Abstract
Aim
To determine the rate of furazolidone resistance of Helicobacter pylori (H. pylori) isolated from gastric biopsy specimens and to explore the relationship between genetic mutations in porD and oorD genes of H. pylori and its resistance to the antibiotic.
Methods
Gastric biopsy was performed in 83 adult patients aged 31-77 years with gastric complaints. H. pylori was isolated from biopsy specimens of 46 patients. E-test and 2-fold agar dilution method were used to determine the rate of H. pylori resistance to furazolidone. The genes porD and oorD from susceptible and resistant isolates were amplified by polymerase chain reaction (PCR), and their PCR products were sequenced.
Results
Resistance to furazolidone was found in 8.7% of H. pylori isolates and 6 mutations were detected in porD and oorD genes of the resistant isolates. Three mutations – G353A, A356G, and C357T – occurred in porD and the other mutations – A041G, A122G, C349A(G) – occurred in oorD genes.
Conclusions
Changes in 6 amino acids may be associated with the resistance of H. pylori to furazolidone.
Helicobacter pylori (H. pylori), a microaerophilic, gram-negative, spiral-shaped bacterium, infects the stomach of more than half of the human population worldwide (1). The prevalence of H. pylori infection greatly varies (2). Eighty percent of middle-aged adults are infected with H. pylori in developing countries, but only 20%-50% in developed countries (3). The organism has been accepted as the etiological agent of many digestive diseases (4). Clinical experience has demonstrated that H. pylori infection is not easy to cure. The primary obstacles to successful treatment are lack of patient compliance with the drug regimens and the development of antibiotic resistance (5).
Metronidazole, a prodrug that can sterilize the gastric epithelium by inducing H. pylori DNA strand breakage, helix destabilization, and unwinding (6), remains a major component of new triple and quadruple therapies for successful treatment for H. pylori infection (7). Monotherapy with metronidazole results in the development of metronidazole resistance, found in more than 50% of H. pylori isolates. Furthermore, metronidazole is also highly mutagenic, and its use may result in the generation of resistance to other clinically used antibiotics, such as clarithromycin (8).
The other prodrug, furazolidone, used for the treatment of H. pylori infection (9), can also induce the resistance in H. pylori but the mechanism of this resistance is still not clear. Since bioactivity of furazolidone is similar to that of metronidazole, Kwon et al (10) presumed that por and oor genetic mutations of H. pylori were involved in furazolidone resistance.
Our aim was to determine the resistance rate of H.pylori to furazolidone in isolates from gastric biopsy specimens taken from the patients from Zhenjiang, Jiangsu province of China, and establish the relationship between possible mutations in H. pylori porD and oorD genes and resistance to furazolidone.
Patients and methods
Patients
A total of 83 patients (48 men and 35 women) aged between 31 and 77 years, who lived in Zhenjiang, were enrolled in the study because of gastric complaints. All patients underwent upper gastrointestinal endoscopy and gastric biopsy. Three biopsy specimens were taken from the antrum of the gastric body or the duodenal mucosa and processed within 3 hours. Two of the samples were used for the rapid detection of urease activity and pathologic diagnosis, and another one was used for biopsy culture. Most patients (n = 74) were diagnosed with gastric ulcer, 8 had gastritis, and one patient was diagnosed with gastric cancer. Of 83 patients, 68 had previously received eradication therapy with metronidazole and clarithromycin.
Biopsy culture
Gastric biopsy samples including NCTC11637 were routinely cultured on Columbia blood agar plates (Oxoid Ltd, London, England) containing 5% fresh sheep blood under microaerobic conditions at 37° for 3 days. Strains were identified according to colony morphology, Gram stain, urease, catalase, oxidase, and biochemical properties (11).
Minimal inhibitory concentration of furazolidone
The minimal inhibitory concentration (MIC) for isolated H. pylori strains was defined as the concentration of a drug needed to inhibit 50% of H. pylori growth and determined by 2-fold agar dilution and E-test. Agar dilution plates were prepared with Jeller-Hinton (MH) agar as the base medium. Sheep blood was added to the MH base medium at a concentration of 5%. Furazolidone solution was prepared in sterile distilled water and was added to 5% sheep blood-MH base medium to achieve serial concentration of 0.5-3.0 µg/mL of furazolidone per mL (12). Fresh H. pylori isolates (3 days culture) were prepared in saline to an optical density. With a Steers-type replicating device, 1-2 mL of the adjusted inoculum was delivered to the agar plates. All plates were incubated under microaerobic conditions for 3 days. The MIC was defined as the lowest concentration of furazolidone that completely inhibited the growth of the inoculum (13).
