Abstract
Endo-1,4-β-d-glucanases (EGases, EC 3.2.1.4) are enzymes produced in bacteria, fungi, and plants that hydrolyze polysaccharides possessing a 1,4-β-d-glucan backbone. All previously identified plant EGases are E-type endoglucanases that possess signal sequences for endoplasmic reticulum entry and are secreted to the cell wall. Here we report the characterization of a novel E-type plant EGase (tomato Cel3) with a hydrophobic transmembrane domain and structure typical of type II integral membrane proteins. The predicted protein is composed of 617 amino acids and possesses seven potential sites for N-glycosylation. Cel3 mRNA accumulates in young vegetative tissues with highest abundance during periods of rapid cell expansion, but is not hormonally regulated. Antibodies raised to a recombinant Cel3 protein specifically recognized three proteins, with apparent molecular masses of 93, 88, and 53 kDa, in tomato root microsomal membranes separated by sucrose density centrifugation. The 53-kDa protein comigrated in the gradient with plasma membrane markers, the 88-kDa protein with Golgi membrane markers, and the 93-kDa protein with markers for both Golgi and plasma membranes. EGase enzyme activity was also found in regions of the density gradient corresponding to both Golgi and plasma membranes, suggesting that Cel3 EGase resides in both membrane systems, the sites of cell wall polymer biosynthesis. The in vivo function of Cel3 is not known, but the only other known membrane-anchored EGase is present in Agrobacterium tumefaciens where it is required for cellulose biosynthesis.
Keywords: cell wall polysaccharides, cellulase, Lycopersicon esculentum, plant development
EGases are enzymes that depolymerize polysaccharides containing 1,4-β-d-glucan linkages and are produced by bacteria, fungi, slime molds, snails, and plants (1, 2). Hydrophobic cluster analysis showed that the catalytic cores of this class of enzymes can be ordered into six or more families, with plant EGases belonging to family E (1). Microbial E-type EGases typically possess cellulose-binding domains, which enable them to degrade crystalline cellulose, whereas plant EGases lack these domains and do not appear capable of degrading crystalline cellulose (1, 2). In microbes, cellulases are secreted because their function is to digest external substrates and convert them to food. In plants the function of EGases is presumably to modify the cells’ own cell wall, in which case they also need to be secreted to the cell surface. All plant EGases cloned to date possess typical eukaryotic signal sequences targeting them to the endomembrane system for processing and secretion to the cell exterior. However, EGase activity has also been detected associated with plant microsomal membrane preparations (3, 4). In bean abscission zones, EGase activity comigrated in sucrose density gradients with ATPase activity, and a putative plasma membrane localization was suggested (3, 5).
Most cellular changes occurring during plant growth and development depend on modification of cell wall structure or physical properties, either by incorporation of new cell wall components or by modification of existing cell wall polymers to accommodate changes in cell size, shape, or function. Disassembly of hemicellulosic cell wall components, particularly xyloglucan, occurs during cell expansion and differentiation, organ abscission, and fruit ripening (2). EGases have been shown to act on hemicellulosic polymers possessing a 1,4-β-glucan backbone, such as xyloglucan and individual cellulose chains (6, 7), and an increase in EGase mRNA abundance and enzyme activity has been observed in association with all of the above physiological processes (2). Plant EGases are encoded by multi-gene families, in tomato consisting of at least six members each possessing a distinctive pattern of temporal and tissue-specific expression (8–11). Here we report the cloning of a novel plant EGase possessing a transmembrane domain and show that it is localized on plant Golgi and plasma membranes, suggestive of a function in cell surface polysaccharide modification or perhaps biosynthesis.
MATERIALS AND METHODS
Plant Material.
Tissues from tomato (Lycopersicon esculentum Mill., cv. T5 or Castlemart) were collected from plants grown in the greenhouse or field, except for etiolated hypocotyls, which were grown from seed in darkness. Immediately after collection or treatment all tissues were frozen in liquid N2 and stored at −80°C. RNA was isolated as previously described (8).
Library Screening and PCR.
A degenerate oligonucleotide complementary to the amino acid domain CWERPEDMD conserved in plant EGases was radiolabeled and used to screen a tomato cv. Castlemart red ripe fruit cDNA library (8). The probe hybridized to a single colony containing a cDNA insert of 1,650 bp (which was designated Cel3), distinct from the previously identified tomato EGase cDNAs Cel1 and Cel2. The truncated Cel3 cDNA was subcloned into pBluescriptII SK(+) (Stratagene) and used to screen a tomato root cDNA library in which 22 identical 2,030-bp cDNA clones were identified, one of which was sequenced on both strands.
