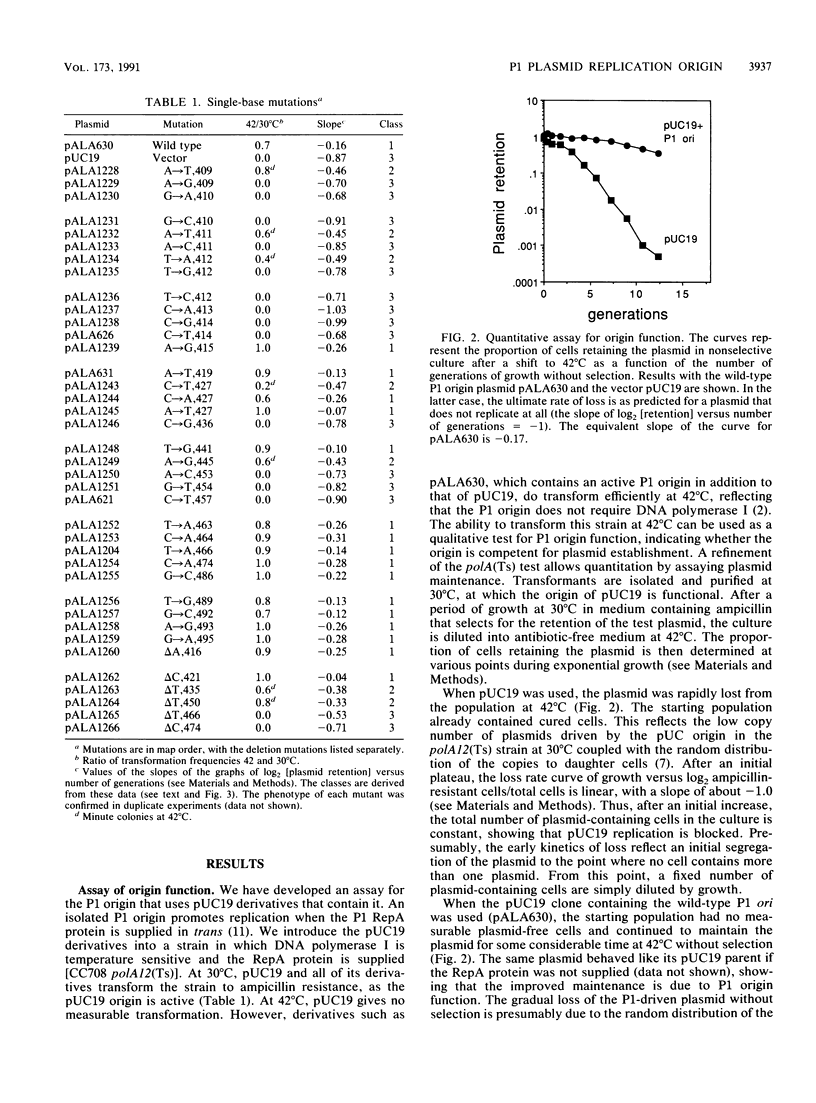
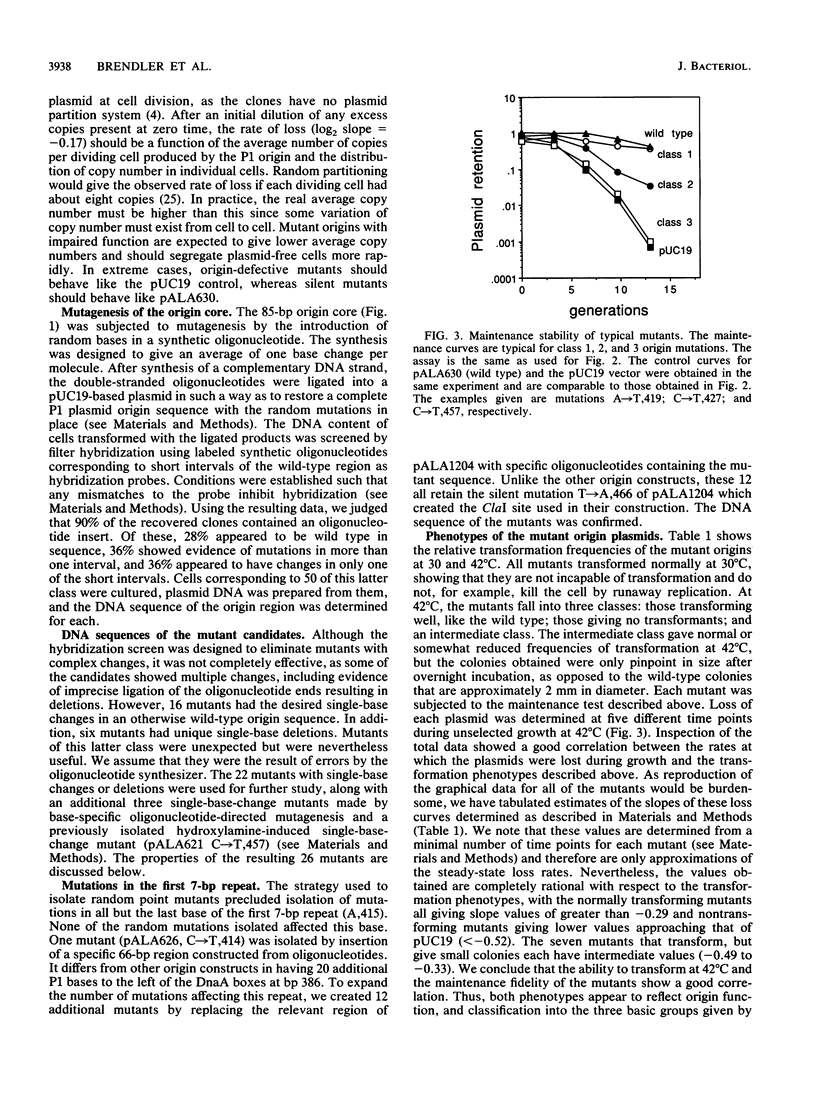
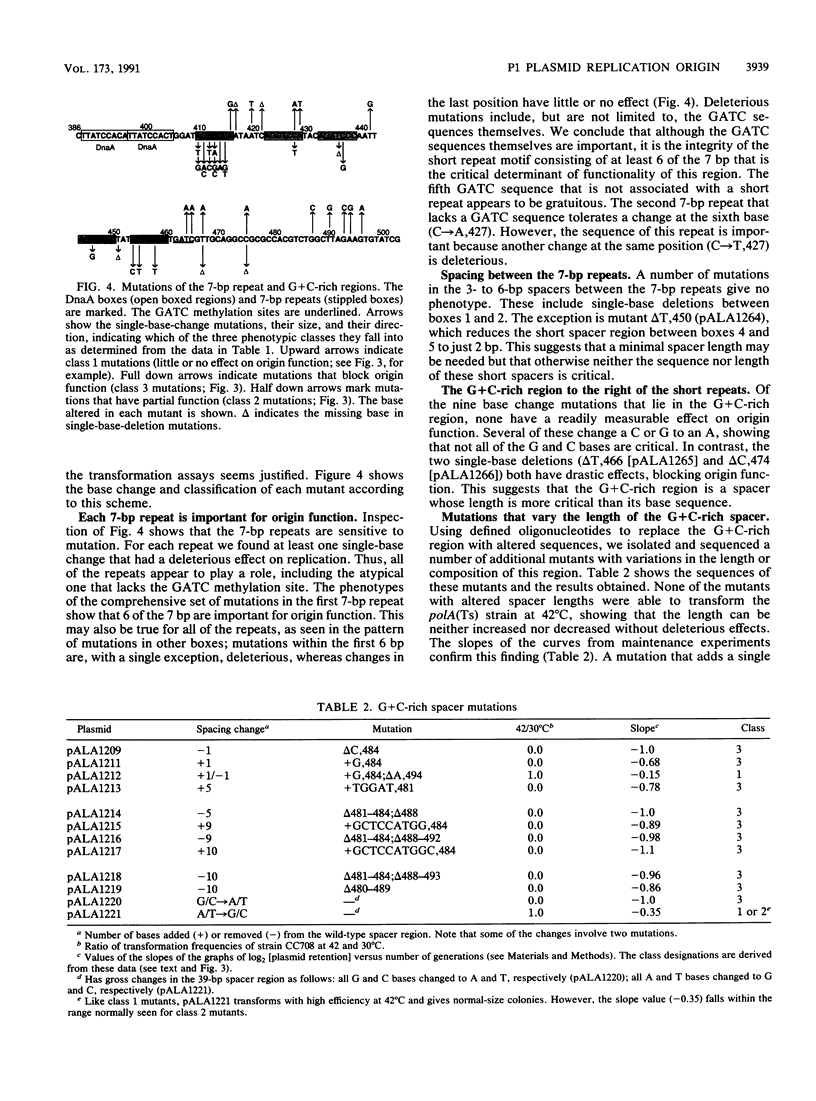
The core of the P1 plasmid replication origin consists of a series of 7-bp repeats and a G+C-rich stretch. Methylation of the GATC sequences in the repeats is essential. Forty different single-base mutations in the region were isolated and assayed for origin function. A single-base change within any 7-bp repeat could block the origin, irrespective of whether GATC bases were affected. The repeats themselves were critical, but the short intervals between them were not. Mutations in the G+C-rich region showed it to be a spacer whose exact length is important but whose sequence can vary considerably. It maintains a precise distance between the 7-bp repeats and binding sites for the P1 RepA initiator protein. It may also serve as a clamp to limit strand separation during initiation.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abeles A. L., Austin S. J. P1 plasmid replication requires methylated DNA. EMBO J. 1987 Oct;6(10):3185–3189. doi: 10.1002/j.1460-2075.1987.tb02630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeles A. L., Friedman S. A., Austin S. J. Partition of unit-copy miniplasmids to daughter cells. III. The DNA sequence and functional organization of the P1 partition region. J Mol Biol. 1985 Sep 20;185(2):261–272. doi: 10.1016/0022-2836(85)90402-4. [DOI] [PubMed] [Google Scholar]
- Abeles A. L. P1 plasmid replication. Purification and DNA-binding activity of the replication protein RepA. J Biol Chem. 1986 Mar 15;261(8):3548–3555. [PubMed] [Google Scholar]
- Abeles A. L., Reaves L. D., Austin S. J. A single DnaA box is sufficient for initiation from the P1 plasmid origin. J Bacteriol. 1990 Aug;172(8):4386–4391. doi: 10.1128/jb.172.8.4386-4391.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeles A. L., Reaves L. D., Austin S. J. Protein-DNA interactions in regulation of P1 plasmid replication. J Bacteriol. 1989 Jan;171(1):43–52. doi: 10.1128/jb.171.1.43-52.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeles A. L., Snyder K. M., Chattoraj D. K. P1 plasmid replication: replicon structure. J Mol Biol. 1984 Mar 5;173(3):307–324. doi: 10.1016/0022-2836(84)90123-2. [DOI] [PubMed] [Google Scholar]
