Abstract
Aim
To present a compendium of off-ladder alleles and other genotyping irregularities relating to rare/unexpected population genetic variation, observed in a large short tandem repeat (STR) database from Bosnia and Serbia.
Methods
DNA was extracted from blood stain cards relating to reference samples from a population of 32 800 individuals from Bosnia and Serbia, and typed using Promega’s PowerPlex®16 STR kit.
Results
There were 31 distinct off-ladder alleles were observed in 10 of the 15 STR loci amplified from the PowerPlex®16 STR kit. Of these 31 alleles, 3 have not been previously reported. Furthermore, 16 instances of triallelic patterns were observed in 9 of the 15 loci. Primer binding site mismatches that affected amplification were observed in two loci, D5S818 and D8S1179.
Conclusion
Instances of deviations from manufacturer’s allelic ladders should be expected and caution taken to properly designate the correct alleles in large DNA databases. Particular care should be taken in kinship matching or paternity cases as incorrect designation of any of these deviations from allelic ladders could lead to false exclusions.
Commercial kits for forensic short tandem repeat (STR) multiplexes include allelic ladders that assist the user in allele designation by co-migrating with any commonly expected allelic variants in the population (1). In the selection of loci and other steps in the development of commercial STR kits, huge amounts of effort are put into screening population genetic variation to ensure that the locus varies in a simple and easy-to-interpret manner, and that allele frequencies are known for statistical interpretation. Allelic ladders are based on variation observed in the development process, but, inevitably, when large population samples are typed, variants are encountered that were not represented in the developmental screening. This is particularly true when the populations under testing differ from those that were studied during kit development.
Practitioners of forensic STR typing need to be aware of the possibility of rare variants, so the variants can be recognized and dealt with properly in casework interpretation. If designated correctly, variant alleles can sometimes greatly increase the power of discrimination in DNA comparisons (2). In this regard, it is important that the forensic science community share information on the occurrence of these variants. In some contexts, these variants may actually occur rather commonly, and public documentation of their occurrence can save individual investigators much time and effort (3).
Unexpected or “anomalous” genetic variation that can complicate STR typing takes a number of forms, with a variety of consequences on the testing results. New length variants not represented on the allelic ladder can be due to insertion/deletions of full repeat units, or to “microvariants” differing due to the insertion/deletion of single bases or partial repeats (2,4-6). In some cases, larger or smaller off-ladder alleles may fall within the allelic ladder of an adjacent locus (5,6), with the potential for significant confusion. Triallelic patterns (7-9) can be due to length mutations that occur and segregate during an individual’s development, or to localized duplication of a locus, or to chromosomal trisomy. Sequence variation, rather than length variation, can also have effects, particularly in the case of sequence differences in the amplification primer binding sites. Primer mismatches can result in complete amplification failure and cause null alleles, or can lower the peak height of affected alleles (10-18). Any of these anomalies can cause problems during interpretation of results if the analysts are not familiar with these occurrences.
The International Commission on Missing Persons (ICMP) was founded in 1996 to address the issue of persons missing as a result of the conflicts that occurred during the breakup of the former Yugoslavia during the 1990s. DNA profiles are obtained from reference samples collected from living relatives which are entered into a DNA reference database (currently greater than 80 000 unique profiles). Likewise, DNA profiles generated from skeletal remains recovered from grave sites are entered into a missing persons DNA database (currently containing more than 12 000 unique profiles). Both databases are screened against each other on a daily basis, resulting in possible kinship matches.
We report here a new compendium of all off-ladder variants/microvariants, primer binding site mutations, and instances of triallelic patterns that have been observed in a subset of the ICMP blood sample reference database representing ~ 32 800 individuals from Bosnia and Serbia, including Kosovo.
