Abstract
Aim
To present a novel strategy for mapping quantitative trait loci (QTL), using human metapopulations. The strategy is based on the expectation that in geographic clusters of small and distinct human isolates, a combination of founder effect and genetic drift can dramatically increase population frequency of rare QTL variants with large effect. In such cases, the distribution of QT measurements in an “affected” isolate is expected to deviate from that observed in neighboring isolates.
Methods
We tested this hypothesis in 9 villages from a larger Croatian isolate resource, where 7 Mendelian disorders have been previously reported. The values of 10 physiological and biochemical QTs were measured in a random sample of 1001 individuals (100 inhabitants of each of 9 villages and 101 immigrant controls).
Results
Significant over- or under- representation of individuals from specific villages in extreme ends of standardized QT measurement distribution was found 10 times more frequently than expected by chance. The large majority of such clusters of individuals with extreme QT values (34/36, 94.4%) originated from the 6 villages with the most pronounced geographic isolation and endogamy.
Conclusion
Early epidemiological assessment supports the feasibility of the proposed strategy. Clusters of individuals with extreme QT values responsible for over-representation of single villages can usually be linked to a larger pedigree and may be useful for further QTL mapping, using linkage analysis.
The common feature of Mendelian diseases is that their characteristic phenotype is caused by a rare mutation in a single gene in the genome. Therefore, the segregation of affected individuals in families follows simple Mendelian predictions (1). The catalogue of known Mendelian diseases is regularly published, with some 8000 diseases or syndromes listed and new ones continually added to this number (2). The last decade saw great successes in identifying genetic variants underlying several thousands of these diseases (3-5). This success was facilitated by the fact that causal genetic mutation is both necessary and sufficient for the development of the disease, which is the key property of Mendelian diseases. This ensures good correlation between disease phenotypes and underlying genotypes (high “penetrance” and “detectance,” ie, the probabilities of observing the disease phenotype given the disease genotype, and vice versa), which is an important requirement for the success of gene mapping using pedigree-based approach (6,7).
Most Mendelian diseases usually present at an early age and with a number of clinically apparent phenotypic changes. Such a spectrum of phenotypes, initially described as a distinct clinical syndrome, reflects the multiple roles the affected gene products have in human development and metabolism. As the human genome harbors some 25 000 predicted genes and an unknown number of conserved functional elements and regulatory regions, perhaps many more than 8000 Mendelian diseases should be expected. Many genes, however, may interact with each other within common biochemical pathways, thus limiting the number of possible phenotypic outcomes of their mutations. However, the diagnosis of Mendelian diseases is typically based on noticing visually apparent disease phenotypes.
These phenotypes all have in common that they represent measurable human biological quantitative traits. Some of them (eg, blood pressure, body mass index, cholesterol levels, and blood glucose) have recently been identified by the World Health Organization as the main contributors to disease burden in developed countries (8). An understanding of their genetic regulation is therefore of great current interest (9,10). The genes underlying human quantitative traits (QT) may actually be easier to detect than those predisposing common complex diseases, as quantitative traits represent just a fraction of the many recognized risk factors underlying common complex diseases of late onset (11). In this paper, we present and test a novel strategy for finding very rare genetic variants with large effect on QTs in human populations, ie, genes underlying "invisible Mendelian diseases." The proposed approach relies on specific population genetic properties of geographically clustered and isolated human populations, often referred to as metapopulations, which allow for increased frequency of large effect genetic variants underlying quantitative trait distributions that would have extremely small frequencies in large outbred populations.
Croatia has 15 Adriatic Sea islands with a population greater than 1000. The villages on the islands have unique population histories and have preserved their isolation from other villages and outside world through many centuries. The history, demography, and genetic structure of these villages have been investigated for more than 50 years. The research, mainly carried out by the Institute for Anthropological Research in Zagreb, Croatia, resulted in over 100 publications in international journals (12-14). On some of the islands, monogenic (Mendelian) diseases and rare genetic variants were found in unexpectedly high frequencies (Table 1) (15-27).
Table 1.
Overview of the evidence of extremely rare mutations present in unusually high frequencies in specific Croatian island isolates
| Type of research | Island | Reference | |
|---|---|---|---|
| Reports on autochthonous Mendelian diseases: | |||
| dwarfism | Krk | 15-17 | |
| albinism | Krk | 17 | |
| progressive spastic quadriplegia | Krk | 17 | |
| familial cognitive dysfunction | Susak | 18,19 | |
| familial congenital hip dislocation | Lastovo | 20 | |
| familial ovarian cancer | Lastovo | 21,22 | |
| keratoderma palmoplantaris transgrediens | Mljet | 23 | |
| Reports of high population frequencies of extremely rare genetic variants: | |||
| deleted/triplicated alpha-globin gene | Silba | 24 | |
| PGM1*W3 phosphoglucomutase-1 variant | Olib | 25 | |
| mtDNA haplogroup F | Hvar | 26 | |
| Y-chromosome haplogroup P* | Hvar | 27 | |
The studies of population genetic structure, along with reports of at least seven autochthonous Mendelian diseases and four highly unusual rare genetic variants, represent strong evidence that each small human isolate may harbor extremely rare variant(s) that were brought to common frequencies by genetic drift. Some of these variants cause the reported Mendelian diseases, especially if causal mutations are recessive and excessively "exposed" by inbreeding, while others may have large effects on quantitative traits. Recent studies in these populations showed a significant positive effect of inbreeding on a quantitative trait (hypertension) and on the prevalence of a number of late-onset complex diseases (28,29), suggestive of the presence of a major class of rare recessive variants underlying those phenotypes in these Croatian isolates.
