Abstract
Aim
To test the hypothesis that phenotypic diversity in isolated human populations is decreased in comparison with general outbred population because of reduced genetic and environmental diversity. To demonstrate this in populations for which reduced genetic and environmental diversity had already been established, by studying the amount of variation in plasma lipid levels.
Methods
Fasting plasma lipid levels (cholesterol, triglycerides, low density lipoprotein [LDL], and high density lipoprotein [HDL]) were measured in randomly selected 300 inhabitants from 2 isolated human populations, the island of Rab and the neighboring islands of Vis and Lastovo, Croatia. The populations were chosen based on previous analyses of genetic diversity and lifestyle patterns, which were shown to be both less diverse and more uniform than the general Croatian population. We studied whether the 25’-75’ and 5′-95’ interpercentile ranges in observed values were consistently smaller in 2 samples of 300 examinees from isolated populations in comparison with nearly 6000 examinees from an earlier study who were demographically targeted to represent the larger Croatian population.
Results
General population had much wider range of observed values of triglycerides and HDL than both isolated populations. However, both isolated populations exhibited greater extent of variation in the levels of LDL, while the ranges of cholesterol values were similar.
Conclusion
Although reduced genetic and environmental diversity in isolated human populations should necessarily reduce the variance in observed phenotypic values, it appears that specific population genetic processes in isolated populations could be acting to maintain the variation. Departure from the Hardy-Weinberg equilibrium due to consanguinity, sub-structuring and differentiation within the isolates, and increased rate of new mutations could theoretically explain this paradox.
Isolated human populations were proposed to have major advantages in mapping genes for complex diseases and quantitative traits due to their reduced genetic and environmental diversity, which should reduce the underlying complexity and facilitate the identification of the genes controlling these traits (1). Subsequently, reduced genetic diversity in isolated human populations has been convincingly demonstrated (2). Reduced environment and lifestyle diversity in isolated human populations was also demonstrated in studies by many anthropologists over the past decades (3). The population genetic theory links phenotypic variation to its genetic and environmental component through the concept of heritability (4). Based on this concept, we should intuitively expect that reduced genetic and environmental diversity in isolated human populations should necessarily reduce the variance in observed phenotypic values. However, although this is an assumption in most studies in isolated human populations, we could find no studies where the presumed reduction in phenotypic variation was convincingly demonstrated in comparison to an appropriate general outbred population.
Levels of lipids in plasma represent a good candidate for traits that could be studied to demonstrate such phenotypic reduction. They are among the major risk factors for cardiovascular diseases, and it is known that many environmental and genetic factors affect their concentrations in the general population (5). Twin studies revealed substantial heritability for the plasma lipid levels, although comparisons of the twins reared together with those reared apart suggested that the effects of environment were also substantial (6). Besides, the effect of genetic factors on the lipid levels mostly decreases with age (5). Individual variability in lipid levels and response to therapeutic intervention is probably explained by the interaction between genetic and environmental factors (5). Recently, a number of candidate genes influencing plasma lipid levels have been identified (eg, ApoE, lipoprotein lipase, ApoC3, ADH3), and their interactions with environmental factors explored (5). Furthermore, the ApoB, TNFalpha, and low-density lipoprotein (LDL) receptor candidate genes seem to influence lipid profiles in youth, independently of environmental factors (7). Also, phenotypic correlations among levels of various plasma lipids may be due to pleiotropic genes (eg, between LDL-C and ApoB and also between triglycerides and HDL-C) (8). Recent studies investigating the effects of common genetic variants on the determination of levels of lipids in plasma have shown that they contribute minimally to the variance in values observed in the population, which highlights the polygenic and multifactorial control of these important quantitative traits through multiple rare variants (9,10). An understanding is emerging on the role of multiple genes with weak associations, which interact with environmental factors and determine levels of lipids in plasma (11).
The aim of this study was to test the hypothesis that phenotypic diversity in isolated human populations is indeed decreased in comparison to general outbred population, because of reduced genetic and environmental diversity, on an example of serum lipid levels.