- Austin S. J., Mural R. J., Chattoraj D. K., Abeles A. L. Trans- and cis-acting elements for the replication of P1 miniplasmids. J Mol Biol. 1985 May 25;183(2):195–202. doi: 10.1016/0022-2836(85)90212-8. [DOI] [PubMed] [Google Scholar]
- Austin S., Friedman S., Ludtke D. Partition functions of unit-copy plasmids can stabilize the maintenance of plasmid pBR322 at low copy number. J Bacteriol. 1986 Nov;168(2):1010–1013. doi: 10.1128/jb.168.2.1010-1013.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. A model for initiation at origins of DNA replication. Cell. 1988 Sep 23;54(7):915–918. doi: 10.1016/0092-8674(88)90102-x. [DOI] [PubMed] [Google Scholar]
- Chattoraj D. K., Snyder K. M., Abeles A. L. P1 plasmid replication: multiple functions of RepA protein at the origin. Proc Natl Acad Sci U S A. 1985 May;82(9):2588–2592. doi: 10.1073/pnas.82.9.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Fuller R. S., Funnell B. E., Kornberg A. The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell. 1984 Oct;38(3):889–900. doi: 10.1016/0092-8674(84)90284-8. [DOI] [PubMed] [Google Scholar]
- Fuller R. S., Kornberg A. Purified dnaA protein in initiation of replication at the Escherichia coli chromosomal origin of replication. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5817–5821. doi: 10.1073/pnas.80.19.5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen E. B., Yarmolinsky M. B. Host participation in plasmid maintenance: dependence upon dnaA of replicons derived from P1 and F. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4423–4427. doi: 10.1073/pnas.83.12.4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S., Brooks J. E., Masurekar M. Sequence specificity of the P1 modification methylase (M.Eco P1) and the DNA methylase (M.Eco dam) controlled by the Escherichia coli dam gene. J Mol Biol. 1978 Dec 15;126(3):367–380. doi: 10.1016/0022-2836(78)90046-3. [DOI] [PubMed] [Google Scholar]