Isolation of DNA
H. pylori genomic DNAs were isolated as described previously by Majewski and Goodwin (14). In brief, H. pylori was diluted in phosphate-buffered saline (PBS), vortexed, and centrifuged at 13000 × g for 5 minutes. The supernatant was discarded, and the pellet was resuspended in 100 μL of Tris-EDTA (EDTA) buffer (pH 8.0, 1 mmol/L Tris-HCl, and 0.1 mmol/L EDTA), 500 μL of guanidine thiocyanate, 0.5 mol/L EDTA, pH 8.0, and 10% sodium lauryl sulfate (GES). The suspension was incubated for 5 minutes at room temperature, then placed on ice for 2 minutes. The mixture was added into 250 μL of ammonium acetate (7.5 mol/L) and placed on ice for 5 minutes. Then, 650 μL of chloroform solution was added and the mixture vortexed. After centrifugation at 13 000 × g for 5-minute, the supernatant was transferred into a new tube (calculate volume), diluted in 0.54 × dimethyl-carbinol, vortexed, and left at room temperature for 5 minutes. Centrifugation at 13 000 × g for 5 minutes was repeated and the supernatant discarded. The pellet was washed twice with 500 mL of 70% alcohol. The DNA samples were stored at 4°C in 100-200 μL of Tris-EDTA buffer until use.
Polymerase chain reaction amplification of porD and oorD
The porD and oorD genes were amplified by polymerase chain reaction (PCR) in a DNA thermal cycler. The amplified fragments of porD and oorD had 592 bp and 458 bp, respectively (Table 1). The PCR conditions consisted of 1 cycle at 95°C for 5 minutes followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 48°C (porD) or 44°C (oorD) for 45 s, and extension at 72°C for 1 minute. The final step was extension at 72°C for 10 minutes. The PCR products were purified and sequenced (Shanghai GeneCore BioTechnologies Co. Ltd, Shanghai, China). The sequences were analyzed with the Blast engine for the alignment of two given sequences (http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi) and ExPASy for the translation of a nucleotide (DNA/RNA) sequence to a protein sequence (http://au.expasy.org/tools/dna.html).
Table 1.
Polymerase chain reaction (PCR) primers used to amplify portions of genes of H. pylori isolated from gastric biopsy specimens from 46 patients
| Primer pair | Encoded protein (gene) | Nucleotide sequences | PCR fragment size (bp) |
|---|---|---|---|
| POR-A | Pyruvate oxidoreductase (porD) | 5′-GCAAGAAGTCATTGACGC-3′ | 592 |
| POR-B | 5′-GGGGTGATAGGATAGGCT-3′ | ||
| OOR-A | 2-oxoglutarate oxidoreductase (oorD) | 5-TTTAGCACAAAGGAGAATG-3′ | 458 |
| OOR-B | 5′-AACTTGGCGTAATAGGAT-3′ |
Results
Antimicrobial susceptibility
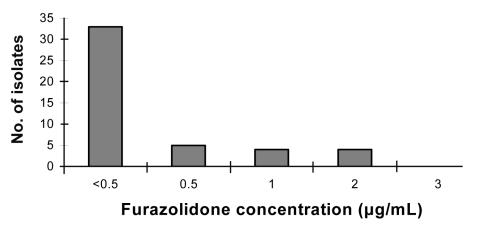
H. pylori was isolated from gastric biopsy specimens from 46 patients of 83 included in the study. Of 46 H. pylori isolates, 42 were susceptible to furazolidone at MIC <2 µg/mL, whereas the remaining 4 were resistant to the drug at MIC >2 µg/mL. Furazolidone resistance was found in patients who had been treated previously with metronidazole. The similar results were found by using E-test. In addition, we found the resistance of 4 isolates at a low level of furazolidone (2-3 µg/mL) (Figure 1).
Figure 1.