Subjects and methods
This study aimed to address a simple but very specific question, which requires a careful study design. In the first step we chose 4 plasma lipid levels: total cholesterol, triglycerides, high-density lipoprotein (HDL), and low-density lipoprotein (LDL) as phenotypes that need to be measured. All of them are determined by both genetic and environmental factors, which was convincingly shown in many previous studies (5-11).
The second step was to define an expected distribution of measurements of the four chosen phenotypes with appropriate ranges in the outbred general population. This was already performed during the First Croatian Health Project (1995-1997) when all 4 plasma lipid levels of interest were measured. The project gathered a sample of about 6000 examinees from 30 regions of Croatia. This sample was targeted to exactly correspond to the total population of the Republic of Croatia, according to the last available population census, by age, gender, and region of residence. Field methods and the description of the performed measurements were described in great detail elsewhere (12).
The third step was to choose isolated human populations where measurements of the same phenotypes were made using comparable field and laboratory methods. We chose the field study which collected data on 1001 examinees from the isolated populations of the islands of Rab, Vis, and Lastovo between 2001 and 2003. The study was conducted by the same team, consisting of the investigators from the Institute for Anthropological Research in Zagreb and medical doctors affiliated to School of Public Health from Zagreb. Serum samples were collected in the same way as in the First Croatian Health Project. Laboratory analyses of serum lipid levels were performed in a laboratory of a highly trained professional who was also involved in measuring the lipid levels during the First Croatian Health Project (12). Field methods of the study in isolated populations and the description of the performed laboratory measurements were described in great detail elsewhere (13), and they are highly comparable to the First Croatian Health Project.
The fourth step was to form two sub-samples from a larger sample of 1001 examinees from the isolate populations, who were chosen randomly to be representatives of their respective 10 village populations. We formed two sub-samples, one with greater and one with lower degree of reduction in genetic and environmental diversity, so that the consistency and the dose-response relationship criteria could be applied to the results to infer causal relationship. This design enabled us to test the hypothesis that the phenotypic variance is decreased in both isolates in comparison with general population in all 4 lipid levels (consistency), and that the reduction of phenotypic variance is greater in the isolate with lower genetic and environmental diversity (dose-response relationship).