Additions to the 5′ end were made in two stages using PCR amplification, with 45 ng of a plasmid preparation from a tomato hypocotyl cDNA library as template. The first set of PCR amplification used primers CEL3B (complementary to nucleotides 863–885) and CEL3J (5′-AA(AG)I(GC)IATI(CT)TITT(CT)T(AT)(CT)GA(AG)GGICA(AG)(AC)G-3′). Amplification was performed (12), then  of the products amplified using CEL3J and CEL3K (complementary to nucleotides 709–732). A band of 508 bp was cloned into the vector pCRII (Invitrogen), and four clones were sequenced on both strands. Each clone contained the expected overlap with the root library clone, plus 153 bp of new sequence. A second set of PCR amplification was carried out using the anchor primer AnX (5′-GGAATTCATCGATGGAT(C)17-3′) and CEL3L (complementary to nucleotides 322–347), followed by AnX and CEL3M (complementary to nucleotides 296–316). The largest product (350 bp) was cloned into pCRII, and five clones were sequenced on both strands. Each clone contained the expected overlap with the CEL3J–CEL3K product, plus 253 bp of new sequence. Based on the nine sequenced PCR clones, a consensus sequence was compiled.
of the products amplified using CEL3J and CEL3K (complementary to nucleotides 709–732). A band of 508 bp was cloned into the vector pCRII (Invitrogen), and four clones were sequenced on both strands. Each clone contained the expected overlap with the root library clone, plus 153 bp of new sequence. A second set of PCR amplification was carried out using the anchor primer AnX (5′-GGAATTCATCGATGGAT(C)17-3′) and CEL3L (complementary to nucleotides 322–347), followed by AnX and CEL3M (complementary to nucleotides 296–316). The largest product (350 bp) was cloned into pCRII, and five clones were sequenced on both strands. Each clone contained the expected overlap with the CEL3J–CEL3K product, plus 253 bp of new sequence. Based on the nine sequenced PCR clones, a consensus sequence was compiled.
The deduced amino acid sequence of Cel3 was aligned with the deduced amino acid sequences of other plant EGase mature proteins (i.e., after removal of signal sequences) using clustalv, then phylogeny was determined using paup and employing a heuristic search with 100 replicates and global (tree bisection and reconnection) branch swapping, as described (2).
DNA and RNA Gel Blot Analysis.
Restriction-digested genomic DNA was probed with a radiolabeled cDNA probe consisting of nucleotides 1058–2175 of the Cel3 cDNA clone, and washed at moderate stringency (11). Total RNA isolated from stem tissue was selected for poly(A)+ using oligo(dT) Dynabeads (Dynal, Great Neck, NY). An RNA gel blot was hybridized with an antisense riboprobe synthesized as described below for ribonuclease protection assays, and washed at moderate stringency.
Antibody Production.
Cel3 cDNA was digested with BstXI and HindIII and the DNA fragment encoding amino acids 292–615 of the deduced protein cloned into the NdeI and BamHI restriction sites of the expression vector pET-16b (Novagen) using synthetic linkers. This construct was introduced into Escherichia coli strain BL21(DE3) harboring repressor plasmid pACYC (Novagen), and a single colony was grown at 37°C to an A600 of 0.6. Aliquots of this culture were inoculated into Luria–Bertani medium containing 50 μg/ml carbenicillin and kanamycin and grown to an A600 of 0.6, then induced by adding 1 mM isopropyl β-d-thiogalactoside and grown for 14 h. Bacteria were pelleted, and disrupted by sonication, and insoluble pellets were extracted with binding buffer containing 6 M urea. Urea-soluble extracts were applied to a column of His-Bind Sepharose 6B (Novagen), washed, then eluted with 1 M imidazole. Strips of polyacrylamide gel containing approximately 1.5 mg SDS/PAGE-purified recombinant protein were freeze-dried, powdered, mixed with 2 ml sterile PBS and 2 ml Freund’s complete adjuvant, and emulsified. The antigen was injected into several subcutaneous sites of two rabbits. Booster injections in Freund’s incomplete adjuvant were carried out at 9 and 27 days, and terminal bleeds were performed at 57 days. Cel3 antibodies were affinity-purified from Cel3 antiserum by incubating 2 μl of serum in 1 ml Tris-buffered saline Tween (TBST) with a strip of Immobilon transfer membrane (Millipore) to which had been blotted 1 mg gel-purified recombinant protein. Antibody bound to the recombinant protein was eluted in 0.2 M HCl/glycine, pH 2.2, then neutralized.
Membrane and Protein Preparation.
Roots (29 g) of tomato seedlings grown hydroponically over Hoagland’s solution under white light for 8 days were chopped with a razor blade then homogenized in 2 vol of ice-cold homogenization medium containing protease inhibitors (13), and the homogenate was filtered through cheesecloth. The filtered homogenate was centrifuged at 10,000 × g, and the supernatant was layered above a cushion of 58% (wt/vol) sucrose then centrifuged at 80,000 × g for 30 min. Microsomal membranes were collected from the homogenate/sucrose cushion interface and diluted to below 15% sucrose, and 7 ml was loaded on top of a 31-ml linear 15–50% sucrose gradient (13). The gradient was centrifuged at 100,000 × g for 4 h, then 1.5-ml fractions were collected from the top.