Population and Methods
Study design
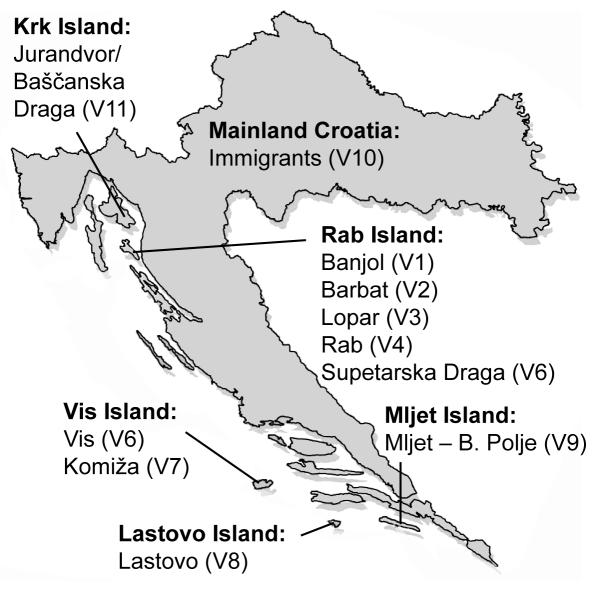
Figure 1 shows the geographic location of the main inhabited islands and the villages chosen for this study.
Figure 1.
Geographic location of the investigated islands of Rab, Vis, Lastovo, and Mljet. Villages from 1 to 9 (V1-V9) are study populations. Immigrants into the islands originate from mainland Croatia (V10).
Assuming a polygenic nature of QTs and exponential distribution of effect size (30,31), the limited number of founders would initially introduce an unknown number of rare mutations (variants specific for their individual genomes) into the gene pool of the isolate they had established. Through such a “founder effect,” the frequency of those variants would thus increase from extremely rare in the general population (eg, f<10-8) to common in that particular isolate (f>10-2). We present a hypothetical case of a geographic cluster of four such isolated populations that share similar environment and lifestyle (eg, villages on an island) (Figure 2). The founder effect would probably not have the power to significantly change the distribution of QT values in any individual village from the one expected in the general population of their origin (Figure 2). However, over the course of time, genetic drift would randomly increase the population frequencies of some of those variants of interest, as long as they do not substantially affect individual fitness. This would provide an opportunity for genetic variants of large effect on QT (eg, blood pressure, cholesterol, and glucose levels) to become very common in particular isolates. They would not be removed from the gene pool by selection, as the resulting chronic late-onset diseases (eg, stroke, coronary heart disease, cancer, or type 2 diabetes mellitus) would develop during the post-reproductive period, although perhaps at an earlier age than expected in the majority of affected cases in the population.
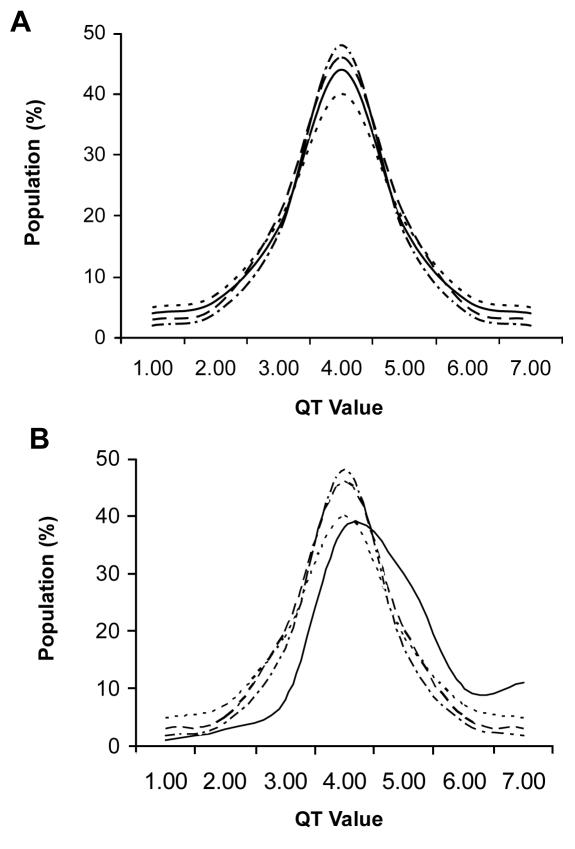
Figure 2.
A hypothetical case of a geographic cluster of four population isolates (A-D), and the distribution of QT measurements in each population (value range 1.00-7.00): (A) a founder effect in isolate A increased the population frequency of a rare variant with large effect on measured quantitative trait from 10-8 in general population to 10-2 in isolate A, but no effect can yet be detected; (B) subsequent genetic drift further increased its frequency over time and the presence of inbreeding exposed the phenotype (recessive case); full line – isolate A; dotted line – isolate B; dashed line – isolate C; dot-dash line – isolate D.
When an introduced variant with large effect on a QT reaches high population frequency due to the action of genetic drift (eg, f>10-1), it should result in a shift of the QT distribution from the values expected in the general population and in surrounding isolate populations.
