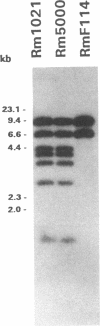
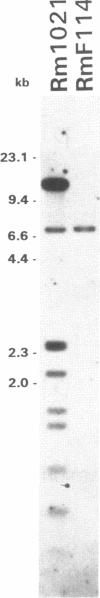
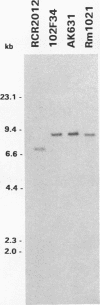
Abstract
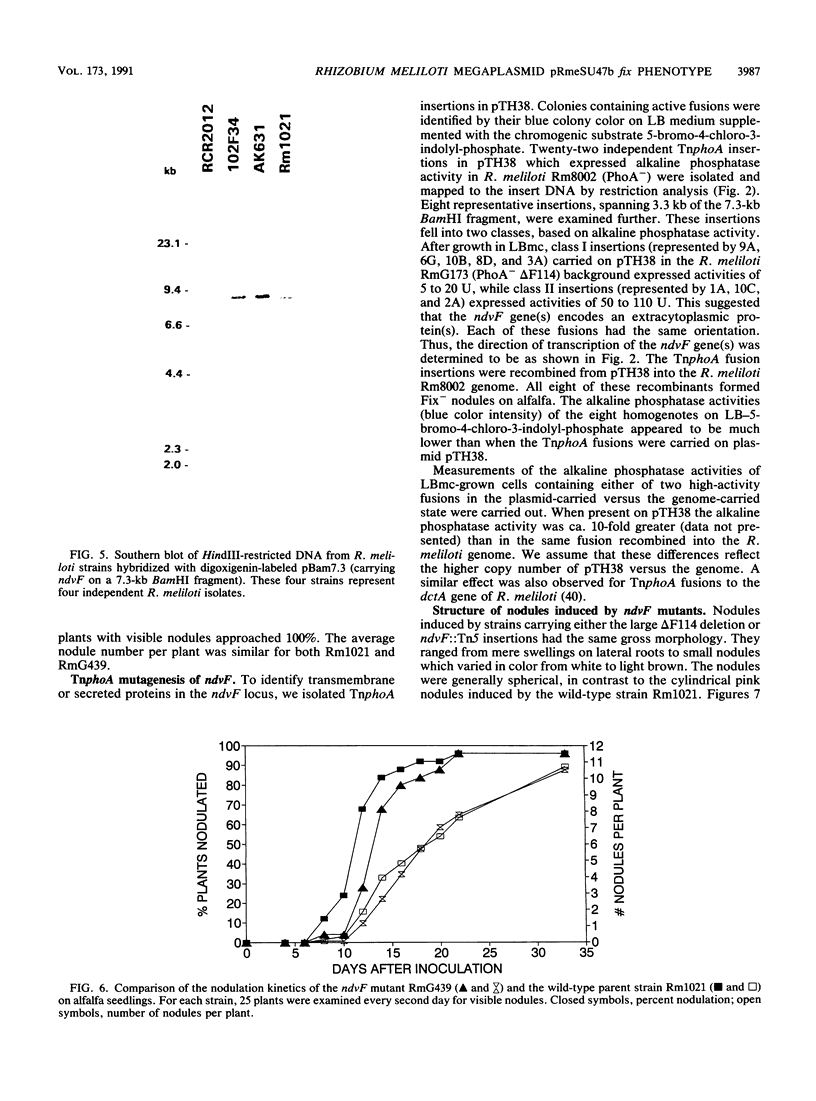
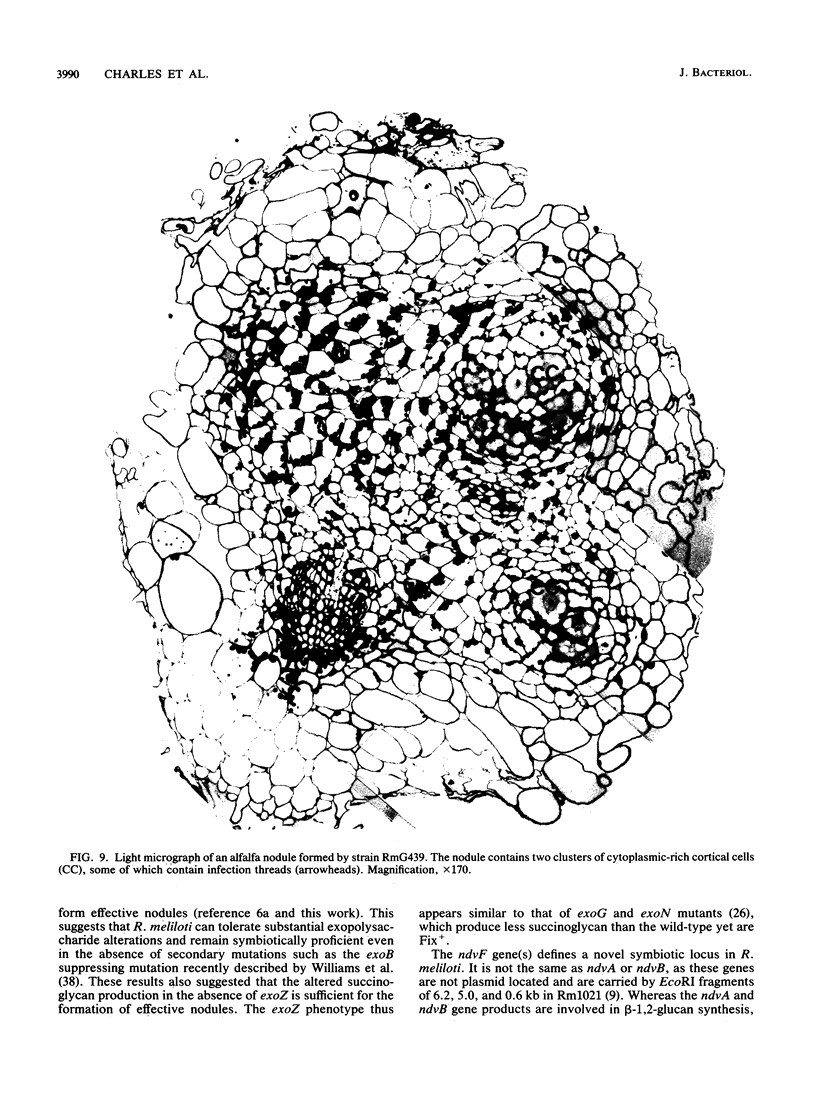
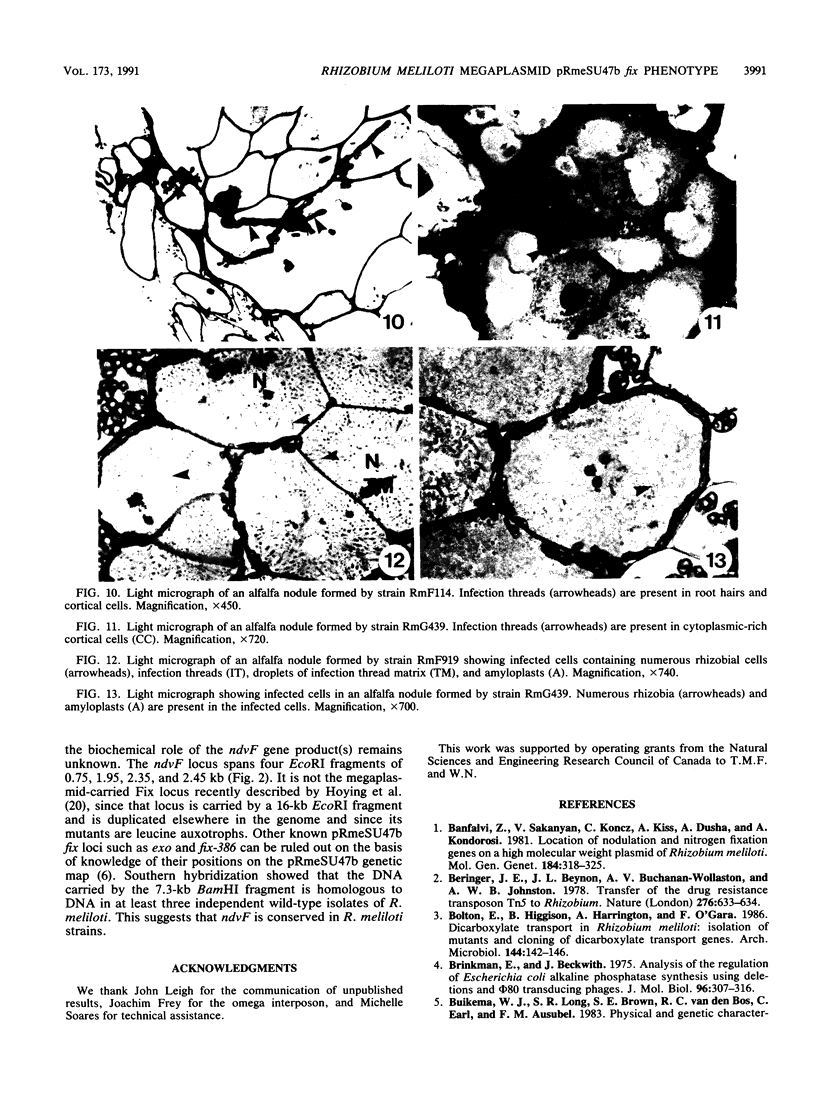
Rhizobium meliloti strains carrying either of two overlapping deletions (delta 5408 and delta F114) of the megaplasmid pRmeSU47b form nodules on alfalfa which fail to fix N2 (Fix-). Strains carrying these deletions also fail to fluoresce on media containing calcofluor, indicating a defect in synthesis of the acidic exopolysaccharide (Exo-) of R. meliloti. We have isolated cosmid clones (pTH21 and pTH22) which complement the Fix- but not the Exo- phenotype of the strains carrying the delta 5408 and delta F114 deletions. In addition, cosmid clones which complement the Exo- phenotype fail to complement the Fix- phenotype of these deletions; thus, the Exo- phenotype is not related to the Fix- phenotype. A 5-kb region within a 7.3-kb BamHI restriction fragment was found to be required for complementation of the Fix- phenotype of the delta 5408 and delta F114 deletion strains. Tn5 insertions in the 5-kb region generated a Fix- phenotype when recombined into the wild-type genome. We have designated this locus ndvF, for nodule development. TnphoA mutagenesis of this region generated active alkaline-phosphatase gene fusions, indicating that ndvF encodes extracytoplasmic protein(s). Induction of nodules by the ndvF mutants was delayed by 2 to 3 days compared with induction by the wild-type strain. Light