Material retained in the cheesecloth (mainly cell wall debris) was extracted with gradient buffer (13) containing 1 M NaCl and protease inhibitors on ice for 2.5 h then centrifuged at 100,000 × g for 2 h, and the supernatant was used as cell wall proteins. The 80,000 × g supernatant described above was centrifuged at 100,000 × g for 2 h and the supernatant was used as soluble proteins.
Trypsin treatments were carried out on microsomal membrane vesicles incubated for 1 h at 29°C in 250 mM sucrose and 25 mM Hepes⋅KOH (pH 7.5), with or without 0.1% Triton X-100 and 500 units of trypsin.
SDS/PAGE and Western Blots.
Density gradient fractions (95 μl) were precipitated with TCA and subjected to electrophoresis on 10% polyacrylamide SDS gels before transferring to Immobilon transfer membrane. Western blots were blocked with 3% BSA in TBST, then incubated in high-salt Tween (13) with affinity-purified Cel3 antibody at a dilution of 1:7,500 overnight, or antibodies to plasma membrane ATPase, the endoplasmic reticulum (ER)-localized protein BiP, or the 58-kDa subunit of the tonoplast ATPase. Blots were washed, incubated with goat anti-rabbit antibody coupled to alkaline phosphatase, rewashed, then color-developed as described (13).
CM-Cellulase Activity.
Viscosity loss was measured at 25°C using size 200 Cannon-Manning semi-micro viscometers with 100 μl of sucrose density gradient fractions in 0.5% (wt/vol) CM-cellulose (high viscosity, Sigma) and 20 mM phosphate-citrate (pH 6.0) with or without 0.1% (vol/vol) Triton X-100. One unit of activity was defined as that causing a 1% decrease in viscosity in 1 h.
Ribonuclease Protection Assays and Hormone Treatments.
Template for probe synthesis was the truncated Cel3 cDNA subcloned into pBluescript, digested with EcoRV at nucleotide 1329 and in the plasmid polylinker to remove the poly(A) tail, then religated and linearized with XbaI. Assays were performed as described, and Cel3 mRNA was estimated against a standard curve by phosphorimagery (11). Flower abscission zones were obtained from flower explants treated with air or with air containing 10 μl/l ethylene for the indicated time (8). The excised upper halves of young etiolated hypocotyls were treated for 24 h with solutions of indoleacetic acid (IAA) or gibberellin. Young etiolated seedlings grown in pots of soil were treated with air or ethylene for 24 h, then the upper half of each hypocotyl was collected (11).
RESULTS
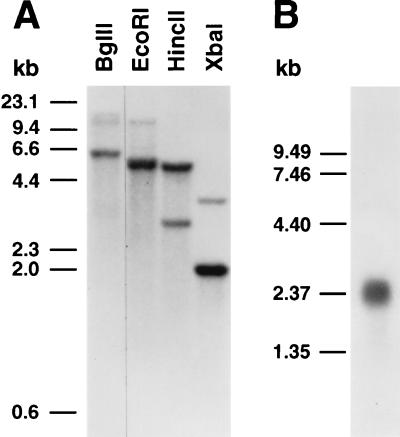
The Cel3 cDNA clone consisted of 148 bp of 5′-untranslated sequence, 1851 bp of coding sequence, 287 bp of 3′-untranslated sequence, and approximately 150 bp of poly(A) tail. The coding region encoded a deduced protein of 617 amino acids, considerably longer than that of other plant EGases (Fig. 1A), with a deduced molecular mass of 68.5 kDa and a pI of 8.96. The deduced Cel3 protein lacked a typical eukaryotic signal sequence, and instead possessed a very highly charged region at the N terminus (21 of the first 58 amino acid residues being charged), followed by a predominantly hydrophobic region of 39 amino acid residues (Fig. 1 B and C). This hydrophobic domain was flanked on the N-terminal side by four consecutive, positively charged amino acid residues and on the C-terminal side by five consecutive positively charged residues. Such a structure is typical of membrane-spanning domains (14). The remainder of the deduced protein possessed seven potential N-glycosylation sites (N-X-S/T) and aligned with the deduced amino acid sequences of the mature proteins of other tomato EGases (Fig. 1A). The C terminus of Cel3 is rich in Pro (10 of the last 16 amino acids), a characteristic of the linker regions connecting different domains in microbial cellulases (1). A phylogenetic comparison of the deduced Cel3 protein with the deduced sequences of 12 full-length plant EGase mature proteins showed that plant EGases fall into three main branches (Fig. 1D). Tomato Cel3 was the most divergent of all plant EGases and formed a separate, distinct branch with no close relatives. However, DNA gel blot analysis suggested that tomato Cel3 belongs to a small gene family probably consisting of two members (Fig. 2A). RNA gel blot analysis indicated that the Cel3 mRNA was of about 2.4 kb (Fig. 2B).
Figure 1.