Methods and materials
Samples, tabulation, and frequency estimation of variant alleles
Blood stain cards have been obtained from family members of missing persons from Bosnia and Serbia (approximately 80% from Bosnia, 14% from Kosovo, and 6% from Serbia), and 32 800 of these were carefully evaluated for the presence of variant alleles for this study. For the purposes of kinship testing, multiple family members are sought for each missing person, so this population database does not contain only unrelated individuals. It has not been feasible to sort the data so that only unrelated individuals are represented and this complicates the calculation of allele frequencies. On average in the ICMP database, missing persons are represented by approximately three family members, so this database might, to some approximation, be considered to represent approximately 11 000 “individuals,” or 22 000 total alleles. When variant alleles were tabulated, they were checked to determine if they occur in multiple related family members; variant alleles segregating within multiple family members were counted as only a single occurrence. To estimate allele frequencies (relating to the chance that a randomly selected allele from the population will match the variant), the number of unique (unrelated) occurrences was divided by 22 000 “unrelated” alleles.
DNA extraction from S&S IsoCode® cards
DNA from reference samples was directly isolated from blood stain cards using a non-organic extraction. S&S IsoCode® (Schleicher & Schuell, Dassell, Germany) cards bind possible polymerase chain reaction (PCR) inhibitors while DNA is eluted with water. 1/8-inch punches from the dried blood stained cards were punched into 96-well plates using a Wallac DBS puncher (Perkin Elmer, Waltham, MA, USA). The punches were washed twice with 200 µL of sterile water, and then the DNA was eluted in 100 µL sterile water at 95°C for 30 minutes.
STR typing
For the former Yugoslavia ICMP generally utilizes the commercial STR kit PowerPlex16® (“PP16”) System (Promega Corporation, Mannheim, Germany) which simultaneously amplifies 15 separate STRs (D3S1358, THO1, D21S11, D18S51, Penta E, D5S818, D13S317, D7S820, D16S539, CSF1PO, Penta D, vWA, D8S1179, TPOX, and FGA) in three different dye colors and the sex determining marker Amelogenin. ICMP also uses Identifiler® (Applied Biosystems, Foster City, CA, USA), PowerPlex16® Monoplex System primers (Promega Corporation) and its own internally designed mini amplicon kits (16).
PCR reactions using PP16 or Monoplex primers were setup according to the manufacturer’s guidelines for each kit except for the following changes: reaction volumes were halved and 2.5 U of AmplliTaq Gold® DNA polymerase (Applied Biosystems) were added per reaction. A total of 1-1.2 µL of extracted DNA was added per reaction. PCR reactions using Identifiler® were setup according to the manufacturer’s guidelines.
Amplification
Reference samples were amplified with PP16, Identifiler or Monoplex primers on either the GeneAmp® PCR system 9700 (Applied Biosystems) or Eppendorf Mastercycler thermocyclers (Eppendorf, Hamburg, Germany) according to manufacturer’s guidelines with the following changes: the cycle number was extended to 32 cycles for PP16 and Monoplex reactions, 30 cycles for Identifiler® and the final extension increased from 30 minutes to 45 minutes for PP16 and Monoplex only.
Electrophoresis and analysis
Fragment separation was performed using ABI PRISM® 3100 Genetic Analyzer (Applied Biosystems) using POP-4 polymer and the collection software, version 1.0 or 1.1. Data was sized using ABI’s GeneScan®, version 3.7, (Applied Biosystems) and alleles designated using ABI’s Genotyper®, version 3.7 NT (Applied Biosystems), using the commercial macro PowerTyper 16 Macro V.2 (www.promega.com) or the Identifiler Macro (Applied Biosystems).
Variant Allele Confirmation
All off-ladder alleles were confirmed either by re-extraction and amplification (in some instances using Monoplex primers), unless, in some cases, they were determined to occur in multiple family members with multiple verifying amplifications already present.
Cases of presumed primer binding site mutations were re-amplified with PP16, and also amplified with the ABI Identifiler kit. The hypothesis of primer mismatch was considered confirmed if amplification with different primer pairs restored amplification of the allele in question.
All samples containing triallelic patterns were confirmed by re-extraction and amplification.