The theoretical outcome will depend on whether the rare variant enriched in population frequency is dominant or recessive. If recessive, which is considerably more likely for the very rare variants with large deleterious effects, inbreeding practices common to isolate populations due to limited mate choice should increase the frequency of homozygous individuals and expose the phenotypic effect. The latter mechanism has helped to reveal phenotypes and to map genes underlying numerous Mendelian diseases in human isolate populations (32-34). The expected end result is a slight deviation of QT value distribution from the mean, with an additional mode at the extreme end of that distribution representing a cluster of inbred individuals in the isolate population that carry both copies of a recessive variant (Figure 2b). If the mode of inheritance is dominant, however, then the whole upper (or lower) tail of the distribution of the values in this village would be expected to be shifted, which should result in substantial deviation of QT value distribution from the mean, but without an additional mode at the extreme end.
An important consequence of this scenario (Figure 2b), and the basis for the proposed strategy, is the expected excess of persons from the single isolate population (village) within the upper (or lower) tail of the value distribution observed when all the studied isolate populations are pooled together. This should hold if the appropriate standardization of all QT measurements (by age, gender, and other possible covariates) is performed across all studied isolates. The power of such analysis increases with the number of isolate populations in the study. We used this rationale to test statistically whether there is an excess of individuals from specific villages in the extreme ends of the distributions of the 10 selected QTs. We also investigated whether those clusters occur more frequently than expected by chance, and in the villages with the most pronounced geographic isolation and endogamy.
After identifying the individuals within each isolated village who are responsible for the excess in the extremes of the QT distribution of interest, some of them may carry a rare variant with a major effect on QT. To further support that, a reconstruction of their pedigrees should point to their increased relatedness or individual inbreeding. In an ideal case, most of those cases will cluster in families or show increased inbreeding and an apparent Mendelian segregation of the trait. Positional cloning of a responsible variant should then be attempted using linkage analysis, as has been successfully achieved for many Mendelian diseases with clinically apparent phenotypes, with homozygosity mapping being an extremely powerful approach in these cases. It may only take studying several individuals from a single pedigree to map the gene of interest (Table 2).
Table 2.
Outline of the proposed strategy for mapping human quantitative trait loci of large effect using clusters of small human isolate populations
| Step | Procedure |
|---|---|
| Step 1 | Characterizing a resource of several neighboring isolate populations (geographic or cultural) that share similar environment and lifestyle (a “metapopulation” – see Table 1 for examples). |
| Step 2 | Measuring a number of quantitative traits (QT) in all villages in a random sample of inhabitants, using the same standardized methods, observers, equipment, techniques, and laboratory procedures. |
| Step 3 | After data entry, standardizing distributions of QT's by gender and age in each population isolate. Assessing the levels of isolation by defining endogamy in each village as a percentage of examinees’ parents (or grandparents) born in the same village. |
| Step 4 | Testing for the presence of a rare recessive variant with a large effect on QT brought to common frequency in a particular isolate. For each QT, ranking all standardized individual values found in examinees from all isolate populations. In case of a significant clustering of examinees from a single village in the extreme tails of the observed values, proceed to step 5. |
| Step 5 | Investigating whether 10-15 examinees with the most extreme standardized QT value who originate from the same isolate are linked to the same large pedigree and/or are inbred by studying genealogical records. |
| Step 6 | Attempting to repeat QT measurements in a specific isolate population of interest in all available members of such pedigree and to obtain DNA for genetic studies at the same time. This is an attempt to eliminate false-positive "cases" before linkage analysis is performed. |
| Step 7 | Investigating whether the segregation of extreme QT values within families is consistent with Mendelian laws, and establishing with which model of inheritance it is consistent. |
| Step 8 | Performing linkage analysis following the same procedures that proved successful for a large number of rare Mendelian diseases found in isolate populations (for Mal-de-Meleda example from Mljet island see ref. 23); |
| Step 9 | When a genomic region with LOD score (log odds of linkage against no linkage) greater than 3.0 is identified, checking electronically available information for functional candidates and replication of previous reported linkage. |
Sample selection
In 1999, scientists from four research institutions in Zagreb, Croatia, and Edinburgh, Scotland, outlined the strategy for the "1001 Dalmatians" research program. The program aims to study multiple small isolated populations that share a similar environment and lifestyle and are situated on the Adriatic islands, Croatia (14). In 2001, 9 settlements were chosen to present a wide range of differing ethnic histories, fluctuations in population size, admixture and bottleneck events, on the basis of known founding times, accessibility of genealogical records, and population willingness to participate in research program. The geographic location of the nine selected villages is presented in Figure 1, and their population histories in Table 3.
Table 3.