Sequence and phylogenetic analysis. (A) Deduced amino acid sequence of tomato Cel3 aligned with tomato Cel1 and Cel2 mature proteins. Amino acid identity is indicated by asterisks and conserved changes by dots. Potential N-glycosylation sequences in Cel3 are overlined. (B) Schematic representation of tomato Cel3 protein structure. Cel3 consists of a charged N terminus (shaded box), a transmembrane domain (solid box), and the catalytic core (hatched box). (C) Hydropathic profile of Cel3 deduced protein. (D) Phylogenetic comparison of plant EGase deduced proteins after removal of signal sequences. Sequences and GenBank accession numbers: avocado, Cel1 (M17634M17634); bean, BAC1 (M57400M57400); elder, JET1 (X74290X74290); pea, EGL1 (L41046L41046); peach, Cel1 (X96853X96853); pepper, Cel1 (X87323X87323), Cel2 (X97190X97190), and Cel3 (X97189X97189); poplar, Cel1 (D32166D32166); tomato, Cel1 (U13054U13054), Cel2 (U13055U13055), and Cel4 (also called TPP18; U20590U20590).
Figure 2.
DNA and RNA gel blot analysis. (A) DNA gel blot of 10-μg aliquots of genomic DNA digested with the indicated restriction enzyme. The gel blot was hybridized with a 32P-labeled 1117-nucleotide Cel3 cDNA fragment in 50% formamide, 0.9 M NaCl (6× SSPE) at 36°C, with final wash in 0.075 M NaCl (0.5× SSC) at 53°C (27°C below Tm). (B) RNA gel blot of 3 μg of stem poly(A)+ RNA hybridized with an antisense Cel3 riboprobe in 50% formamide, 0.75 M NaCl (5× SSPE) at 59°C, with final wash in 0.015 M NaCl (0.1× SSC) at 49°C (23°C below Tm).
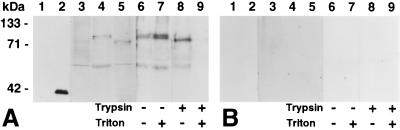
Affinity-purified antibody raised against a recombinant Cel3 fusion protein detected a single protein of 40 kDa in induced E. coli, which was coincident with the recombinant Cel3 protein and was not present in uninduced E. coli (Fig. 3). The structure of the deduced Cel3 protein suggested that it would be anchored to an ER-derived membrane in tomato cells. To test this possibility, the subcellular distribution of protein reactive with Cel3 antibody was examined in soluble proteins and in microsomal membrane preparations (Fig. 3). Cel3 antiserum reacted with a single band of 53 kDa in soluble proteins, and with 53- and 82-kDa bands in proteins released from cell walls by high salt concentrations. With microsomal membranes, two protein doublets were detected, corresponding to proteins of 93 and 88 kDa, and 53 and 52 kDa. None of these proteins could be released from microsomal membranes by sonication with or without 0.1 M Na2CO3 or by 0.5 M KI (data not shown), and they appeared to be on the inside of microsomal vesicles because they were not susceptible to protease digestion in the absence of Triton X-100 (Fig. 3, lane 8). After trypsin treatment of membrane vesicles, the 93- and 88-kDa proteins showed a slight reduction in size and appeared to run coincident with one other. This decrease in molecular mass is presumably due to the cleavage of the N-terminal domain, which spans the membrane and is present on the outside of the membrane vesicles.
Figure 3.
Western blots of uninduced and induced E. coli proteins (100 ng per lane) and tomato proteins (50 μg per lane) probed with Cel3 antibody (1:7,500) affinity-purified against recombinant Cel3 protein blotted to transfer membrane (A) or against transfer membrane alone blocked with BSA (B). Lanes: 1, uninduced E. coli; 2, E. coli induced with isopropyl β-d-thiogalactoside; 3, tomato soluble proteins; 4, tomato microsomal membranes; 5, tomato cell wall proteins; 6–9, microsomal membranes treated with no trypsin, no Triton (lane 6), no trypsin, 0.1% Triton (lane 7); 500 units of trypsin, no Triton (lane 8); 500 units of trypsin, 0.1% Triton (lane 9).