Results
Off-ladder alleles
A total of 275 off-ladder alleles were observed in a subset of ICMP blood reference database consisting of 32 800 individuals. Of these 275, 176 off-ladder alleles were from unrelated individuals (Table 1). Of these 176, 31 distinct variant alleles were identified within 10 of the 15 loci amplified by the PP16 kit (Table 1). The 10 loci include TH01, D21S11, D18S51, Penta E, D13S317, D7S820, D16S539, CSF1PO, Penta D, and FGA. In one instance, with D13S317, the off-ladder allele was shifted up in size creating the unusual and troublesome situation of falling in an allelic bin for the adjacent larger locus with the same fluorescent label in the multiplex electropherograms.
Table 1.
Observed variant alleles
| Locus | Allele | No. of observations | Frequency |
|---|---|---|---|
| TH01 | 8.3 | 11 | 0.0500 |
| 10.3 | 1 | 0.0045 | |
| D21S11 | 33.1 | 4 | 0.0182 |
| 34.1 | 3 | 0.0136 | |
| D18S51 | 15.2 | 2 | 0.0091 |
| Penta E | 16.4 | 1 | 0.0045 |
| 17.4 | 1 | 0.0045 | |
| 18.2* | 4 | 0.0182 | |
| D13S317 | 6 | 22 | 0.0999 |
| 16 | 16 | 0.0727 | |
| 17 | 1 | 0.0045 | |
| D7S820 | 7.3 | 24 | 0.1090 |
| 9.1 | 15 | 0.0681 | |
| 9.3 | 1 | 0.0045 | |
| 11.1 | 3 | 0.0136 | |
| 15 | 11 | 0.0500 | |
| D16S539 | 12.1 | 2 | 0.0091 |
| 16 | 1 | 0.0045 | |
| CSF1PO | 10.1 | 1 | 0.0045 |
| 16 | 1 | 0.0045 | |
| 17* | 1 | 0.0045 | |
| Penta D | 11.2 | 1 | 0.0045 |
| 12.1 | 18 | 0.0818 | |
| 12.2 | 1 | 0.0045 | |
| 14.1 | 1 | 0.0045 | |
| 18 | 12 | 0.0545 | |
| 19* | 1 | 0.0045 | |
| FGA | 20.1 | 11 | 0.0500 |
| 22.3 | 4 | 0.0182 | |
| 26.2 | 1 | 0.0045 | |
| 27.2 | 1 | 0.0045 |
*Alleles not previously reported (STRbase).
Primer binding site mismatches
Two loci exhibited presumed primer binding site mismatches that affected PP16 typing results (Table 2). Both D5S818 and D8S1179 each had several cases in which a silent (null) allele was produced by the effect of the binding site mismatch. For the D5S818 locus, all cases involved a silent 10 allele except for 1 case, which involved a silent 11 allele. All cases involving D8S1179 involved a silent 14 allele. Of the two loci, D5S818 had the higher frequency of primer binding site mismatch alleles, with a frequency of 0.045%.
Table 2.
Primer binding site mutations observed
| Locus | Allele | No. observations | Kit affected | Frequency (%) |
|---|---|---|---|---|
| D5S818 | 10 | 9 | PP16 | 0.0409 |
| D5S818 | 11 | 1 | PP16 | 0.0045 |
| D8S1179 | 14 | 4 | PP16 | 0.0182 |
Triallelic patterns
A total of 15 three allele patterns were observed in our study (Table 3), involving the following nine loci: TH01, D21S11, D18S51, Penta E, CSF1PO, Penta D, D8S1179, vWA, TPOX and FGA. The loci with the highest rate of triallelic bands include D21S11 (3 cases), D18S51 (3 cases), and FGA (2 cases). No single pattern was observed more than once in unrelated individuals. A single case was observed in which siblings shared the same three alleles at TPOX.
Table 3.