microscopy of nodules elicited by strains carrying the large 150-kb delta F114 deletion, a 12-kb deletion removing ndvF, or an individual ndvF::Tn5 insertion mutation demonstrated that many nodules contained few infected cortical cells, indicating that nodule development was blocked early in the infection process, before the release of bacteria from the infection threads.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brickman E., Beckwith J. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and phi80 transducing phages. J Mol Biol. 1975 Aug 5;96(2):307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- Bánfalvi Z., Sakanyan V., Koncz C., Kiss A., Dusha I., Kondorosi A. Location of nodulation and nitrogen fixation genes on a high molecular weight plasmid of R. meliloti. Mol Gen Genet. 1981;184(2):318–325. doi: 10.1007/BF00272925. [DOI] [PubMed] [Google Scholar]
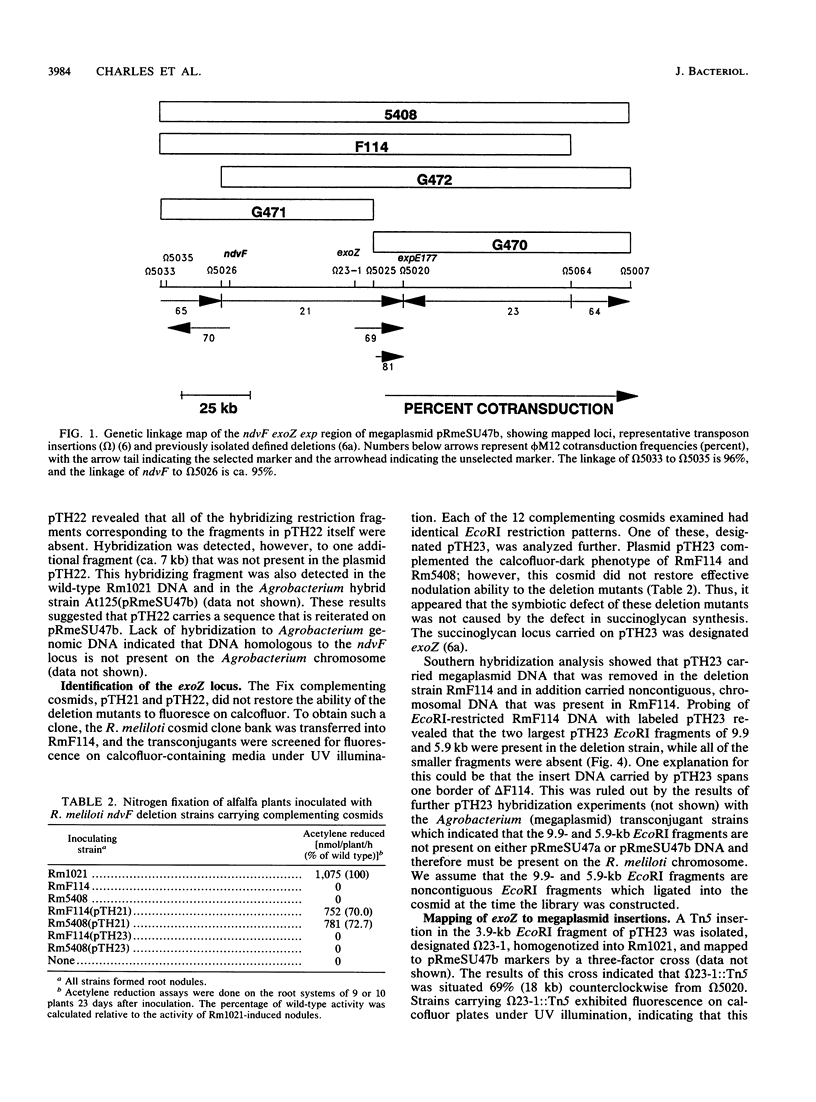
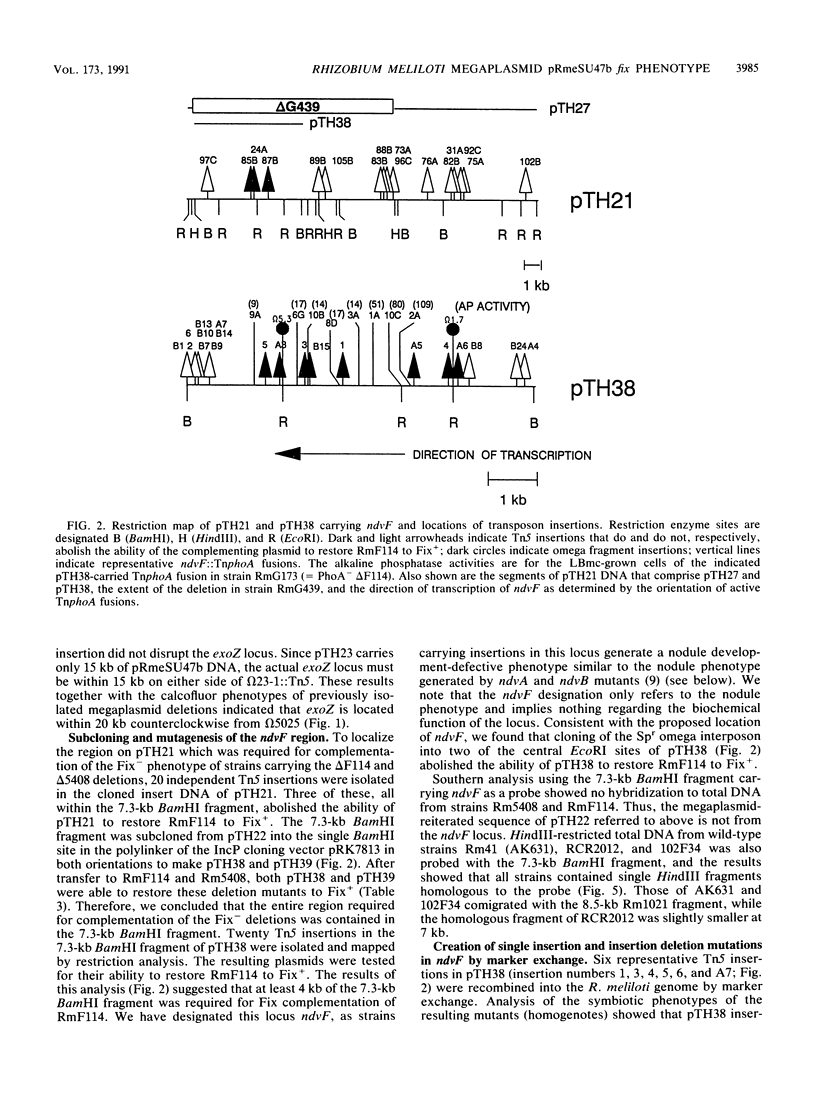
- Charles T. C., Finan T. M. Analysis of a 1600-kilobase Rhizobium meliloti megaplasmid using defined deletions generated in vivo. Genetics. 1991 Jan;127(1):5–20. doi: 10.1093/genetics/127.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles T. C., Finan T. M. Genetic map of Rhizobium meliloti megaplasmid pRmeSU47b. J Bacteriol. 1990 May;172(5):2469–2476. doi: 10.1128/jb.172.5.2469-2476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos G. F., Walker G. C., Signer E. R. Genetic manipulations in Rhizobium meliloti utilizing two new transposon Tn5 derivatives. Mol Gen Genet. 