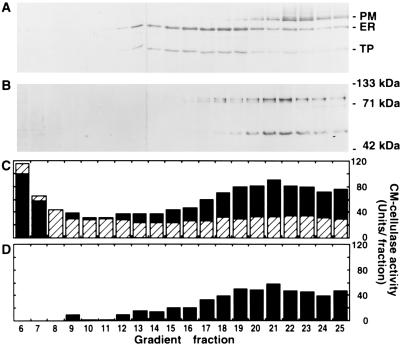
When subjected to sucrose density gradient centrifugation, root microsomal membranes were separated according to density, with lighter tonoplast membranes near the top of the gradient, followed by overlapping peaks of ER and Golgi membranes, and denser plasma membranes near the bottom of the gradient as indicated by the distribution of enzymic markers diagnostic for each membrane (Fig. 4). When these density gradient fractions were subjected to SDS/PAGE and a Western blot of the proteins reacted with Cel3 antibody, membrane-associated proteins of 93, 88, and 53 kDa were detected (Fig. 5B). The 93-kDa protein was detected throughout the lower part of the gradient, from fractions 14 to 25. Its distribution with a slight peak in fraction 18 and a major peak in fractions 21 and 22 was consistent with the protein being localized on both Golgi and plasma membranes. The 88-kDa protein was detected in fractions 14–22 with a peak in fraction 18. This was consistent with the 88-kDa protein being localized on an internal cell membrane, and its distribution in the gradient most closely corresponded to Golgi membranes. The 53-kDa protein was detected in fractions 17–25 with a peak in fractions 21–22, coincident in the gradient with the plasma membrane marker vanadate-sensitive ATPase activity (Fig. 4B) and immunodetection of the plasma membrane ATPase protein (Fig. 5A). In the absence of Triton, a low baseline of CM-cellulase activity was present throughout the density gradient (Fig. 5C), with higher activity near the top of the gradient presumably being due to soluble proteins that did not enter the sucrose gradient. In the presence of Triton a significant detergent activation of activity was observed as a broad peak of CM-cellulase activity toward the bottom of the gradient with a peak in fraction 21 (Fig. 5D). This was consistent with the apparent localization of Cel3-reactive proteins on the inside of membrane vesicles. The distribution of CM-cellulase activity was correlated with the distribution of both Golgi-associated (93 and 88 kDa) and plasma membrane-associated (93 and 53 kDa) Cel3 proteins, indicating that all the immunodetected proteins are probably enzymically active. The 93- and 88-kDa proteins are consistent in size with a heavily glycosylated Cel3 gene product. Whether the 53-kDa protein is another EGase related to Cel3, perhaps the homologous gene detected by Southern analysis (Fig. 2A), or is a highly processed product of the 93- or 88-kDa proteins remains to be elucidated. Although the antibody was affinity-purified against recombinant protein before use, it is also possible that detection of the 53-kDa band is due to cross-reactivity of the antibody with other EGase proteins, most of which are 50–55 kDa in size (2) and would be found in the soluble fraction of cell extracts.
Figure 4.
Distribution of membrane marker enzymes in a sucrose density gradient used to separate tomato root microsomal membranes. (A) Total protein determined using Bradford assay and sucrose concentration measured by refractometry. (B) NADH:cytochrome c reductase (ER) and vanadate-sensitive ATPase (plasma membrane). (C) Bafilomycin-sensitive ATPase (tonoplast) and latent UDPase (Golgi). Marker enzyme and protein assays performed as described (13).
Figure 5.
Western blots of tomato root microsomal membrane density gradient fractions probed with antibodies to plasma membrane ATPase (100 kDa), ER-localized BiP (70 kDa), and 58-kDa subunit of tonoplast ATPase (58 kDa) (A); or affinity-purified Cel3 antibody (B). (C) CM-cellulase activity determined using viscometry in the absence (striped bars) or presence (solid bars) of 0.1% Triton X-100. (D) Triton-activated CM-cellulase activity calculated from C.
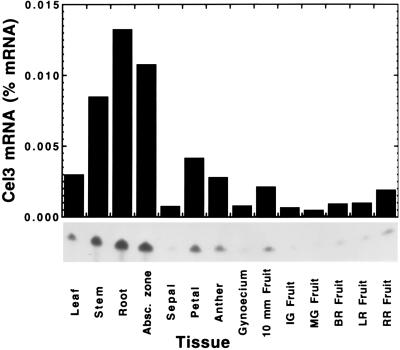
To gain insight into the potential physiological function of a plasma membrane or Golgi membrane-anchored EGase, the pattern of expression of Cel3 was examined using ribonuclease protection assays in a number of tissues and in response to hormone treatments. Cel3 mRNA accumulated to substantial levels in stems, roots, and young, immature abscision zones, and was present at moderate amounts in expanded leaves, petals, anthers, and expanding fruit (Fig. 6). Cel3 mRNA was virtually absent during the later stages of fruit expansion and during fruit ripening.
Figure 6.
Cel3 mRNA abundance in various tomato vegetative, flower, and fruit tissues. Cel3 mRNA abundance was determined against a standard curve using ribonuclease protection assays.