Because of high level of genetic differentiation in the villages from which the examinees were chosen, demonstrated by Vitart et al (14), it was important to choose two samples that will fulfill the following requirements: to contain a number of examinees large enough (at least 300 each) to allow firmer conclusions when compared with a sample of 6000 examinees from the general population; to contain examinees from the villages that have shared genetic ancestry and similar genetic structure, and where gene diversity was proven to be reduced in comparison with the large outbred population; and to have a demonstrated reduction in environment and lifestyle variability in comparison with the large outbred population.
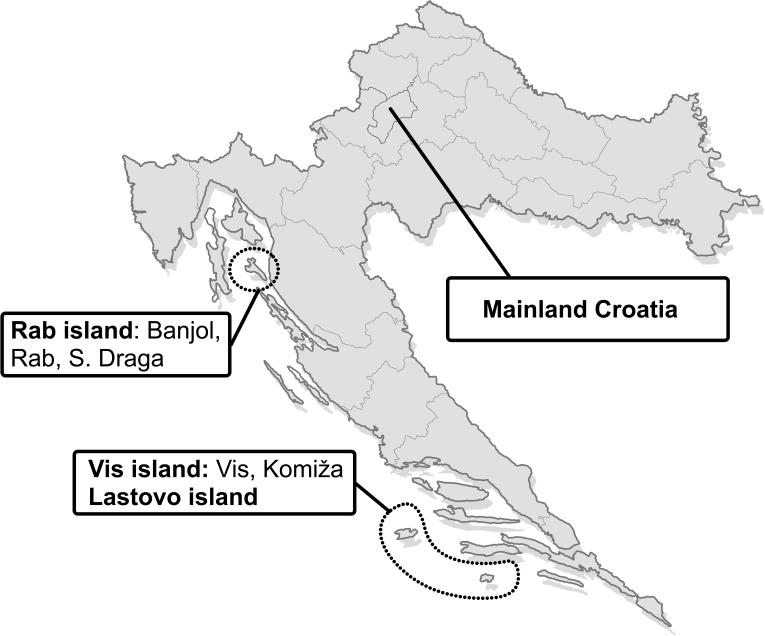
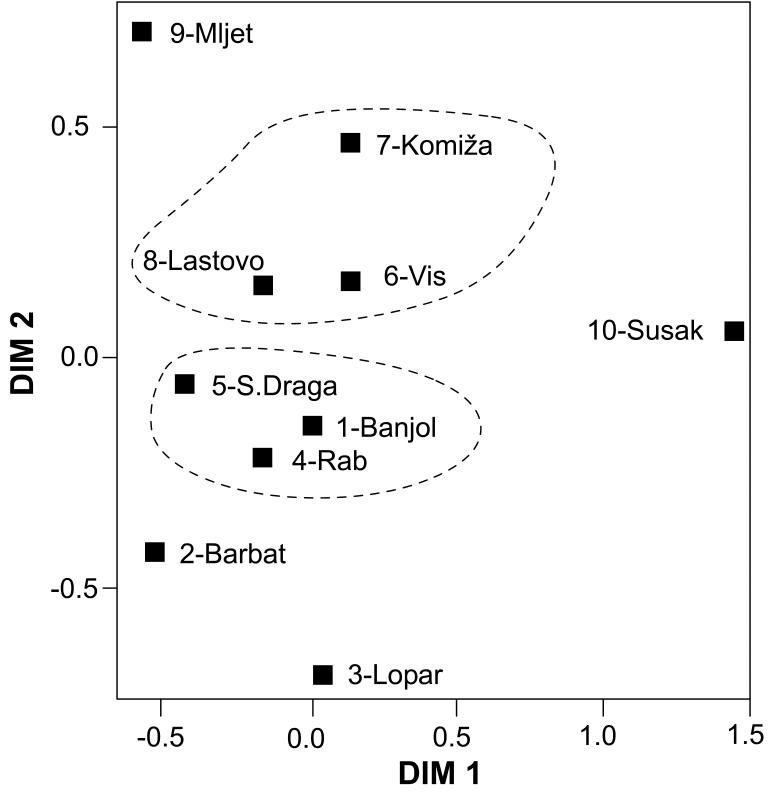
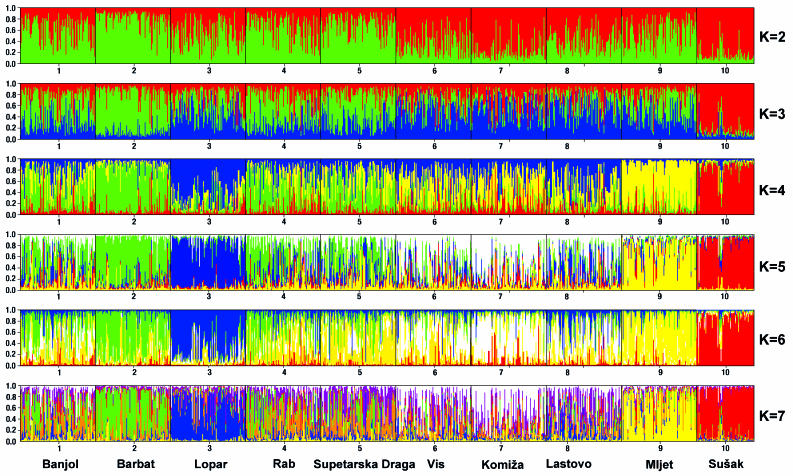
Eventually, we compared a sample of 6000 examinees representative of the population of Croatia, a sample of 300 examinees from Rab island (villages Rab, Banjol, and Supetarska Draga), and a sample of 300 examinees from two neighboring islands of Vis and Lastovo (villages Vis, Komiža, and Lastovo) (Figure 1). The justification for this choice was based on the findings from a previous genetic study (14) based on 26 autosomal short tandem repeat polymorphisms (Figure 2 and 3). The analysis of genetic distances between the 10 villages showed that Rab-Banjol-Supetarska Draga and Vis-Komiža-Lastovo were 2 distinct clusters of villages in terms of genetic similarity, while all the other villages were more genetically distant from each other (Figure 2). Model-based clustering of the examinees based on their genomic information and their prior assignment to distinct villages, implemented using the STRUCTURE program (pritch.bsd.uchicago.edu/software/structure2_1.html), further supported this clustering. It is clear that the genetic background of the villages from each of the two proposed clusters was very similar (Figure 3), while the other studied villages diverged significantly and exhibited reduced genetic variation. Thereby, it was also apparent that the genetic diversity of the Vis-Komiža-Lastovo cluster of 300 individuals was reduced in comparison with the genetic diversity of Rab-Banjol-Supetarska Draga cluster. The gene diversity of both clusters was significantly reduced in comparison with the general Croatian population (14). The diversity in the “Vis cluster”, which is more isolated and further from the coast, was more reduced than in the “Rab cluster”, which is situated in the vicinity of the mainland (14). The details of the analyses shown in Figures 2 and 3 were provided in a previous study (14).