Triallelic patterns observed
| Locus | No. observed | Alleles | Type | Confirmation |
|---|---|---|---|---|
| TH01 | 1 | 7/8/9 | I | 2 reps PP16 |
| D21S11 | 3 | 29/30.2/31.2 | I | 2 reps PP16 |
| 29/31/32 | I | 6 reps PP16 | ||
| 28/30.2/32.2 | II | 3 reps PP16 | ||
| D18S51 | 3 | 13/14/21 | I | 3 reps PP16 |
| 16/17/18 | I | 3 reps PP16 | ||
| 14/15/16 | I | 2 rep PP16 | ||
| Penta E | 1 | 13/16/17 | I | 2 reps PP16 |
| CSF1PO | 1 | 10/12/13 | I | 2 reps PP16 |
| Penta D* | 1 | 9/10/10 | II | 3 reps PP16 |
| D8S1179 | 1 | 14/16/17 | I | 2 reps PP16 |
| vWA | 1 | 15/16/20 | I | 3 rep PP16, 1 rep Identifiler |
| TPOX | 1 | 8/11/12 | II | 5 reps PP16 |
| FGA | 2 | 20/21/22 | I | 2 reps PP16 |
| 23/25/26 | I | 2 reps PP16 |
*This locus exhibited diallelic pattern consistent with 2 copies of 1 allele and 1 copy of the other allele. This represents the same individual with the triallelic pattern 28/30.2/32.2 in D21S11, likely resulting from an extra copy of the 21st chromosome donated by the father.
Discussion
In the total of 31 distinct off-ladder alleles, 16 instances of triallelic patterns and 14 instances of primer binding site mutations were observed in profiles amplified using the PowerPlex®16 STR kit.
Of the 31 distinct off-ladder alleles observed, 14 were seen only in a single individual or family, each with a nominal frequency of 0.0045% (Table 1). Of course, it is not reasonable to consider that frequencies can be reliably known from such rare observations, and the fact that our source population contains multiple sets of related individuals makes the frequency estimates all the more of an approximation. Nine of the off-ladder variants represented full repeat insertions or deletions placing them outside the size range of the allelic ladder. The remaining 22 were off-ladder as partial repeats or microvariants. The most commonly observed variant allele was the 7.3 allele of D7S820, which was observed 24 times in unrelated individuals, representing a 0.11% frequency in our population. The STRBase web site (ref. 3, http://www.cstl.nist.gov/biotech/strbase/) lists seven different reports of this D7S820 variant. This was followed closely by the 6 allele from D13S317 which was observed 22 times in unrelated individuals representing a 0.10% frequency in our population. This allele has been separately reported ten times in STRBase.
Of the 31 distinct off-ladder alleles listed, 3 were not reported as previously being observed (STRbase). Interestingly, 2 of these were found in the Penta loci, perhaps due to the fact that these loci are not frequently being tested in larger databases such as the UK (19) or CODIS databases (http://www.fbi.gov/hq/lab/codis/index1.htm). The 18.2 allele of Penta E was observed 4 times in unrelated individuals while the 19 allele in Penta D was observed once. We also report a 17 allele in the locus CSF1PO. The Penta D locus had the greatest number of unique off-ladder alleles. A total of 6 alleles did not fall on ladder, including the “19” allele, which has not been reported before (STRbase). D7S820 followed closely with 5 off-ladder alleles.