Basic demographic parameters recorded for each studied village
| Village
(V) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Demographic parameter | Banjol (V1) | Barbat (V2) | Lopar (V3) | Rab (V4) | Supetarska Draga (V5) | Vis (V6) | Komiža (V7) | Lastovo (V8) | Mljet (V9) |
| Years since the foundation of the settlement | 1600 | 1450 | 1600 | 3000 | 950 | 3000 | 640 | 1200 | 1200 |
| Number of episodes of major admixture events* | 3 | 2 | 3 | 4 | 1 | 4 | 0 | 0 | 0 |
| Time since the last recorded admixture event (years) | 350 | 350 | 350 | 350 | 350 | 350 | 640 | 1200 | 1200 |
| Maximum achieved population size | 1971 | 1300 | 1500 | 5000 | 1164 | 4300 | 3572 | 1602 | 2106 |
| Year in which maximum size was achieved | 2001 | 1950 | 1400 | 1400 | 2001 | 1910 | 1910 | 1931 | 1948 |
| Last bottleneck event (in years before present)† | 550 | 550 | 550 | 550 | 550 | 25 | 25 | 25 | 25 |
| Percentage of reduction in population size during the last bottleneck event | 60 | 60 | 60 | 95 | 60 | 53 | 44 | 32 | 43 |
| Demographic trend 1 (percentage of pop. in 2001 vs 1750) | 340 | 402 | 657 | 55 | 333 | 127 | 585 | 76 | 101 |
| Demographic trend 2 (percentage of pop. in 2001 vs 1875) | 229 | 280 | 505 | 62 | 162 | 58 | 68 | 83 | 77 |
| Demographic trend 3 (percentage of pop. 2001 vs 1925) | 167 | 110 | 208 | 64 | 116 | 55 | 46 | 58 | 57 |
| Population size (in year 2001) | 1971 | 1205 | 1191 | 554 | 1164 | 1776 | 1523 | 835 | 1111 |
| Percentage of consanguineous population‡ | 7 | 13 | 10 | 4 | 5 | 12 | 27 | 3 | 11 |
| Percentage of grand-parental endogamy§ | 42 | 90 | 98 | 39 | 47 | 88 | 91 | 72 | 94 |
*Events in history when a major influx of immigrants of different genetic background (at least 10% of the population) occurred within a single generation.
†Event in history where a reduction in population size of at least 50% occurred within a single generation.
‡Defined as having parents related to a degree of second cousins or closer.
§Proportion of all grandparents of the examinees who were born in the same village as the examinees.
Fieldwork to collect the data of interest was performed during 2002 and 2003 by a team of researchers from the Zagreb University School of Medicine and the Institute for Anthropological Research in Zagreb, Croatia. In each of the 9 villages, a random sample of 100 adult inhabitants was collected. Sampling was based on computerized randomization of the most complete and accessible population registries in each village, which included medical records (Mljet and Lastovo islands), voting lists (Vis island), or household numbers (Rab island). An additional sample of 101 examinees was recruited from volunteering second-generation immigrants into all 9 villages to form a genetically diverse control population sharing the same environment. Ethical approval was obtained from appropriate research ethics committees in Croatia and Scotland. Informed written consent was obtained from all participants in the study.
Measures of health-related quantitative traits of interest
Blood pressure (mmHg), height (mm), and weight (kg) were measured by a single observer in local health centers and dispensaries between 8 and 11 AM, following standard procedures (35). Blood pressure was measured on the right forearm in a sitting position. Two measurements of both systolic and diastolic blood pressure were taken 5 minutes apart in each individual, the mean value was used for analysis. Height and body mass were measured using the single anthropometer (Hospitalija, Zagreb, Croatia). Biochemical analyses of creatinine, uric acid, high density lipoprotein, low density lipoprotein and total cholesterol, tryglicerides, and blood glucose were done from fasting blood samples taken from the examinees between 7 and 9 AM Blood samples were extracted into 10 mL clotted blood tubes (BD Vacutainer Systems, Franklin Lakes, NJ, USA) for further biochemical analyses. Plasma and serum were rapidly frozen and stored at -20°C in 200 µL aliquots, using standardized sample handling procedures. They were then transported frozen within a maximum of 3 days to the single biochemical laboratory based in Zagreb. The laboratory was chosen because it has been internationally accredited for performing this type of analysis and included in internal quality assessment by Roche and Olympus, and in external monitoring programs by Croatian referent center for biochemical analyses and by an international organization (RIQAS) that performs quality control (36).
Statistical analysis
The aim of statistical analysis was to standardize individual measurements of all 10 measured QTs in all 1001 examinees and make them comparable across the villages. Then, in a merged sample of 1001 examinees, a search for the presence of over- or under-representation of individuals from the single village populations (V1-V10) in the most extreme 10% of values was performed. In such analysis the values of measurements of each QT were ranked from 1 to 1001, and the top and bottom 100 values were investigated for an excess of individuals from the single village. Exactly 10 persons from each village would be expected in top and bottom 10% by chance. Based on a simple χ2 statistics for 10 independent samples with 100 randomly chosen individuals, the presence of 17 or more individuals from the same village in the top 10% values by rank among all 1001 examinees would indicate statistically significant over-representation, while 3 or fewer would indicate statistically significant under-representation of a specific village. Multiple comparisons performed in the same way for 10 different quantitative traits would be expected to result in 2-3 false positive clusters by chance, but we expected to demonstrate that the number of clusters was dramatically greater than expected by chance.
To make this analysis as informative as possible, two different models of standardization of QT measurements were applied. Data on age, gender, height, weight, and village of origin (V1–V10) were analyzed in a logistic regression with QT measurement as outcome (37). Age and gender were forced in all prediction models, irrespective of whether they were formally significant. Model 1 allowed age, gender, height, and weight as predictors, whereas Model 2 allowed only age and gender as predictors.
