Abstract
Aim
To assess genotyping with microsatellite-based markers of the olive (Olea europaea L.) for potential application of olive as legal case evidence, with regard to the degree of variability within the Croatian olive genomic pool and to the effectiveness of the chosen set of microsatellite-based markers in revealing olive divergence.
Methods
The total of 44 autochthonous Croatian olive specimens were subjected to genotyping with 16 previously described and developed microsatellite-based markers. According to previous morphological analyses, 44 specimens were classified into 30 cultivars with the exception of an additional, previously unassigned specimen.
Results
Genotyping of 44 specimens distinguished a total of 44 different genotype profiles by 16 microsatellite-based loci. Average expected heterozigosity amounted to 0.758, which points to significant diversity of Croatian olives.
Conclusion
Croatian olive genotyping showed strong varietal discrimination up to the single tree and considerable potential application of olive as evidence in investigation of crime, accident, and suicide circumstances.
Forensic botany is the study of plants and plant material with the purpose of presenting the plant evidence in court. It includes a number of disciplines, such as plant anatomy and systematics palynology, plant ecology, limnology, plant chemistry, and plant molecular biology (1). In spite of its high potential in assessing the legal case evidence, only a few cases of plant forensic investigation applying DNA profiling, when a suspect was linked to the crime scene, were described (2,3). Plant DNA profiling serves to identify the origin of detected plant material connected to a crime, suicide, or accident, and hence, it may contribute to identifying the location(s) where the event took place (primary scene), recent location of the body, whether a victim had been transferred or moved (secondary scene), and whether a suspect was present at a crime or accident scene (4,5). DNA profiling is also employed in solving the issues of narcotics and drug enforcement, as well as of unauthorized commercialization of some plants.
Microsatellite-based genotyping, due to its great reproducibility and high degree of certainty in assigning the origin of a biological material that serves as legal case evidence, represents one of the most reliable DNA profiling methods in forensic investigation (6).
Microsatellites, short tandem repeats (STR) or simple sequence repeats (SSR) consist of a number of tandemly repeated short DNA sequences (1-6 base pairs long). They are distributed throughout the eukaryotic genome. In addition, microsatellites are multiallelic due to their high intraspecies variability and are easily amenable to polymerase chain reaction (PCR)-based analysis. Both characteristics make them the DNA markers of choice for human DNA profiling analyses. However, microsatellite-based markers found their way of wider application in different branches of animal and plant sciences.
Olea europaea L. is a diploid, outcrossing species. Cultivated olives have been reproduced mainly by vegetative propagation and sporadically by cross-breeding, which resulted in the creation of a number of varieties due to accidental crosses between cultivated forms or between wild and cultivated forms, but also due to accumulation of mutations, along with local selection of outstanding individuals. Hence, most olive cultivars have a local origin. More than 2000 cultivars have been documented in the Mediterranean region by means of their morphology (7).
At present, microsatellite-based DNA sequences are the most appropriate genetic markers used in olive cultivar characterization and classification. Many microsatellites have been isolated from olives and their respective primer pairs have been developed (8-14).
Due to their mainly local origin, specific olive cultivars are indigenous to specific geographical areas. In addition, the same cultivars grown in different environmental conditions have different genotype profiles. Both olive characteristics ensure their relevance in the assessment of the location of origin of the olive sample in question.
Olive trees are abundant in Croatia. In order to assess the application potential of Croatian olive DNA profiling in forensic investigations, we genotyped the total of 44 specimens that comprise 30 cultivars and their 13 varieties, as well as one unassigned olive specimen.
Material and methods
Plant material
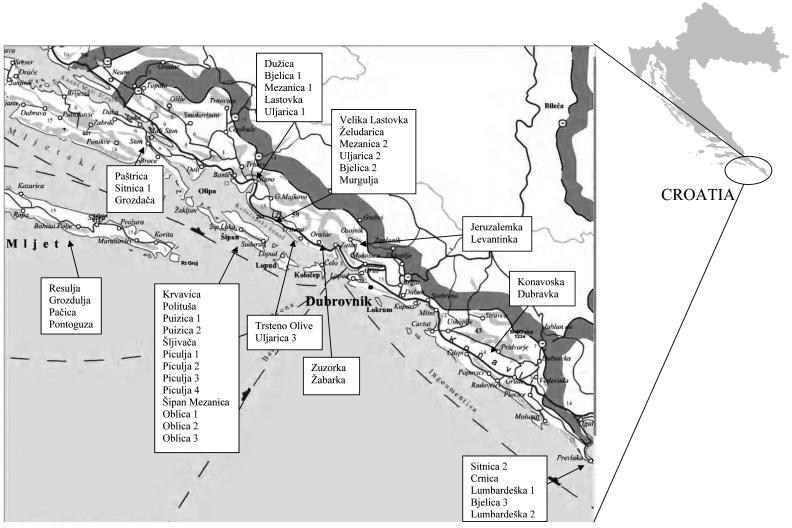
Forty-four autochthonous Croatian olive specimens Olea europaea subsp. europaea var. europaea (30 cultivars and their 13 varieties, as well as one unassigned olive specimen) from the south of Croatia, the native area of their cultivation, were selected for this study (Figure 1).