1986 Sep;204(3):485–491. doi: 10.1007/BF00331029. [DOI] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dylan T., Ielpi L., Stanfield S., Kashyap L., Douglas C., Yanofsky M., Nester E., Helinski D. R., Ditta G. Rhizobium meliloti genes required for nodule development are related to chromosomal virulence genes in Agrobacterium tumefaciens. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4403–4407. doi: 10.1073/pnas.83.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan T. M., Hartweig E., LeMieux K., Bergman K., Walker G. C., Signer E. R. General transduction in Rhizobium meliloti. J Bacteriol. 1984 Jul;159(1):120–124. doi: 10.1128/jb.159.1.120-124.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan T. M., Hirsch A. M., Leigh J. A., Johansen E., Kuldau G. A., Deegan S., Walker G. C., Signer E. R. Symbiotic mutants of Rhizobium meliloti that uncouple plant from bacterial differentiation. Cell. 1985 Apr;40(4):869–877. doi: 10.1016/0092-8674(85)90346-0. [DOI] [PubMed] [Google Scholar]
- Finan T. M., Kunkel B., De Vos G. F., Signer E. R. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986 Jul;167(1):66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan T. M., Oresnik I., Bottacin A. Mutants of Rhizobium meliloti defective in succinate metabolism. J Bacteriol. 1988 Aug;170(8):3396–3403. doi: 10.1128/jb.170.8.3396-3403.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrai T., Vincze E., Bánfalvi Z., Kiss G. B., Randhawa G. S., Kondorosi A. Localization of symbiotic mutations in Rhizobium meliloti. J Bacteriol. 