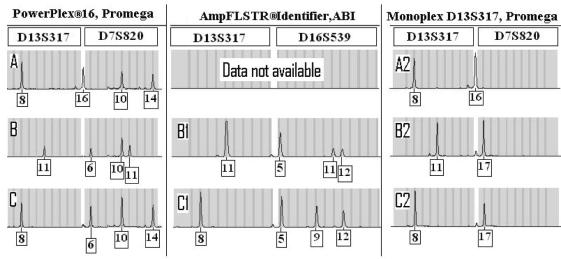
It sometimes occurs that increases or decreases in length can take variant alleles into the size range of an adjacent locus; several examples have been published in the literature (5,6) or STRbase. This is most problematic when the allele falls within an allelic bin from the adjacent locus. Generally, multiplex kits are designed to avoid this problem by having adjacent loci “out of phase” with respect to full repeat units, so that off-ladder alleles from one locus will not fall into an allele bin of the adjacent locus. However, with PP16 we encountered a kinship case with two off-ladder alleles in different individuals, one of which binned as an allele of the adjacent locus (Figure 1). Here, in one person, an off-ladder D13S317 allele 16 falls directly between the D13S317 and D7S820 ladders. In this person’s two siblings another off-ladder D13S317 allele 17 falls within the bin of D7S820 allele 6. To reach these conclusions, amplifications were also done using Identifiler and with primers for D13S317 in Monoplex. The Monoplex amplifications proved that the peaks were off-ladder alleles of D13S317. Moreover, the Identifiler amplification indicated that the problem of locus crossover of off-ladder alleles is not confined to PP16. In Identifiler, the locus adjacent to D13S317 is D16S539, and the off-ladder D13S317 allele 17 bins as a D16S539 allele 5. As a final interesting note to this kinship case, it is evident through comparison to other relatives (not shown) that the D13S317 allele 16 (present in a single individual in this family) is a result of a mutation from the father’s 17 allele. This intersection of unusual observations (presence of an extremely rare off-ladder allele, and an intergenerational mutation event) invites the speculation that the large 17 allele may be unusually prone to mutation via strand slippage.
Figure 1.
Amplifications from 3 siblings (A, B, C) manifesting off-ladder D13S317 alleles 16 or 17, using PP16 (left), Identifiler (center), or D13S317 Monoplex amplification primers from Promega (right). For each amplification type, the D13S317 locus is at the left side and the adjacent larger locus at the right side (D7S820 for PP16 kit or Promega Monoplex; and D16S539 for Identifiler). Note that the 17 allele of D13S317 for both PP16 and Identifiler falls on bin with the first allele in the adjacent larger locus in each kit.
In both PP16 and Identifiler the bins of the D13S317 locus are “in phase” by full repeat units with the adjacent larger locus. The distance between last allele from D13S317 and first allele from either D7S820 (PP16) or D16S539 (Identifiler) is exactly 8 base pairs. In the case listed above, both D13S317, D7S820 and D16S539 were heterozygous, which flagged an apparent triallelic pattern for closer inspection. But what if D13S317 were heterozygous and either D7S820 (in PP16) or D16S539 (in Identifiler) were homozygous? This could complicate analysis, as the non-D13S317 locus would appear heterozygous, while D13S317 would appear homozygous. In response to this potential for mistyping, our entire database was screened for any samples having an allele 15 in D13S317 (the last allele in the ladder) and/or allele 6 from D7S820 (the first allele in the ladder). In total, 18 cases were found in which a 16 allele was submitted for D13S317 (falling between the allelic ladders). A total of 302 samples were found in which a 15 allele from D13S317 was observed, while 61 samples were found in which a 6 allele was observed for D7S820. All profiles were rechecked for misinterpretation of allele calls. Cases where D13S317 was homozygous and D7S820 heterozygous, or vice-versa, were closely reviewed. In certain cases Monoplex primers were used to check the original call. In all cases, the original designation was confirmed and no improper allele designations had been made.
Primer binding site “mutations” (sequence variants that mismatch with amplification primers) have been observed in most loci found in commercial STR kits (10-18). When these cause a drop out in amplification, this can be difficult to detect, particularly with case samples where peak heights can vary by more than the usually expected ratios for heterozygotes and homozygotes. Sometimes the mismatch causes only a decrease in amplification, rather than complete drop-out. This can cause pronounced peak imbalance (11,18), and if heterozygous alleles are within one repeat of each other, this can be misinterpreted as stutter. Null alleles due to primer mismatch can be more readily detected in paternity testing or kinship matching due to apparent inconsistent allele sharing among relatives.