Minimum inhibitory concentration of furazolidone for Helicobacter pylori isolates from gastric biopsy specimens obtained from 46 patients.
Amplicons and sequence analysis
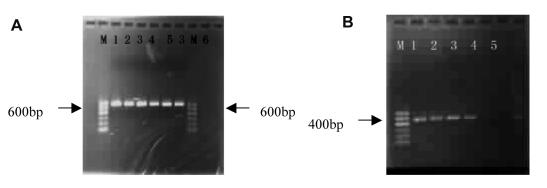
The porD and oorD amplicons had 592 bp and 458 bp, respectively (Figure 2). By sequencing porD and oorD, we detected 6 amino acid changes in resistant isolates. Among these mutations, three (G353A, A356G, and C357T) occurred in porD gene and these mutations led to three amino acid changes; Glu→ Asp, Ala→Thr, and Thr→Val, respectively (Figure 3). The other three – A041G, A122G, and C349A(G) – were found in oorD gene, causing three amino acid substitutions: Thr→Ala, Ile→Val and Asn→Lys, respectively (Figure 4). In addition, a synonymous substitution C357G at position 357 in porD was detected in intermediate susceptible strains to furazolidone (MIC = 1-2 µg/mL) (Figure 3). This substitution might suggest a transitional form of H. pylori from a susceptive one to that resistant to furazolidone.
Figure 2.
Agarose gel electropherogram of porD (A) and oorD (B). A. Markers (from the left): NCTC11637, ZJ0021, ZJ0023, ZJ0027, ZJ0024, ZJ0021, and negative control. B. Markers (from the left): NCTC11637, ZJ0021, ZJ0024, ZJ0003, and negative control.
Figure 3.
Analysis of porD mutations from furazolidone-resistant strains of Helicobacter pylori showed that 353 glutamate was replaced by aspartate, 356 alanine by threonine, and 357 threonine by valine.
Figure 4.
Analysis of oorD mutations from furazolidone-resistant strains of Helicobacter pylori showed that 041 threonine was replaced by alanine, 122 isoleucine by valine, and 349 asparagine by lysine.
Discussion
Our results show that the mutations of porD and oorD in H. pylori may be associated with resistance to furazolidone. The physiology and metabolism of H. pylori are not well understood (15). H. pylori has enzymes of the Entner-Doudoroff and pentose-phosphate pathways (16). However, the major routes for the generation of acetyl coenzyme A (acetyl-CoA) and succinyl-CoA are via pyruvate:flavodoxin oxidoreductase (POR) and 2-oxoglutarate:acceptor oxidoreductase (OOR), respectively. These two electron acceptors may be ferredoxin (Fd) or flavodoxin (FldA). Both porD and oorD encode integral ferredoxin-like subunits (17). Furazolidone is a nitrofuran antibiotic. For nitrofurans, the spectrum of bioactivity and the mechanism of activation mainly depend on the redox potential of the 5-nitro groups. Furazolidone shows relatively high redox potentials (-250 to -270 mA) and is reductively activated by a wide range of nitroreductases (18). Putative H. pylori nitroreductases include FdxB, FrxA, RdxA, OorD, and PorD. Kwon DH et al (19) found that the inactivation of fdxB, frxA, and rdxA genes did not cause a lethal effect on H. pylori, but that both porD and oorD seem to be necessary for survival of H. pylori. They also discovered that RdxA, FrxA, and FdxB could not deoxidize furazolidone. The rdxA, frxA, and fdxB genes, which were knocked out, did not result in the resistance of H. pylori to furazolidone (10). Therefore, Kwon et al (19) suggested that por and oor genes might be crucial for the resistance of H. pylori to furazolidone.
The extensive use of furazolidone results in increasing resistance rate of H. pylori. In addition, the resistance rate seems to vary from one geographic region to another (20). For example, Shan et al (21) reported H. pylori resistance rate to furazolidone of up to 40% in Shanghai, in Southeastern China. On the other hand, Liu et al (22) reported that all H. pylori isolates were susceptible to furazolidone in Shijiazhuang city, in Northern China. Our results showed that H. pylori resistance rate was 8.7% in Zhenjiang, which is located in the south of Yangtze River, supporting the previous observation of geographic variations in H. pylori resistance rate. The major reasons for this difference may be due to different lifestyle in different regions. For example, people living in southeast China eat lot of fish. Since furazolidone is often used as an antiseptic in fishponds, it can lead to the accumulation of the drug in fish and increased exposure of the population living in that part of China, leading to the observed high H. pylori resistance rate to furazolidone.