1983 Feb;153(2):635–643. doi: 10.1128/jb.153.2.635-643.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey J., Krisch H. M. Omega mutagenesis in gram-negative bacteria: a selectable interposon which is strongly polar in a wide range of bacterial species. Gene. 1985;36(1-2):143–150. doi: 10.1016/0378-1119(85)90078-2. [DOI] [PubMed] [Google Scholar]
- Friedman A. M., Long S. R., Brown S. E., Buikema W. J., Ausubel F. M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982 Jun;18(3):289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- Geremia R. A., Cavaignac S., Zorreguieta A., Toro N., Olivares J., Ugalde R. A. A Rhizobium meliloti mutant that forms ineffective pseudonodules in alfalfa produces exopolysaccharide but fails to form beta-(1----2) glucan. J Bacteriol. 1987 Feb;169(2):880–884. doi: 10.1128/jb.169.2.880-884.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J., Walker G. C. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell. 1989 Feb 24;56(4):661–672. doi: 10.1016/0092-8674(89)90588-6. [DOI] [PubMed] [Google Scholar]
- Hoying J. B., Behm S. M., Lang-Unnasch N. Cloning and characterization of Rhizobium meliloti loci required for symbiotic root nodule invasion. Mol Plant Microbe Interact. 1990 Jan-Feb;3(1):18–27. doi: 10.1094/mpmi-3-018. [DOI] [PubMed] [Google Scholar]
- Jones J. D., Gutterson N. An efficient mobilizable cosmid vector, pRK7813, and its use in a rapid method for marker exchange in Pseudomonas fluorescens strain HV37a. Gene. 1987;61(3):299–306. doi: 10.1016/0378-1119(87)90193-4. [DOI] [PubMed] [Google Scholar]