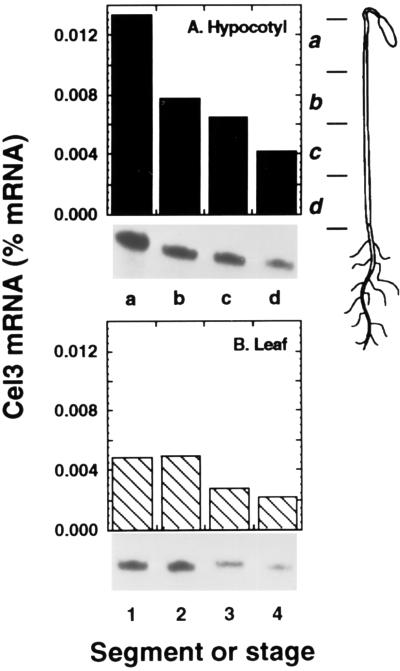
Cel3 mRNA accumulated to its highest levels in young expanding tissues, but declined in less rapidly growing tissues (Fig. 7). In etiolated hypocotyls, zone a, which contains the zone of rapid cell expansion, possessed a substantially greater level of Cel3 mRNA than other regions of the hypocotyl (Fig. 7A). In progressively more mature regions of the hypocotyl (zones b–d), the abundance of Cel3 mRNA progressively declined. Similarly, young expanding leaves accumulated higher levels of Cel3 mRNA, which declined as leaves became fully expanded (Fig. 7B).
Figure 7.
Cel3 mRNA abundance in young expanding tissues. (A) Zones of etiolated hypocotyl, as indicated in the Inset. (B) Expansion of young green leaves. Stages: 1, 1.0 × 0.3 cm; 2, 2.5 × 1.0 cm; 3, 3.5 × 1.5 cm; 4, 5.5 × 3.0 cm.
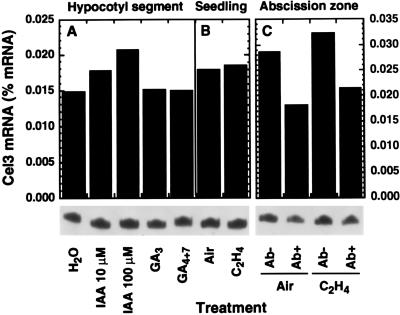
The association of Cel3 mRNA with regions of rapid cell growth suggested that its expression may be hormonally regulated, and this was tested by assaying Cel3 mRNA levels following a variety of hormone treatments. However, incubation of excised hypocotyl segments in solutions of IAA, gibberellin (GA)3, or GA4+7 for 24 h caused no significant change in Cel3 mRNA abundance (Fig. 8A). Accumulation of Cel3 mRNA was also not significantly affected by incubation of excised segments in IAA, naphthaleneacetic acid, or 2,4-dichlorophenoxyacetic acid (2,4-D) for shorter times, including the period when auxin-induced cell enlargement was occurring (data not shown). Treatment of entire young etiolated seedlings with ethylene (Fig. 8B) or with high concentrations of 2,4-D (11; data not shown) produced a cessation of hypocotyl longitudinal growth and caused radial swelling, but did not evoke changes in Cel3 mRNA abundance. Treatment of flower explants with ethylene to hasten abscission also had no effect on the abundance of Cel3 mRNA in flower abscission zones (Fig. 8C). Indeed, during abscission of tomato flowers, Cel3 mRNA abundance in flower abscission zones declined by about one-third.
Figure 8.
Effect of hormone treatments on Cel3 mRNA abundance. (A) Etiolated hypocotyl segments incubated for 24 h in solutions of IAA, 10 μM GA3, or 10 μM GA4+7. (B) Young etiolated seedlings exposed to air or 10 μl/l ethylene for 24 h. (C) Abscission zones of flower explants treated with air or 10 μl/l ethylene, then nonabscised (Ab−) and abscised (Ab+) zones were collected separately.
DISCUSSION
The deduced amino acid sequence of Cel3 is highly divergent from that of other plant or microbial E-type EGases and, within the higher plant EGases, it forms a separate phylogenetic branch. The deduced structure of all other plant EGases for which full-length sequences exist shows the presence of N-terminal hydrophobic signal sequences that target cotranslational import to the ER, and several have been confirmed to be secreted to the cell wall (2). In contrast, the deduced amino acid sequence of Cel3 indicates that it lacks a typical cleavable signal peptide and consists of three domains: an N-terminal region rich in charged amino acids, a hydrophobic membrane-spanning domain flanked by positive charges, and a C-terminal catalytic domain. This structure is characteristic of integral membrane proteins (14, 15). The charged N-terminal domain is composed of 58 amino acid residues, of which 11 are negatively charged (Asp or Glu) and 10 are positively charged (Lys, Arg, or His). This is followed by a predominantly hydrophobic domain of 39 amino acid residues, flanked on the N-terminal side by four consecutive Lys residues and on the C-terminal side by one Arg, one Lys, and three His residues. A positive difference in the number of Arg+Lys residues on the N-terminal side relative to that on the C-terminal side predicts that the topology of the protein is with the N terminus cytoplasmic and the C terminus facing the exterior (15), indicating that the Cel3 gene product is a type II integral membrane protein. The catalytic domain of Cel3 aligns with the deduced amino acid sequences of other plant EGase mature proteins and possesses all the amino acid domains characteristic of plant EGases. The Cel3 catalytic domain contains seven potential sites for N-glycosylation, and the mature protein may thus be highly glycosylated.