- Hill D. E., Oliphant A. R., Struhl K. Mutagenesis with degenerate oligonucleotides: an efficient method for saturating a defined DNA region with base pair substitutions. Methods Enzymol. 1987;155:558–568. doi: 10.1016/0076-6879(87)55036-4. [DOI] [PubMed] [Google Scholar]
- Kingsbury D. T., Helinski D. R. DNA polymerase as a requirement for the maintenance of the bacterial plasmid colicinogenic factor E1. Biochem Biophys Res Commun. 1970 Dec 24;41(6):1538–1544. doi: 10.1016/0006-291x(70)90562-0. [DOI] [PubMed] [Google Scholar]
- McNeil J. B., Smith M. Saccharomyces cerevisiae CYC1 mRNA 5'-end positioning: analysis by in vitro mutagenesis, using synthetic duplexes with random mismatch base pairs. Mol Cell Biol. 1985 Dec;5(12):3545–3551. doi: 10.1128/mcb.5.12.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Monk M., Kinross J. Conditional lethality of recA and recB derivatives of a strain of Escherichia coli K-12 with a temperature-sensitive deoxyribonucleic acid polymerase I. J Bacteriol. 1972 Mar;109(3):971–978. doi: 10.1128/jb.109.3.971-978.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström K., Austin S. J. Mechanisms that contribute to the stable segregation of plasmids. Annu Rev Genet. 1989;23:37–69. doi: 10.1146/annurev.ge.23.120189.000345. [DOI] [PubMed] [Google Scholar]
- Oliphant A. R., Nussbaum A. L., Struhl K. Cloning of random-sequence oligodeoxynucleotides. Gene. 1986;44(2-3):177–183. doi: 10.1016/0378-1119(86)90180-0. [DOI] [PubMed] [Google Scholar]
- Pal S. K., Mason R. J., Chattoraj D. K. P1 plasmid replication. Role of initiator titration in copy number control. J Mol Biol. 1986 Nov 20;192(2):275–285. doi: 10.1016/0022-2836(86)90364-5. [DOI] [PubMed] [Google Scholar]
- Prentki P., Chandler M., Caro L. Replication of prophage P1 during the cell cycle of Escherichia coli. Mol Gen Genet. 1977 Mar 28;152(1):71–76. doi: 10.1007/BF00264942. [DOI] [PubMed] [Google Scholar]