Sisson et al (23) discovered that partially inhibitory (near MIC) concentrations of furazolidone were somewhat mutagenic for H. pylori, suggesting that the porD and oorD mutations of resistant and intermediately susceptible strains might have relations with furazolidone bioactivity. In the present study, we found that both porD and oorD of resistant strains had three mutations each. These mutations in both genes may contribute to the resistance of H. pylori to furazolidone, which is a findind consistent with observations by Kwan et al (19).
In conclusion, we found a relatively high rate of H. pylori resistance to antibiotics in in Zhenjiang, China. This finding is in accordance with previous observations that H. pylori resistance significantly varies with geographic region in China. Furthermore, our results showed the association between mutations in porD and oorD genes of H. pylori and its resistance to furazolidone. These findings may provide useful information to a clinician administering antibiotics for the treatment of H. pylori infection. However, further research is needed to understand the exact resistance mechanism of H. pylori to furazolidone.
Acknowledgment
We thank Dr Huang Xinxiang for his technical support and Dr Wang Shengjun for helpful suggestions and technical assistance on determination of MICs. This work was supported by Grants BS2004021 and 333 project from Jiangsu province.
References
- 1.van Amsterdam K, van Vliet AH, Kusters JG, van der Ende A. Of microbe and man: determinants of Helicobacter pylori-related diseases. FEMS Microbiol Rev. 2006;30:131–56. doi: 10.1111/j.1574-6976.2005.00006.x. [DOI] [PubMed] [Google Scholar]
- 2.Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347:1175–86. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- 3.Thomas JE, Dale A, Harding M, Coward WA, Cole TJ, Weaver LT. Helicobacter pylori colonization in early life. Pediatr Res. 1999;45:218–23. doi: 10.1203/00006450-199902000-00010. [DOI] [PubMed] [Google Scholar]
- 4.Williamson JS. Helicobacter pylori: current chemotherapy and new targets for drug design. Curr Pharm Des. 2001;7:355–92. doi: 10.2174/1381612013397979. [DOI] [PubMed] [Google Scholar]
- 5.McLoughlin RM, O'Morain CA, O'Connor HJ. Eradication of Helicobacter pylori: recent advances in treatment. Fundam Clin Pharmacol. 2005;19:421–7. doi: 10.1111/j.1472-8206.2005.00340.x. [DOI] [PubMed] [Google Scholar]
- 6.Leiros HK, Kozielski-Stuhrmann S, Kapp U, Terradot L, Leonard GA, McSweeney SM. Structural basis of 5-nitroimidazole antibiotic resistance: the crystal structure of NimA from Deinococcus radiodurans. J Biol Chem. 2004;279:55840–9. doi: 10.1074/jbc.M408044200. [DOI] [PubMed] [Google Scholar]
- 7.Kim JG. Treatment of Helicobacter pylori infection. Korean J Gastroenterol. 2005;46:172–80. [in Korean]. [PubMed] [Google Scholar]
- 8.Sisson G, Jeong JY, Goodwin A, Bryden L, Rossler N, Lim-Morrison S, et al. Metronidazole activation is mutagenic and causes DNA fragmentation in Helicobacter pylori and in Escherichia coli containing a cloned H. pylori RdxA(+) (Nitroreductase) gene. J Bacteriol. 2000;182:5091–6. doi: 10.1128/jb.182.18.5091-5096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisig JN, Silva FM, Rodriguez TN, Hashimoto CL, Barbuti RC. A furazolidone-based quadruple therapy for Helicobacter pylori retreatment in patients with peptic ulcer disease. Clinics. 2005;60:485–8. doi: 10.1590/s1807-59322005000600010. [DOI] [PubMed] [Google Scholar]