In our study we observed probable primer binding mismatch at two loci, D5S818 and D8S1179. This is not the first time a mutation at D5S818 has been reported for PP16. Several groups have identified a possible polymorphic nucleotide in the reverse primer binding site of the 10 allele (10,17). It is interesting to note that most (9 out of 10) of the instances of D5S818 primer mismatch observed in our work also involves the 10 allele. It seems likely that this is the same identical-by-descent variant in both cases. Our single observation of an 11 allele with the mismatch may be due to repeat length mutation of the “mutant” 10 allele.
Most cases of reported polymorphic nucleotides in the primer binding site of D8S1179 do not involve the PP16 kit (10,13-15). However, we report 4 instances of primer binding site mutations at D8S1179 which involve the 14 allele in the PP16 kit. One of these cases was previously reported by our laboratory in a mini amplicon concordance study (16). It is important to note that point mutations are not isolated to PP16, but occur in most other STR kits also (10,13-15).
Triallelic patterns are classed as two predominant types (8) (Figure 2). In Type I, somatic mutation of one allele occurs during an individual’s development, resulting in a chimera with some cells containing the original allele and others the mutant allele. Type I patterns are characterized by uneven peak heights for the two variants of the affected allele that sum to the height of an unmutated allele. In Type II, a localized duplication event, or chromosomal aneuploidy (8,20), is responsible for multiple alleles with peaks usually appearing of equal height. Localized duplication on chromosomes is more frequent on the Y-chromosome than autosomes (21). In our study, 12/15 triallelic patterns seen were of Type I.
Figure 2.
Examples of triallelic patterns. (A) Type I pattern generally associated with somatic mutation during development. (B) Type II pattern generally associated with an additional copy of the locus.
In several instances we were able to track the inheritance of duplicated alleles responsible for Type II triallelic patterns. When localized duplication events occur, it is likely that the two resulting alleles would be tightly linked, and therefore inherited together in progeny (8). In one of the cases we observed, two siblings had the same Type II pattern at TPOX. A third sibling was homozygous. It would seem that two of the three siblings inherited the same duplication event from one of the parents. Since the parents were not typed this cannot be confirmed.
Type II patterns can also be the result of autosomal trisomy. Many cases involving autosomal trisomy can be detrimental to the health of the developing fetus. However, certain cases of aneuploidy involving chromosome 13, 18, and the most common, 21, result in live births (22). STRs have been employed to diagnose possible trisomies (20,23). We observed one case where the profile exhibits a Type II pattern with alleles at 28, 30.2, and 32.2 at the D21S11 locus, located on the 21st chromosome (data not shown). It is possible that this occurred due to a localized duplication in which two repeats were lost. However, it is also possible that that this could be the result of autosomal trisomy. The father (28/32.2) and mother (29/30.2) have all the alleles seen in the affected child (28/30.2/32.2). It could be that non-disjunction occurred in the father’s sperm during meiosis I. This would lead to two out of four gametes having two copies of chromosome (28/32.2) instead of one. If one of these combined with the mother’s egg (30.2), the result would be aneuploidy in the progeny with a 28/30.2/32.2 at the D21S11 locus. Paternal non-disjunction giving rise to trisomy 21 is rare (22,24) but could be the cause of the banding pattern in this case.
Further insight into this case was gained from examination of Penta D, also located on chromosome 21. Penta D did not exhibit triallelic banding pattern, but it did indicate a diallelic pattern characteristic of a “double dose” of one of the paternal alleles (20,23). In all three amplifications the first allele of Penta D (a 9 allele) had significantly lower peak height than the second allele (a 10 allele), resulting in an average fluorescent intensity ratios of 1.7:1. The father is 10/10 and the mother is 9/12 for Penta D. If this case were to fit the paternal non-disjunction, then the father would be contributing two 10 alleles, while the mother would donate the 9 allele. Therefore the 10 allele would be ~ twice the height of the 9 allele, which is reflected in the fluorescent intensity ratios. This case would seem to fit a paternal error in meiosis. The health of the individual in question is unknown.