Cel3 antiserum was raised to the C-terminal half of the catalytic domain, a hydrophilic region that lacks most of the amino acid domains conserved between plant EGases. Affinity-purified Cel3 antibody detected a 53-kDa protein in soluble and cell wall extracts, and reacted strongly with 93-, 88-, and 53-kDa microsomal membrane proteins. An 82-kDa protein detected in cell wall extracts is consistent in size with the 93-kDa protein lacking the N-terminal-charged and membrane-spanning domains and thus becoming released from the plasma membrane to the cell wall. In density gradients the 53-kDa protein comigrated with plasma membrane markers, the 88-kDa protein comigrated with Golgi membrane markers, and the 93-kDa protein cofractionated with both plasma membrane and Golgi markers. CM-cellulase activity correlated with the distribution of the 93-kDa protein or with the 88- and 53-kDa proteins together rather than with either the 88- or 53-kDa proteins alone, suggesting that each of the immunodetected proteins is likely to be enzymically active.
In mammalian cells, Golgi membrane-bound glycosyltransferases are also type II integral membrane proteins, and the signal for the retention of these enzymes on Golgi membranes appears to reside in the hydrophobic membrane-spanning domain (16). Progressively increasing the length of this domain allows increasing transport of these enzymes from the Golgi to the plasma membrane (17). In tomato the 88-kDa protein is retained on Golgi membranes, whereas the 93-kDa protein appears to be inefficiently retained and is also detected on plasma membranes. The predicted size of the Cel3-encoded polypeptide is 68.5 kDa, but could be substantially larger if all seven potential N-glycosylation sites are utilized.
Plant EGases participate in several physiological functions. Increased EGase expression has been observed during organ abscission (8, 18), fruit ripening (8, 19), and the growth of vegetative tissues (20, 21). Ethylene increases the mRNA accumulation of EGases expressed during organ abscission and fruit ripening, while auxin promotes the mRNA accumulation of EGases expressed during growth of vegetative tissues. Cel3 mRNA was detected at high levels in vegetative tissue, but its mRNA abundance was not increased by treatment with auxin, ethylene, or gibberellin. In vegetative tissues the highest levels of Cel3 mRNA were found in the most actively growing cells, in the expanding zones of young hypocotyls, and in young expanding leaves. As tissues became mature and cell expansion declined, the abundance of Cel3 mRNA also declined. Thus, Cel3 expression is ubiquitous, but higher levels of expression are correlated with regions of relatively rapid growth.
Membrane-associated EGase enzyme activity has been previously reported in plants. In bean plant tissues, including abscission zones, stems, petioles, leaves, and roots, a proportion of the total CM-cellulase activity was associated with membranes and was activated by Triton X-100 (5). Sedimentation of microsomal membranes in sucrose density gradients found that the bulk of this activity migrated with K+-stimulated ATPase activity, suggesting a plasma membrane localization (3, 5). In pea hypocotyl membranes separated by density gradient centrifugation, a low level of EGase activity was detected coincident with markers for Golgi membranes (22). In bean abscission zones the membrane-associated EGase was identified as a glycoprotein with a pI of 4.5, and its activity declined substantially during ethylene-promoted abscission (4). Cel3 mRNA abundance also declined substantially during abscission and may correspond to the membrane-associated EGase previously described in bean abscission zones.
Only one other membrane-anchored EGase has been previously described. In Agrobacterium tumefaciens a gene encoding a membrane-anchored EGase (celC) is part of the cellulose synthesis operon, and its disruption by transposon insertion showed that celC is required for cellulose synthesis (23). Although the function of the celC membrane-anchored EGase is not known, it was proposed to participate in the transfer of lipid-linked glucan oligosaccharides to cellulose chains (24). Similarly, in the cellulose-synthesizing bacterium Acetobacter xylinum, a gene encoding an EGase is part of the operon required for cellulose synthesis, but in this case the EGase appears to be secreted rather than membrane-anchored (25). Cellulose synthesis in plants occurs on the plasma membrane but probably does not involve lipid-linked intermediates as has been proposed in Agrobacterium (26). Nevertheless, in pea epicotyl slices, treatment with purified pea EGase increased incorporation of labeled substrate into newly synthesized cellulose, perhaps by creating primer ends for glucosyl transfer (27). Similarly, in the synthesis of xyloglucan in the Golgi apparatus, the growing glucosyl chains are thought to be elongated from primers (28). Interestingly, glucan synthase activities are found to be associated with both plasma membranes and Golgi membranes (29), and CM-cellulase activity and proteins reacting with Cel3 antibody were found on both of these membranes. The role of the Cel3 gene product in glucan synthesis, transglucosylation, or cell wall assembly or loosening remains to be determined. However, attempts to suppress Cel3 gene expression by expression of a constitutive antisense Cel3 transgene have been unsuccessful, suggesting that the gene product may be vital for normal plant growth.