Figure 1.
Geographic location of the studied population samples (Croatian general population, “Rab cluster,” and “Vis cluster”).
Figure 2.
Representation in two-dimensional space of genetic distances between villages from Croatian islands based on allele frequencies at 26 autosomal short tandem repeat (STR) markers (14). DIM 1 and DIM 2 represent the loadings of specific villages (relative to each other) on the two main principal components that explained the highest percent of variance of the system based on genetic distance matrix between the villages.
Figure 3.
Population structure in 10 villages on Croatian islands based on 26 short tandem repeat markers. Results from the clustering method implemented by the STRUCTURE program for inferring population structure. In each run, each separate cluster is represented by different color. Each individual is represented by a line, which is partitioned into colored segments according to the individual’s estimated membership fractions in each cluster (14).
Two previously conducted studies also demonstrated a reduced environmental and lifestyle variation in Croatian islands in terms of lesser variability in weekly nutrition choices, lesser distribution of the population by occupation status, and by basic socio-economical index in comparison with the general outbred population (13,15). In this way, we convincingly demonstrated that the two random samples of 300 examinees from isolated populations (“Rab cluster” and “Vis cluster”) had considerably reduced genetic and environmental variability in comparison with the general Croatian population sample, while the methods of measurement of phenotypes of interest between the three samples were as similar as realistically possible. This should ensure comparability of the distribution of measurements and show whether the phenotypic variance is indeed reduced in isolated populations, regardless of their age or gender structure, as these are comparative advantages for mapping genes underlying complex traits in isolated human populations.
We tested the hypothesis by presenting the percentiles of measurement distributions (5′, 25’, 50’, 75’, and 95’) in the three populations and assessing whether there was a consistency and dose-response relationship in the differences of 5′-95’ and 25’-75’ value ranges in four serum lipid measurements. Statistical significance of the differences between the observed range in the sample and the expected range in the Croatian general population were determined using Microsoft Excel χ2 statistics.
Results
We studied the differences in 5′-95’ and 25’-75’value ranges in 4 serum lipid measurements in 3 population samples, looking for consistency of observations and dose-response relationship. The final sample from the “Rab cluster” consisted of 300 examinees, 300 examinees from the “Vis cluster”, while the Croatian sample had between 4889 and 5768 examinees, depending on the success rate of determination of lipid levels in samples brought from the field after de-freezing.
In both isolated populations, values of triglycerides and HDL exhibited significantly narrower ranges in comparison with the general Croatian population, but without dose-response effect (the range was wider in the “Vis cluster” than in the “Rab cluster”). However, cholesterol levels exhibited narrower but not statistically significant ranges in the general population. General population showed significantly narrower ranges of LDL levels than Vis island (Table 1).
Table 1.