In conclusion, our study offers a detailed discussion on a compendium of unusual variant alleles detected in a large database of nuclear STR profiles from the former Yugoslavia, based on the PP16 multiplex kit, with particular emphasis on how the variants were diagnosed and the types of interpretation issues they can cause. Continued public dissemination of this information will assist practitioners in an awareness of anomalies that may be encountered due to variants such as off-ladder alleles, primer binding site mismatches, and triallelic patterns. All variant/microvariant and triallelic patterns discussed in this paper will be uploaded to STRbase and we encourage other laboratories to contribute such examples to the public domain. Documentation that particular variants have been previously identified can assist greatly in interpretation and presentation of results. Particular care should be taken in kinship matching or paternity cases, as incorrect designation of any of these deviations from allelic ladders could lead to false exclusions.
Acknowledgments
The authors acknowledge the remarkable contributions of more than 200 members of ICMP staff, past and present, without whom this work would not have been accomplished. We particularly acknowledge the leadership and/or support of Jon Davoren, Adnan Rizvić, Kathryne Bomberger, and Adam Boys. The ICMP acknowledges the generous support provided by the governments of Canada, Denmark, Finland, France, Germany, Greece, the Holy See, Iceland, Ireland, Italy, the Netherlands, Norway, Sweden, Switzerland, the United Kingdom, and the United States, as well as the European Union and the C.S. Mott Foundation. ICMP also wishes to thank the effort from its staff, both past and present, who contributed to the data.
References
- 1.Griffiths RA, Barber MD, Johnson PE, Gillbard SM, Haywood MD, Smith CD, et al. New reference allelic ladders to improve allelic designation in a multiplex STR system. Int J Legal Med. 1998;111:267–72. doi: 10.1007/s004140050167. [DOI] [PubMed] [Google Scholar]
- 2.Allor C, Einum DD, Scarpetta M. Identification and characterization of variant alleles at CODIS STR loci. J Forensic Sci. 2005;50:1128–33. [PubMed] [Google Scholar]
- 3.Ruitberg CM, Reeder DJ, Butler JM. STRBase: a short tandem repeat DNA database for the identity testing community. Nucleic Acids Res. 2001;29:320–2. doi: 10.1093/nar/29.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizuno N, Sekiguchi K, Sato H, Kasai K. Variant alleles on the penta E locus in the PowerPlex 16 kit. J Forensic Sci. 2003;48:358–61. [PubMed] [Google Scholar]
- 5.Heinrich M, Felske-Zech H, Brinkmann B, Hohoff C. Characterization of variant alleles in the STR systems D2S1338, D3S1358 and D19S433. Int J Legal Med. 2005;119:310–3. doi: 10.1007/s00414-005-0554-8. [DOI] [PubMed] [Google Scholar]
- 6.Grubwieser P, Mühlmann R, Niederstätter H, Pavlic M, Parson W. Unusual variant alleles in a commonly used short tandem repeat loci. Int J Legal Med. 2005;119:164–6. doi: 10.1007/s00414-004-0508-6. [DOI] [PubMed] [Google Scholar]
- 7.Lukka M, Tasa G, Ellonen P, Moilanen K, Vassiljev V, Ulmanen I. Triallelic patterns in STR loci used for paternity analysis: Evidence for a duplication in chromosome 2 containing the TPOX STR locus. Forensic Sci Int. 2006;164:3–9. doi: 10.1016/j.forsciint.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Clayton TM, Guest JL, Urquhart AJ, Gill PD. A genetic basis for anomalous band patterns encountered during DNA STR profiling. J Forensic Sci. 2004;49:1207–14. [PubMed] [Google Scholar]