Results
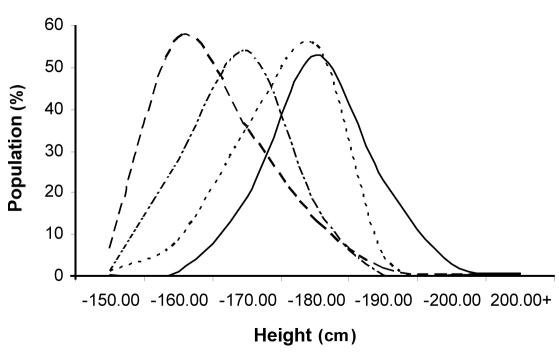
To demonstrate the proposed principle on a clinically apparent QT (in this example, human height) using actual data from this isolate resource, we presented the distributions of male and female population by height in Jurandvor and Baščanska Draga villages on Krk island (Figure 1; V11) and in our sample from 9 other villages (including the V10 with immigrants). We used the example of villages Jurandvor and Baščanska Draga (V11) because a variant in PROP1 gene has recently been identified as a cause of dwarfism, which is prevalent in the villages (16). We presented a reported population distribution by height in 258 examinees from V11 (15) alongside the distribution of 1001 examinees in villages V1-V10 combined (Figure 3). This presentation was made solely to demonstrate how the presence of the causal PROP1 mutation in V11 population could have been identified using the strategy we propose, as the cases affected by dwarfism from V11 would have been in large excess in the lower end of the height distribution of all the 11 villages combined.
Figure 3.
Effect of PROP-1 mutation on height in Jurandvor/Baščanska Draga (V11) population isolate in comparison to other investigated population isolates (V1-V10). Dashed line – Jurandvor/Baščanska Draga (V11), females (n=151); full line – other villages (V1-V10), males (n=455); dotted line – Jurandvor/Baščanska Draga (V11), males (n=107); dot-dash line – other villages (V1-V10), females (n=546).
In Table 4, we present the comparison between the chosen villages for all the investigated quantitative traits. Based on this comparison, we tried to identify over- or under-represented villages in the extremes of the distributions of 10 measured QTs under the two models of measurement standardization. Statistical computation how many of the 10 villages would be expected as positive or negative outliers purely by chance showed that between 2 and 3 such cases would be expected for all 10 traits in Table 4. Under Model 1, the number of clusters is 34, ie, an order of magnitude greater. The exclusion of height and weight as predictors (model 2) changed this result further only marginally, by increasing it to 36 such clusters.
Table 4.
Villages (V1-V10) with unusually large or small numbers of individuals having extreme values of the 10 quantitative traits
| No. of
individuals from the same village in |
||||||||
|---|---|---|---|---|---|---|---|---|
| bottom 10%
of all recorded values* |
top 10% of
all recorded values* |
|||||||
| Parameter† | 0 or 1 (P<0.01) | 2 or 3 (P<0.05) | 17 or 18 (P<0.05) | 19+ (P<0.01) | 0 or 1 (P<0.01) | 2 or 3 (P<0.05) | 17 or 18 (P<0.05) | 19+ (P<0.01) |
| Model 1 (predictors: age, sex, height, weight): | ||||||||
| SBP | V3,V4 | V8 | V3 | |||||
| DBP | V6 | V3,4,7 | V8,V9 | V6 | ||||
| BMI | ||||||||
| log (LDL) | V6,V7 | V2,V8 | V8 | V6,V7 | ||||
| log (triglycerides) | V6 | |||||||
| log (total cholesterol) | V6 | V8 | V8 | V6 | ||||
| log (creatinine) | V6 | V8 | ||||||
| urate | V9 | |||||||
| log (serum glucose) | V8 | V1 | V2 | V6,V8 | ||||
| HDL | V1 | V9 | V8 | |||||
| Model 2 (predictors: age, sex): | ||||||||
| SBP | V3 | V3 | ||||||
| DBP | V3,4,7 | V8,V9 | ||||||
| BMI | ||||||||
| log (LDL) | V6,V7 | V2,V8 | V8 | V6,V7 | ||||
| log (triglycerides) | V8 | V6 | ||||||
| log (total cholesterol) | V6 | V8 | V8 | V7 | V6 | |||
| log (creatinine) | V6,9 | V3 | V8 | V8 | ||||
| urate | V8,V9 | |||||||
| log (serum glucose) | V8 | V3 | V2 | V8 | ||||
| HDL | V1 | V6 | V9 | V8 | ||||
*Ten expected per each village.
†Abbreviations: SBP – systolic blood pressure; DBP – diastolic blood pressure; BMI – body mass index; log – logarithmically transformed variable; LDL – low density lipoprotein; HDL – high density lipoprotein.
However, when the “village of residence” was also included as a predictor in Model 1, along with age, gender, height, and weight, the number of observed clusters decreased from 34 to only 6 (not shown in Table 4). This result pointed to a dramatic "village effect," implying true differences between the individual villages in the distributions of most measured QTs. If the observed "village effect” was mainly environmentally influenced, then we should not expect clusters of individuals with extreme values to originate almost exclusively from the villages with high endogamy, and rarely, or never, from those with low endogamy or immigrant group. However, under the second model (Table 4), 34 of 36 identified clusters (94.4%) originated from the 6 villages with the highest endogamy and long-term isolation (V2-3 and V6-9, Table 3). Moreover, 12 of them (33.3%) were associated with the geographically most isolated village (V8) in which two rare Mendelian disorders have previously been described (Table 1), and a case of ichthyosis recently encountered (I.R., unpublished data). The 4 most geographically isolated villages – Vis, Komiža, Lastovo, and Mljet – together accounted for 27/36 (75%) of the clusters. Also, the group of immigrants from V10, genetically representing outbred mainland population, appeared in none of the extremes, as expected under our hypotheses. This suggests that the "village effect" may largely represent true population genetic differences in frequencies of rare alleles with large effect on the measured QTs.