Percentiles of measurement distributions of four plasma lipid levels in the sample of Croatian general population, “Rab cluster,” and “Vis cluster” of isolated villages
| Percentile
of data distribution |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Plasma lipids* | Sample | 5′ | 25’ | 50’ | 75’ | 95’ | 25’-75’ range | P | 5′-95’ range | P |
| Cholesterol | Croatia (n = 5768) | 3.80 | 4.80 | 5.64 | 6.50 | 8.08 | 1.70 | 4.28 | ||
| Rab island (n = 300) | 4.00 | 5.00 | 5.90 | 6.80 | 8.40 | 1.80 | 0.366 | 4.40 | 0.524 | |
| Vis island (n = 300) | 4.10 | 5.10 | 5.90 | 7.10 | 8.71 | 2.00 | 0.092 | 4.61 | 0.112 | |
| Triglycerides | Croatia (n = 5759) | 0.56 | 0.90 | 1.34 | 2.14 | 4.30 | 1.24 | 3.74 | ||
| Rab island (n = 300) | 0.70 | 1.00 | 1.10 | 1.60 | 2.91 | 0.60 | <0.001 | 2.21 | <0.001 | |
| Vis island (n = 300) | 0.60 | 1.00 | 1.25 | 1.70 | 3.10 | 0.70 | 0.003 | 2.50 | 0.021 | |
| HDL | Croatia (n = 4920) | 0.64 | 0.90 | 1.14 | 1.40 | 1.86 | 0.50 | 1.22 | ||
| Rab island (n = 300) | 0.85 | 0.97 | 1.10 | 1.18 | 1.26 | 0.21 | <0.001 | 0.41 | <0.001 | |
| Vis island (n = 300) | 0.76 | 0.91 | 1.10 | 1.21 | 1.30 | 0.30 | 0.018 | 0.54 | 0.003 | |
| LDL | Croatia (n = 4889) | 2.03 | 2.94 | 3.69 | 4.47 | 5.83 | 1.53 | 3.80 | ||
| Rab island (n = 300) | 2.30 | 3.20 | 4.00 | 5.10 | 6.50 | 1.90 | 0.065 | 4.20 | 0.053 | |
| Vis island (n = 300) | 2.40 | 3.30 | 4.10 | 5.30 | 6.91 | 2.00 | 0.032 | 4.51 | 0.013 | |
*Abbreviations: LDL – low-density lipoprotein; HDL – high-density lipoprotein, P – statistical significance of the χ2 test.
Discussion
The results of this study suggest that the assumption that phenotypic variations would be reduced in isolated populations in comparison with the general outbred populations is not necessarily true, even if the reduced genetic and environmental diversity was clearly demonstrated. This is counter-intuitive and requires more careful consideration of the possible underlying mechanisms, as there is no simple and immediate explanation why in both isolated populations the distribution of cholesterol and LDL shows greater dispersal around the median in comparison with the general population, even with demonstrated reduction in genetic and environmental diversity (lack of consistency), and why there is no dose-response relationship.
Further review of literature suggested possible theoretical explanations for this unexpected finding. It appears that among isolated populations, in which the genetic diversity is being lost through genetic drift, some specific population genetic mechanisms could be acting to maintain phenotypic variation and thereby also maintain the fitness of the entire population. First, it has been shown in human, mammal, and plant population isolates that the mechanism of “genetic rescue” becomes important under such circumstances, in which the introduction of very small immigration rates can lead to dramatic changes in phenotypic diversity through outbreeding, the effect of which is enhanced in such cases (16-18).
Furthermore, due to limited mate choice in isolated populations, inbreeding effects may also become very important. Inbreeding does not lead to decrease in gene diversity, apart from highly deleterious recessive mutations that it exposes. However, it affects genotype frequencies, and subsequent departure from Hardy-Weinberg equilibrium due to consanguinity may introduce trait instability, and result in a higher proportion of the extremes in the population (19). This would become immediately apparent in our study as the increase in 5′-95’ range, without similar effect on 25’-75’ range. However, we also found no substantial support for this hypothesis.