- 10.Kwon DH, Lee M, Kim JJ, Kim JG, El-Zaatari FA, Osato MS, et al. Furazolidone- and nitrofurantoin-resistant Helicobacter pylori: prevalence and role of genes involved in metronidazole resistance. Antimicrob Agents Chemother. 2001;45:306–8. doi: 10.1128/AAC.45.1.306-308.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guttner Y, Windsor HM, Viiala CH, Dusci L, Marshall BJ. Nitazoxanide in treatment of Helicobacter pylori: a clinical and in vitro study. Antimicrob Agents Chemother. 2003;47:3780–3. doi: 10.1128/AAC.47.12.3780-3783.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai G, Cheng N, Dong L, Muramatsu M, Xiao S, Wang MW, et al. Bactericidal and morphological effects of NE-2001, a novel synthetic agent directed against Helicobacter pylori. Antimicrob Agents Chemother. 2005;49:3468–73. doi: 10.1128/AAC.49.8.3468-3473.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standard. Performance standards for antimicrobial disk susceptibility tests. Approved standard M2-A7. Wayne (PA): National Committee for Clinical Laboratory Standards; 2000. [Google Scholar]
- 14.Majewski SI, Goodwin CS. Restriction endonuclease analysis of the genome of Campylobacter pylori with a rapid extraction method: evidence for considerable variation. J Infect Dis. 1988;157:465–71. doi: 10.1093/infdis/157.3.465. [DOI] [PubMed] [Google Scholar]
- 15.Srinivasan V, Morowitz HJ. Ancient genes in contemporary persistent microbial pathogens. Biol Bull. 2006;210:1–9. doi: 10.2307/4134531. [DOI] [PubMed] [Google Scholar]
- 16.Wanken AE, Conway T, Eaton KA. The Entner-Doudoroff pathway has little effect on Helicobacter pylori colonization of mice. Infect Immun. 2003;71:2920–3. doi: 10.1128/IAI.71.5.2920-2923.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughes NJ, Clayton CL, Chalk PA, Kelly DJ. Helicobacter pylori porCDAB and oorDABC genes encode distinct pyruvate: flavodoxin and 2-oxoglutarate: acceptor oxidoreductases which mediate electron transport to NADP. J Bacteriol. 1998;180:1119–28. doi: 10.1128/jb.180.5.1119-1128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nivinskas H, Koder RL, Anusevicius Z, Sarlauskas J, Miller AF, Cenas N. Quantitative structure-activity relationships in two-electron reduction of nitroaromatic compounds by Enterobacter cloacae NAD(P)H: nitroreductase. Arch Biochem Biophys. 2001;385:170–8. doi: 10.1006/abbi.2000.2127. [DOI] [PubMed] [Google Scholar]
- 19.Kwon DH, El-Zaatari FA, Kato M, Osato MS, Reddy R, Yamaoka Y, et al. Analysis of rdxA and involvement of additional genes encoding NAD(P)H flavin oxidoreductase (FrxA) and ferredoxin-like protein (FdxB) in metronidazole resistance of Helicobacter pylori. Antimicrob Agents Chemother. 2000;44:2133–42. doi: 10.1128/aac.44.8.2133-2142.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMahon BJ, Hennessy TW, Bensler JM, Bruden DL, Parkinson AJ, Morris JM, et al. The relationship among previous antimicrobial use, antimicrobial resistance, and treatment outcomes for Helicobacter pylori infections. Ann Intern Med. 2003;139:463–9. doi: 10.7326/0003-4819-139-6-200309160-00008. [DOI] [PubMed] [Google Scholar]
- 21.Shanyun, Xu Shuchang, Ye Yuankang. Analysis of Helicobacter pylori resistance rate. Journal of Tongji University. 2004;25:31. [in Chinese]. . Medical Science. [Google Scholar]
- 22.Liu Gaifang Xu Huazhou, Zhang Yuzhen, Zhu Xuemei, Wu Jing. Resistance rate of Helicobacter pylori isolates. Chinese Journal of Practical Internal Medicine. 2005;25:130. [in Chinese]. [Google Scholar]
- 23.Sisson G, Goodwin A, Raudonikiene A, Hughes NJ, Mukhopadhyay AK, Berg DE, et al. Enzymes associated with reductive activation and action of nitazoxanide, nitrofurans, and metronidazole in Helicobacter pylori. Antimicrob Agents Chemother. 2002;46:2116–23. doi: 10.1128/AAC.46.7.2116-2123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]