- Leigh J. A., Signer E. R., Walker G. C. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6231–6235. doi: 10.1073/pnas.82.18.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge P., Roche P., Faucher C., Maillet F., Truchet G., Promé J. C., Dénarié J. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature. 1990 Apr 19;344(6268):781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- Long S. R. Rhizobium genetics. Annu Rev Genet. 1989;23:483–506. doi: 10.1146/annurev.ge.23.120189.002411. [DOI] [PubMed] [Google Scholar]
- Long S., McCune S., Walker G. C. Symbiotic loci of Rhizobium meliloti identified by random TnphoA mutagenesis. J Bacteriol. 1988 Sep;170(9):4257–4265. doi: 10.1128/jb.170.9.4257-4265.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S., Reed J. W., Himawan J., Walker G. C. Genetic analysis of a cluster of genes required for synthesis of the calcofluor-binding exopolysaccharide of Rhizobium meliloti. J Bacteriol. 1988 Sep;170(9):4239–4248. doi: 10.1128/jb.170.9.4239-4248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoil C., Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meade H. M., Long S. R., Ruvkun G. B., Brown S. E., Ausubel F. M. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol. 1982 Jan;149(1):114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel K. D., Vandenbosch K. A., Kulpaca B. Mutations in Rhizobium phaseoli that lead to arrested development of infection threads. J Bacteriol. 1986 Dec;168(3):1392–1401. doi: 10.1128/jb.168.3.1392-1401.