- 9.Rolf B, Wiegand P, Brinkmann B. Somatic mutations at STR loci – a reason for three-allele pattern and mosaicism. Forensic Sci Int. 2002;126:200–2. doi: 10.1016/s0379-0738(02)00054-3. [DOI] [PubMed] [Google Scholar]
- 10.Delamoye M, Duverneuil C, Riva K, Leterreux M, Taieb S, De Mazancourt P. False homozygosities at various loci revealed by discrepancies between commercial kits: implications for genetic databases. Forensic Sci Int. 2004;143:47–52. doi: 10.1016/j.forsciint.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Clayton TM, Hill SM, Denton LA, Watson SK, Urquhart AJ. Primer binding site mutations affecting the typing of STR loci contained within the AMPFlSTR SGM Plus kit. Forensic Sci Int. 2004;139:255–9. doi: 10.1016/j.forsciint.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Budowle B, Masibay A, Anderson SJ, Barna C, Biega L, Brenneke S, et al. STR primer concordance study. Forensic Sci Int. 2001;124:47–54. doi: 10.1016/s0379-0738(01)00563-1. [DOI] [PubMed] [Google Scholar]
- 13.Leibelt C, Budowle B, Collins P, Daoudi Y, Moretti T, Nunn G, et al. Identification of a D8S1179 primer binding site mutation and the validation of a primer designed to recover null alleles. Forensic Sci Int. 2003;133:220–7. doi: 10.1016/s0379-0738(03)00035-5. [DOI] [PubMed] [Google Scholar]
- 14.Han GR, Song ES, Hwang JJ. Non-amplification of an allele of the D8S1179 locus due to a point mutation. Int J Legal Med. 2001;115:45–7. doi: 10.1007/s004140100213. [DOI] [PubMed] [Google Scholar]
- 15.Forrest SW, Kupferschmid TD, Hendrickson BC, Judkins T, Petersen DJ, Scholl T. Two rare novel polymorphisms in the D8S1179 and D13S317 markers and method to mitigate their impact on human identification. Croat Med J. 2004;45:457–60. [PubMed] [Google Scholar]
- 16.Parsons TJ, Huel R, Davoren J, Katzmarzyk C, Miloš A, Selmanović A, et al. Application of novel “mini amplicon” STR multiplexes to high volume casework on degraded skeletal remains. Forensic Science International Genetics. 2007;1:175–9. doi: 10.1016/j.fsigen.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 17.Alves C, Gusmao L, Pereira L, Amorim A. Multiplex STR genotyping: comparison study, population data and new sequence information. In: Brinkman B, Carracedo A, editors. Progress in forensic genetics, vol. 9. Amsterdam: Excerpta Medica, Elsevier; 2003. p. 131-5. [Google Scholar]
- 18.Butler JM, Shen Y, McCord BR. The development of reduced size STR amplicons as tools for analysis of degraded DNA. J Forensic Sci. 2003;48:1054–64. [PubMed] [Google Scholar]
- 19.Werrett DJ. The national DNA database. Forensic Sci Int. 1997;88:33–42. [Google Scholar]
- 20.Findlay I, Tóth T, Matthews P, Marton T, Quirke P, Papp Z. Rapid trisomy diagnosis (21, 18, and 13) using fluorescent PCR and short tandem repeats: applications for prenatal diagnosis and preimplantation genetic diagnosis. J Assist Reprod Genet. 1998;15:266–75. doi: 10.1023/A:1022536309381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Butler JM, Decker AE, Kline MC, Vallone PM. Chromosomal duplications along the Y-chromosome and their potential impact on Y-STR interpretation. J Forensic Sci. 2005;50:853–9. [PubMed] [Google Scholar]
- 22.Hassold TJ, Jacobs PA. Trisomy in man. Annu Rev Genet. 1984;18:69–97. doi: 10.1146/annurev.ge.18.120184.000441. [DOI] [PubMed] [Google Scholar]
- 23.Yan J, Wu J, Li Y, Wang H, Huang Z, Zhou X, et al. A novel diagnostic strategy for trisomy 21 using short tandem repeats. Electrophoresis. 2006;27:416–22. doi: 10.1002/elps.200500349. [DOI] [PubMed] [Google Scholar]
- 24.Petersen MB, Mikkelsen M. Nondisjunction in trisomy 21: origin and mechanisms. Cytogenet Cell Genet. 2000;91:199–203. doi: 10.1159/000056844. [DOI] [PubMed] [Google Scholar]