- Schneider T. D., Stormo G. D. Excess information at bacteriophage T7 genomic promoters detected by a random cloning technique. Nucleic Acids Res. 1989 Jan 25;17(2):659–674. doi: 10.1093/nar/17.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogeli G., Kaytes P. S. Amplification, storage, and replication of libraries. Methods Enzymol. 1987;152:407–415. doi: 10.1016/0076-6879(87)52047-x. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Berger S. L. Screening colonies or plaques with radioactive nucleic acid probes. Methods Enzymol. 1987;152:415–423. doi: 10.1016/0076-6879(87)52048-1. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Miyada C. G. Oligonucleotide probes for the screening of recombinant DNA libraries. Methods Enzymol. 1987;152:432–442. doi: 10.1016/0076-6879(87)52050-x. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Shaffer J., Murphy R. F., Bonner J., Hirose T., Itakura K. Hybridization of synthetic oligodeoxyribonucleotides to phi chi 174 DNA: the effect of single base pair mismatch. Nucleic Acids Res. 1979 Aug 10;6(11):3543–3557. doi: 10.1093/nar/6.11.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S. H., Chattoraj D. K. Replication of mini-P1 plasmid DNA in vitro requires two initiation proteins, encoded by the repA gene of phage P1 and the dnaA gene of Escherichia coli. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3668–3672. doi: 10.1073/pnas.84.11.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosnick M. A., Barnett R. W., Vicentini A. M., Erfle H., Elliott R., Sumner-Smith M., Mantei N., Davies R. W. Rapid construction of large synthetic genes: total chemical synthesis of two different versions of the bovine prochymosin gene. Gene. 1987;60(1):115–127. doi: 10.1016/0378-1119(87)90219-8. [DOI] [PubMed] [Google Scholar]
- Yamaki H., Ohtsubo E., Nagai K., Maeda Y. The oriC unwinding by dam methylation in Escherichia coli. Nucleic Acids Res. 1988 Jun 10;16(11):5067–5073. doi: 10.1093/nar/16.11.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]