Discussion
The results of our study strongly supported our hypothesis that distinct isolates within human metapopulations would deviate in their own unique ways from expected genetic frequencies in large outbred populations. The proposed causes of these deviations in Croatian island isolates are founder effect, genetic drift, and subsequent inbreeding, which may jointly act to expose extremely rare genetic variants of very large effect and therefore cause shifts in frequency distributions of affected quantitative traits in comparison to other isolates or to large general population. Although rare in general population, such variants of large effect may still prove helpful in revealing some common and yet unrecognized physiological pathways that could prove important in pathological mechanisms, and therefore point to novel targets for drug development and future therapy. This approach may be feasible, because a number of recent studies in both model organisms and humans have recognized that the genetic architecture underlying biological quantitative traits may be more complex than previously thought and variants of truly substantial effect on their distribution in the population difficult to identify (38-41).
It has already been demonstrated by anthropological research that distinct villages retained isolation and preserved genetic structure through many centuries, revealing a number of extremely rare variants or haplotypes with common frequencies (12-27). This led to the identification of at least 7 Mendelian diseases in specific islands or even villages (Table 1). In 9 villages from the islands of Rab, Vis, Mljet, and Lastovo, we conducted measurements of 10 quantitative traits and demonstrated numerous apparent clusters of individuals with extreme values originating from particular villages. This was done in a random sample of 100 persons from each village, carefully selected from voting lists or medical records, and using unified methods and a single biochemical laboratory of international standard (42).
The “village effect” in this metapopulation of human genetic isolates, which apparently explains highly prevalent clustering of individuals from the specific villages in the extreme tails of the distributions of trait values, probably results from the joint action of factors related to variation in environment, population genetic structure, and data collection between the villages. Since we made strenuous efforts to minimize differences related to data collection and analysis, we need to investigate the possible role of the other two variables.
Another possible confounding effect that could, at least in theory, be responsible for modifying the values of measured traits are the prescribed medications. We obtained the information about the medications taken from each examinee in this study. The level of medication prescription in these isolated communities is rather low, and about 50% of the examinees do not take any medication regularly. Among the prescribed medications, no single product was being taken regularly by more than 20 persons (ie, only 2% of the sample). The most commonly prescribed groups of drugs were painkillers, antihypertensives, antiarrythmics, and anxyolitics. Among the drugs that could significantly affect the results of our study, antihypertensives were being taken by 8-15% of the population in the studied villages, with no statistically significant differences in age-standardized prevalence among the villages. Other drugs, such as statins to reduce serum lipid levels, were only taken by 1% of the examinees in the sample, whereas glucose regulators were being taken regularly by less than 2% of the examinees. Therefore, given the magnitude of the observed village-specific effects on the clustering of the examinees in the tails of value distributions, we believe that the differences in medication prescription could not have possibly been responsible for the observed results.
Based on these results, an early epidemiological assessment, performed in this study, seems to generally be supportive of the feasibility of the proposed strategy. We hope that further analysis of specific clusters of extreme QT values identified in this study will indeed expose at least a few underlying Mendelian variants of large effect influencing measured QTs of interest. If so, this approach could become a feasible supplementary effort to the Hap-Map project, which is designed to find common variants in the human genome associated with common diseases (43).
We emphasize that the aim of this study was only to serve as a “proof of principle” that the proposed strategy could work and have a considerable power to detect rare variants of large effect. Thereby, we were only interested in clustering of the extreme values in measured traits due to homozygosity of rare and recessive genetic variants of large effect. The affected individuals were therefore expected to have values so large (or small) to always find themselves at the extreme ends of the distribution. Because of this, there is no need for complex statistical procedures to spot these individuals – they should either be there clustering together, or not, and their presence should be immediately apparent as they should be from the same village and frequently share the same surname. In our minds, the value of the proposed strategy is the fact that through a careful study design and choice of study populations, this strategy enables a very simple way of “screening” for possible large effects of rare recessive variants on measured QTs in these populations. All the numbers in this study were deliberately chosen to reveal the simplicity of the approach and make it intuitive to a reader: there are 10 study populations, 100 subjects from each population, and 10 measured quantitative traits; after ranking a total of pooled 1001 individuals by QT value, top 10% and bottom 10% of the subjects are examined according to village of residence, and 10 subjects per village are expected to be found there by chance. Any significant deviation in the representation of a single village could imply a cluster of individuals carrying very rare variants of large effect. This should make the strategy easily applicable in similar settings globally, without the need of statistical genetic or bioinformatic support. It should therefore provide a helpful tool for initial research towards gene mapping to the scientists working in the countries with very limited resources for research, while positive outcomes should then hopefully attract further funding. Through analysis of isolates worldwide, we could begin to catalogue very rare variants of large effect, which may eventually provide more promising clues into the pathogenesis of human diseases and reveal new pathogenic mechanisms that could become therapeutic targets in the future.