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg C., Boistard P., Dénarié J., Casse-Delbart F. Genes controlling early and late functions in symbiosis are located on a megaplasmid in Rhizobium meliloti. Mol Gen Genet. 1981;184(2):326–333. doi: 10.1007/BF00272926. [DOI] [PubMed] [Google Scholar]
- Ruvkun G. B., Ausubel F. M. A general method for site-directed mutagenesis in prokaryotes. Nature. 1981 Jan 1;289(5793):85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- Watson R. J., Chan Y. K., Wheatcroft R., Yang A. F., Han S. H. Rhizobium meliloti genes required for C4-dicarboxylate transport and symbiotic nitrogen fixation are located on a megaplasmid. J Bacteriol. 1988 Feb;170(2):927–934. doi: 10.1128/jb.170.2.927-934.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M. N., Hollingsworth R. I., Klein S., Signer E. R. The symbiotic defect of Rhizobium meliloti exopolysaccharide mutants is suppressed by lpsZ+, a gene involved in lipopolysaccharide biosynthesis. J Bacteriol. 1990 May;172(5):2622–2632. doi: 10.1128/jb.172.5.2622-2632.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yarosh O. K., Charles T. C., Finan T. M. Analysis of C4-dicarboxylate transport genes in Rhizobium meliloti. Mol Microbiol. 1989 Jun;3(6):813–823. doi: 10.1111/j.1365-2958.1989.tb00230.x. [DOI] [PubMed] [Google Scholar]
- Zhan H. J., Levery S. B., Lee C. C., Leigh J. A. A second exopolysaccharide of Rhizobium meliloti strain SU47 that can function in root nodule invasion. Proc Natl Acad Sci U S A. 1989 May;86(9):3055–3059. doi: 10.1073/pnas.86.9.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]