Other population genetic mechanisms that could theoretically explain the observed paradox are sub-structuring and differentiation within the isolates and resulting multimodal distribution of the traits, and also an increase in the rate of new mutations, as a special case of genetic rescue. However, there is no simple way to substantiate this hypothesis, although examples of such occurrences have been previously reported in humans (16).
Finally, it is possible that difference between the HDL and LDL may be related to the underlying genetic architecture of these traits, with LDL exhibiting a substantial amount of non-additive genetic variance, and HDL being mainly determined through additive genetic variance (20). In such setting, a departure from the Hardy-Weinberg equilibrium caused by inbreeding would have stronger effects on the traits controlled mainly by non-additive genetic variance component, such as LDL . This may explain the difference between the distribution of LDL and HDL, and also the stronger effects in the “Vis cluster” (high prevalence of consanguinity) than in the “Rab cluster” (low prevalence of consanguinity). This was predicted to be especially apparent for traits showing antagonistic pleiotropy, such as cholesterol and LDL, ie, favorable increased values in pre-reproductive age and unfavorable in post-reproductive age (18).
This study could benefit from greater sample sizes of isolated populations, but apart from that, we were not able to identify any confounder which would be the result of the study design. The studied populations clearly differed in genetic and environmental diversity according to predictions, the samples are representative of the respective populations, and measurement errors are unlikely to be of a magnitude that could substantially affect the outcomes of the study. We conclude that the amount of phenotypic variation in the isolated populations is not necessarily reduced in comparison to larger general population, and that specific population genetic mechanisms may be operating in isolated populations to maintain the phenotypic diversity and thereby also the fitness of the population.
Acknowledgments
The study was partially supported through the grants from the Ministry of Science, Education, and Sports of the Republic of Croatia (No. 0108330) to Igor Rudan, and the grants from The British Council, The Wellcome Trust, The Royal Society, and Medical Research Council to Harry Campbell and Igor Rudan. The authors collectively thank to very large number of individuals (medical students of the Zagreb University School of Medicine, Croatia; local general practitioners and nurses in study populations; the employees of several other Croatian institutions, including but not limited to the University of Rijeka and Split, Croatia; Croatian Institute of Public Health; Institutes of Public Health in Split and Dubrovnik, Croatia; and the Institute for Anthropological Research in Zagreb, Croatia) for their individual help in planning and carrying out the field work related to the project. Ozren Polašek is supported by the PhD Scholarship from the University of Edinburgh, University of Edinburgh Overseas Research Scheme Scholarship and the International Postgraduate Scholarship from the Ministry of Science, Education, and Sports, Republic of Croatia.
There are no conflicts of interest related to this manuscript.
References
- 1.Wright AF, Carothers AD, Pirastu M. Population choice in mapping genes for complex diseases. Nat Genet. 1999;23:397–404. doi: 10.1038/70501. [DOI] [PubMed] [Google Scholar]
- 2.Shifman S, Darvasi A. The value of isolated populations. Nat Genet. 2001;28:309–10. doi: 10.1038/91060. [DOI] [PubMed] [Google Scholar]
- 3.Arlon P, Mack L, Shalev Z. How people live. New York: Dorling Kindersley Publishing; 2003. [Google Scholar]