Acknowledgements
I.R. was supported by Overseas Research Scheme and the Scholarship from the University of Edinburgh. I.R. and Z.B. were supported by the British Scholarship Trust Fellowship. Z.B. was supported by “Miroslav Čačković” fellowship of the Zagreb University School of Medicine. The study was partially supported through the grants from the EU FP6 project )No. 018947), Ministry of Science, Education, and Sports of the Republic of Croatia (No. 0108330) to I.R., and the grants from The British Council, The Wellcome Trust, The Royal Society, and Medical Research Council to H.C. and I.R. The authors collectively thank to a very large number of individuals (medical students of the Zagreb University School of Medicine, Croatia; local general practitioners and nurses in study populations; the employees of several other Croatian institutions, including but not limited to the University of Rijeka and Split; Croatian Institute of Public Health; Institutes of Public Health in Split and Dubrovnik; and the Institute for Anthropological Research in Zagreb) for their individual help in planning and carrying out the field work related to the project. There are no conflicts of interest related to this manuscript.
References
- 1.Hamosh A, Scott AF, Amberger J, Valle D, McKusick VA. Online Mendelian Inheritance in Man (OMIM). Hum Mutat. 2000;15:57–61. doi: 10.1002/(SICI)1098-1004(200001)15:1<57::AID-HUMU12>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 2.Hamosh A, Scott AF, Amberger J, Bocchini C, Valle D, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2002;30:52–5. doi: 10.1093/nar/30.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet. 2003;33(Suppl):228–37. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- 4.Peltonen L, Palotie A, Lange K. Use of population isolates for mapping complex traits. Nat Rev Genet. 2000;1:182–90. doi: 10.1038/35042049. [DOI] [PubMed] [Google Scholar]
- 5.Weatherall DJ. Phenotype-genotype relationships in monogenic disease: lessons from the thalassaemias. Nat Rev Genet. 2001;2:245–55. doi: 10.1038/35066048. [DOI] [PubMed] [Google Scholar]
- 6.Cardon LR, Bell JI. Association study designs for complex diseases. Nat Rev Genet. 2001;2:91–9. doi: 10.1038/35052543. [DOI] [PubMed] [Google Scholar]
- 7.Zondervan KT, Cardon LR. The complex interplay among factors that influence allelic association. Nat Rev Genet. 2004;5:89–100. doi: 10.1038/nrg1270. [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization. World health Report 2002. Geneva: WHO; 2002. [Google Scholar]
- 9.Barton NH, Keightley PD. Understanding quantitative genetic variation. Nat Rev Genet. 2002;3:11–21. doi: 10.1038/nrg700. [DOI] [PubMed] [Google Scholar]
- 10.Abiola O, Angel JM, Avner P, Bachmanov AA, Belknap JK, Bennett B, et al. The nature and identification of quantitative trait loci: a community's view. Nat Rev Genet. 2003;4:911–6. doi: 10.1038/nrg1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiss KM, Terwilliger JD. How many diseases does it take to map a gene with SNPs? Nat Genet. 2000;26:151–7. doi: 10.1038/79866. [DOI] [PubMed] [Google Scholar]
- 12.Rudan P, Simic D, Smolej-Narancic N, Bennett LA, Janicijevic B, Jovanovic V, et al. Isolation by distance in middle Dalmatia-Yugoslavia. Am J Phys Anthropol. 1987;74:417–26. doi: 10.1002/ajpa.1330740313. [DOI] [PubMed] [Google Scholar]
- 13.Waddle DM, Sokal RR, Rudan P. Factors affecting population variation in eastern Adriatic isolates (Croatia). Hum Biol. 1998;70:845–64. [PubMed] [Google Scholar]
- 14.Rudan I, Campbell H, Rudan P. Genetic epidemiological studies of eastern Adriatic island isolates, Croatia: objective and strategies. Coll Antropol. 1999;23:531–46. [PubMed] [Google Scholar]
- 15.Kopajtic B, Dujmovic M, Kolacio Z, Kogoj-Bakic V. Enclaves of hereditary dwarfism on the island of Krk, Croatia. Coll Antropol. 1995;19:365–71. [Google Scholar]
- 16.Krzisnik C, Kolacio Z, Battelino T, Brown M, Parks JS, Laron Z. The “little people” of the island of Krk – revisited. Etiology of hypopituitarism revealed. J Endocr Genet. 1999;1:9–19. [Google Scholar]
- 17.Zergollern L. A follow-up on Hanhart's dwarfs of Krk. Birth Defects Orig Artic Ser. 1971;7:28–32. [PubMed] [Google Scholar]
- 18.Bohacek N. Tristan da Cunha and Susak. Lijec Vjesn. 1964;86:1412–6. [in Serbian]. [PubMed] [Google Scholar]
- 19.Rudan I, Stevanovic R, Vitart V, Vuletic G, Sibbett L, Vuletic S, et al. Lost in transition – the island of Susak (1951-2001). Coll Antropol. 2004;28:403–21. [PubMed] [Google Scholar]
- 20.Maricevic A. Incidence of congenital hip dislocation in Lastovo 1885-1993. Lijec Vjesn. 1995;117:126–9. [in Croatian]. [PubMed] [Google Scholar]