Acknowledgments
We thank Drs. David J. Meyer, Alex A. Grantz, and Nuria Ferrol for helpful advice; Carmen Gonzalez-Bosch for making a plasmid template for RPAs; and David J. Meyer and Ramon Serrano for the antibodies to BiP and plasma membrane ATPase. This work was supported in part by grants from Unilever/Van den Bergh Foods and U.S. Department of Agriculture–National Research Initiative Competitive Grants Program Grant 91-37304-6508.
ABBREVIATIONS
- EGase
endo-1,4-β-d-glucanase
- ER
endoplasmic reticulum
- IAA
indoleacetic acid
Footnotes
References
- 1.Beguin P. Annu Rev Microbiol. 1990;44:219–248. doi: 10.1146/annurev.mi.44.100190.001251. [DOI] [PubMed] [Google Scholar]
- 2.Brummell D A, Lashbrook C C, Bennett A B. Am Chem Soc Symp Ser. 1994;566:100–129. [Google Scholar]
- 3.Koehler D E, Leonard R T, Vanderwoude W J, Linkins A E, Lewis L N. Plant Physiol. 1976;58:324–330. doi: 10.1104/pp.58.3.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.del Campillo E, Durbin M, Lewis L N. Plant Physiol. 1988;88:904–909. doi: 10.1104/pp.88.3.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis L N, Koehler D E. Planta. 1979;146:1–5. doi: 10.1007/BF00381248. [DOI] [PubMed] [Google Scholar]
- 6.Wong Y-S, Fincher G B, Maclachlan G A. J Biol Chem. 1977;252:1402–1407. [PubMed] [Google Scholar]
- 7.Hayashi T, Wong Y-S, Maclachlan G. Plant Physiol. 1984;75:605–610. doi: 10.1104/pp.75.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lashbrook C C, Gonzalez-Bosch C, Bennett A B. Plant Cell. 1994;6:1485–1493. doi: 10.1105/tpc.6.10.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milligan S B, Gasser C S. Plant Mol Biol. 1995;28:691–711. doi: 10.1007/BF00021194. [DOI] [PubMed] [Google Scholar]
- 10.del Campillo E, Bennett A B. Plant Physiol. 1996;111:813–820. doi: 10.1104/pp.111.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brummell D A, Bird C R, Schuch W, Bennett A B. Plant Mol Biol. 1997;33:87–95. doi: 10.1023/a:1005733213856. [DOI] [PubMed] [Google Scholar]
- 12.Frohman, M. A. (1994) PCR Methods Appl. 4, Suppl., S40–S58. [DOI] [PubMed]
- 13.Ferrol N, Bennett A B. Plant Cell. 1996;8:1159–1169. doi: 10.1105/tpc.8.7.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyd D, Beckwith J. Cell. 1990;62:1031–1033. doi: 10.1016/0092-8674(90)90378-r. [DOI] [PubMed] [Google Scholar]
- 15.von Heijne G. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 16.Nilsson T, Warren G. Curr Opin Cell Biol. 1994;6:517–521. doi: 10.1016/0955-0674(94)90070-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munro S. EMBO J. 1995;14:4695–4704. doi: 10.1002/j.1460-2075.1995.tb00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tucker M L, Sexton R, del Campillo E, Lewis L N. Plant Physiol. 1988;88:1257–1262. doi: 10.1104/pp.88.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christoffersen R E, Tucker M L, Laties G G. Plant Mol Biol. 1984;3:385–391. doi: 10.1007/BF00033386. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura S, Mori H, Sakai F, Hayashi T. Plant Cell Physiol. 1995;36:1229–1235. [PubMed] [Google Scholar]
- 21.Wu S-C, Blumer J M, Darvill A G, Albersheim P. Plant Physiol. 1996;110:163–170. doi: 10.1104/pp.110.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maclachlan G A. Appl Polym Symp. 1976;28:645–658. [Google Scholar]
- 23.Matthysse A G, White S, Lightfoot R. J Bacteriol. 1995;177:1069–1075. doi: 10.1128/jb.177.4.1069-1075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matthysse A G, Thomas D L, White A R. J Bacteriol. 1995;177:1076–1081. doi: 10.1128/jb.177.4.1076-1081.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Standal R, Iversen T-G, Coucheron D H, Fjaervik E, Blatny J M, Valla S. J Bacteriol. 1994;176:665–672. doi: 10.1128/jb.176.3.665-672.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delmer D P, Amor Y. Plant Cell. 1995;7:987–1000. doi: 10.1105/tpc.7.7.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong Y-S, Fincher G B, Maclachlan G A. Science. 1977;195:679–681. doi: 10.1126/science.195.4279.679. [DOI] [PubMed] [Google Scholar]
- 28.Gordon R, Maclachlan G. Plant Physiol. 1989;91:373–378. doi: 10.1104/pp.91.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibeaut D M, Carpita N C. FASEB J. 1994;8:904–915. doi: 10.1096/fasebj.8.12.8088456. [DOI] [PubMed] [Google Scholar]