- 4.Wright A, Charlesworth B, Rudan I, Carothers A, Campbell H. A polygenic basis for late-onset disease. Trends Genet. 2003;19:97–106. doi: 10.1016/s0168-9525(02)00033-1. [DOI] [PubMed] [Google Scholar]
- 5.Ordovas JM, Shen AH. Genetics, the environment, and lipid abnormalities. Curr Cardiol Rep. 2002;4:508–13. doi: 10.1007/s11886-002-0115-4. [DOI] [PubMed] [Google Scholar]
- 6.Heller DA, de Faire U, Pedersen NL, Dahlen G, McClearn GE. Genetic and environmental influences on serum lipid levels in twins. N Engl J Med. 1993;328:1150–6. doi: 10.1056/NEJM199304223281603. [DOI] [PubMed] [Google Scholar]
- 7.Davis CL, Wang X, Snieder H, Treiber FA. Genetic and environmental determinants of lipid profile in black and white youth: a study of four candidate genes. Ethn Dis. 2005;15:568–77. [PubMed] [Google Scholar]
- 8.Feitosa MF, Rice T, Rankinen T, Almasy L, Leon AS, Skinner JS, et al. Common genetic and environmental effects on lipid phenotypes: the HERITAGE family study. Hum Hered. 2005;59:34–40. doi: 10.1159/000084735. [DOI] [PubMed] [Google Scholar]
- 9.Talmud PJ, Hawe E, Robertson K, Miller GJ, Miller NE, Humphries SE. Genetic and environmental determinants of plasma high density lipoprotein cholesterol and apolipoprotein AI concentrations in healthy middle-aged men. Ann Hum Genet. 2002;66:111–24. doi: 10.1017/S0003480002001057. [DOI] [PubMed] [Google Scholar]
- 10.Cohen JC, Kiss RS, Pertsemlidis A, MarcelYLMcPherson R, Hobbs HH.Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science 2004305869–72. [DOI] [PubMed] [Google Scholar]
- 11.Costanza MC, Cayanis E, Ross BM, Flaherty MS, Alvin GB, Das K, et al. Relative contributions of genes, environment, and interactions to blood lipid concentrations in a general adult population. Am J Epidemiol. 2005;161:714–24. doi: 10.1093/aje/kwi103. [DOI] [PubMed] [Google Scholar]
- 12.Turek S, Rudan I, Smolej-Narancic N, Szirovicza L, Cubrilo-Turek M, Zerjavic-Hrabak V, et al. A large cross-sectional study of health attitudes, knowledge, behaviour and risks in the post-war Croatian population (the First Croatian Health Project). Coll Antropol. 2001;25:77–96. [PubMed] [Google Scholar]
- 13.Rudan I, Biloglav Z, Vorko-Jovic A, Kujundzic-Tiljak M, Stevanovic R, Ropac D, et al. Effects of inbreeding, endogamy, genetic admixture, and outbreeding on human health: a “1001 Dalmatians” study. Croat Med J. 2006;47:601–10. [PMC free article] [PubMed] [Google Scholar]
- 14.Vitart V, Biloglav Z, Hayward C, Janicijevic B, Smolej-Narancic N, Barac L, et al. 3000 years of solitude: extreme differentiation in the island isolates of Dalmatia, Croatia. Eur J Hum Genet. 2006;14:478–87. doi: 10.1038/sj.ejhg.5201589. [DOI] [PubMed] [Google Scholar]
- 15.Rudan I, Campbell H, Rudan P. Genetic epidemiological studies of eastern Adriatic island isolates, Croatia: objectives and strategies. Coll Antropol. 1999;23:531–46. [PubMed] [Google Scholar]
- 16.Titus-Trachtenberg EA, Rickards O, De Stefano GF, Erlich HA. Analysis of HLA class II haplotypes in the Cayapa Indians of Ecuador: a novel DRB1 allele reveals evidence for convergent evolution and balancing selection at position 86. Am J Hum Genet. 1994;55:160–7. [PMC free article] [PubMed] [Google Scholar]
- 17.Hogg JT, Forbes SH, Steele BM, Luikart G. Genetic rescue of an insular population of large mammals. . Proc Biol Sci. 2006;273:1491–9. doi: 10.1098/rspb.2006.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards CM. Inbreeding depression and genetic rescue in a plant metapopulation. Am Nat. 2000;155:383–94. doi: 10.1086/303324. [DOI] [PubMed] [Google Scholar]
- 19.Rudan I, Campbell H. Five reasons why inbreeding may have considerable effect on post-reproductive human health. Coll Antropol. 2004;28:943–50. [PubMed] [Google Scholar]
- 20.Ober C, Abney M, McPeek MS. The genetic dissection of complex traits in a founder population. Am J Hum Genet. 2001;69:1068–79. doi: 10.1086/324025. [DOI] [PMC free article] [PubMed] [Google Scholar]