- 21.Rudan I. Ancestral kinship and cancer in Lastovo island, Croatia. Hum Biol. 2001;73:871–84. doi: 10.1353/hub.2001.0090. [DOI] [PubMed] [Google Scholar]
- 22.Rudan I. Inbreeding and cancer incidence in human isolates. Hum Biol. 1999;71:173–87. [PubMed] [Google Scholar]
- 23.Bakija-Konsuo A, Basta-Juzbasic A, Rudan I, Situm M, Nardelli-Kovacic M, Levanat S, et al. Mal de Meleda: genetic haplotype analysis and clinicopathological findings in cases originating from the island of Mljet (Meleda), Croatia. Dermatology. 2002;205:32–9. doi: 10.1159/000063151. [DOI] [PubMed] [Google Scholar]
- 24.Turcinov D, Krishnamoorthy R, Janicijevic B, Markovic I, Mustac M, Lapoumeroulie C, et al. Anthropogenetical analysis of abnormal human alpha-globin gene cluster arrangement on chromosome 16. Coll Antropol. 2000;24:295–301. [PubMed] [Google Scholar]
- 25.Borot N, Arnaud J, Rudan P, Chaventre A, Sevin J. Phosphoglucomutase-1 subtypes in two populations in Adriatic islands: presence of PGM1*W3 (PGM1*7+) allele. Hum Hered. 1991;41:309–15. doi: 10.1159/000154018. [DOI] [PubMed] [Google Scholar]
- 26.Tolk HV, Barac L, Pericic M, Klaric IM, Janicijevic B, Campbell H, et al. The evidence of mtDNA haplogroup F in a European population and its ethnohistoric implications. Eur J Hum Genet. 2001;9:717–23. doi: 10.1038/sj.ejhg.5200709. [DOI] [PubMed] [Google Scholar]
- 27.Barac L, Pericic M, Klaric IM, Rootsi S, Janicijevic B, Kivisild T, et al. Y chromosomal heritage of Croatian population and its island isolates. Eur J Hum Genet. 2003;11:535–42. doi: 10.1038/sj.ejhg.5200992. [DOI] [PubMed] [Google Scholar]
- 28.Rudan I, Smolej-Narancic N, Campbell H, Carothers A, Wright A, Janicijevic B, et al. Inbreeding and the genetic complexity of human hypertension. Genetics. 2003;163:1011–21. doi: 10.1093/genetics/163.3.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rudan I, Rudan D, Campbell H, Carothers A, Wright A, Smolej-Narancic N, et al. Inbreeding and risk of late onset complex disease. J Med Genet. 2003;40:925–32. doi: 10.1136/jmg.40.12.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright A, Charlesworth B, Rudan I, Carothers A, Campbell H. A polygenic basis for late-onset disease. Trends Genet. 2003;19:97–106. doi: 10.1016/s0168-9525(02)00033-1. [DOI] [PubMed] [Google Scholar]
- 31.Falconer DS, Mackay TFC. Introduction to quantitative genetics. 4th ed. Edinburgh: Longman & Harlow; 1996. [Google Scholar]
- 32.Miano MG, Jacobson SG, Carothers A, Hanson I, Teague P, Lovell J, et al. Pitfalls in homozygosity mapping. Am J Hum Genet. 2000;67:1348–51. doi: 10.1016/s0002-9297(07)62966-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Najmabadi H, Neishabury M, Sahebjam F, Kahrizi K, Shafaghati Y, Nikzat N, et al. The Iranian Human Mutation Gene Bank: a data and sample resource for worldwide collaborative genetics research. Hum Mutat. 2003;21:146–50. doi: 10.1002/humu.10164. [DOI] [PubMed] [Google Scholar]
- 34.Medina-Arana V, Barrios Y, Fernandez-Peralta A, Jimenez A, Salido E, Gonzalez F, et al. Tumour spectrum of non-polyposis colorectal cancer (Lynch syndrome) on the island of Tenerife and influence of insularity on the clinical manifestations. Eur J Cancer Prev. 2004;13:27–32. doi: 10.1097/00008469-200402000-00005. [DOI] [PubMed] [Google Scholar]
- 35.Weiner JS, Lourie JA. Human biology – A guide to field methods. Oxford: Blackwell Scientific Publications; 1969. [Google Scholar]
- 36.Riqas-To-MultiQC. User manual. Available from: http://www.multiqc.com/Riqas2MultiQC.pdf Accessed: June 28, 2006.
- 37.Armitage P, Berry G, Matthews JNS. Statistical methods in medical research. Oxford: Blackwell Science; 2002. [Google Scholar]
- 38.Korstanje R, Paigen B. From QTL to gene: the harvest begins. Nat Genet. 2002;31:235–6. doi: 10.1038/ng0702-235. [DOI] [PubMed] [Google Scholar]
- 39.Anholt RR, Dilda CL, Chang S, Fanara JJ, Kulkarni NH, Ganguly I, et al. The genetic architecture of odor-guided behavior in Drosophila: epistasis and the transcriptome. Nat Genet. 2003;35:180–4. doi: 10.1038/ng1240. [DOI] [PubMed] [Google Scholar]
- 40.Dilda CL, Mackay TF. The genetic architecture of Drosophila sensory bristle number. Genetics. 2002;162:1655–74. doi: 10.1093/genetics/162.4.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altshuler D, Clark AG. Genetics. Harvesting medical information from the human family tree. Science. 2005;307:1052–3. doi: 10.1126/science.1109682. [DOI] [PubMed] [Google Scholar]
- 42.Vitart V, Biloglav Z, Hayward C, Janicijevic B, Smolej-Narancic N, Barac L, et al. 3000 years of solitude: extreme differentiation in the island isolates of Dalmatia, Croatia. Eur J Hum Genet. 2006;14:478–87. doi: 10.1038/sj.ejhg.5201589. [DOI] [PubMed] [Google Scholar]
- 43.The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]