Figure 1.
Analyzed olive specimens and native areas of their cultivation. The oval indicates geographical location of olive specimens in Croatia, sampled in this study.
DNA extraction
Total DNA was isolated from young olive leaves and flower buds, following an already published olive DNA isolation method (15), with several modifications. Young olive leaves and buds were washed by 4% sodium hypochlorite and 0.2 g of plant tissue was ground into liquid nitrogen and incubated in 4 mL of prewarmed CTAB buffer [2%(w/v) CTAB, 100 mM Tris-HCl pH 8, 1.4 M NaCl, 40 mM EDTA pH 8, 0.5% SDS, 6% (w/v) PVP, 0.2%(v/v) 2-mercaptoethanol]. The samples were incubated at 65°C for an hour and a half and mixed gently several times. Equal volume of chloroform-isoamyl alcohol [24:1 (v/v)] was added to the mix and centrifuged twice (incubated for 20 minutes and centrifuged at 8000 rpm for 10 minutes). After the RNase digestion, samples were purified by phenol-chloroform-isoamyl alcohol [25:24:1 (v/v)] and further precipitated by 2 M ammonium-acetate and 2/3 (v/v) isopropanol. DNA was further washed by water using Centricon Centrifugal Filter Devices (with YM-100 MW membrane-Amicon; Millipore, Billerica, MA, USA) in order to remove polyphenols and pigments soluble in water.
Quantification of olive DNA was performed by spectrophotometar (Ultrospec 2000, Pharmacia Biotech (Biochrom) Ltd. Cambridge, UK).
Primers and microsatellite-based marker analysis
Olive specimens were characterized with the following 16 microsatellite-based markers: UDO99-008, UDO99-012, UDO99-019, UDO99-024, UDO99-028, UDO99-031, UDO99-039, UDO99-043 (11), ssrOeUA-DCA3, ssrOeUA-DCA8, ssrOeUA-DCA9, ssrOeUA-DCA10, ssrOeUA-DCA14, ssrOeUA-DCA16 (9), EMO2, and EMO3 (12).
Polymerase chain reactions were carried out in a volume of 12.5 μL, containing 1.5 mM MgCl2 for all ssrOeUA markers and EMO2 and EMO3; 2 mM MgCl2 for all UDO99 markers except UDO99-008; and 2.5 mM MgCl2 for UDO99-008 marker; 0.2 mM of each dNTP (Applied Biosystems, Foster City, CA, USA), GeneAmp 10×PCR Buffer II (1.25 μL for ssrOeUA-DCA3, ssrOeUA-DCA14, and ssrOeUA-DCA16; 1.5 μL for ssrOeUA-DCA8, ssrOeUA-DCA9, ssrOeUA-DCA10, EMO2, EMO3, UDO99-019, and UDO99-043; 1.75 μL for UDO99-008, UDO99-012, UDO99-024, UDO99-028, UDO99-031, and UDO99-039 markers; Applied Biosystems), primers, and 0.25 U AmpliTaq Gold DNA polymerase (Applied Biosystems). PCR reactions were performed in Applied Biosystems thermocyclers 9600 and 9700 under the following conditions: a step of 10 minutes at 95°C, followed by 35 cycles of 45 seconds at 94°C, 1 minute at the appropriate annealing temperature of the primer, and 1 minute at 72°C, and a final extension at 72°C for 30 minutes.
PCR products were analyzed in an automated sequencer (ABI Prism 310 Genetic Analyzer, Software v3.2, Applied Biosystems) and fragment lengths were determined using Genescan 500 Liz internal size standard (Applied Biosystems).
All PCR reactions were repeated three times if the results were perfectly concordant, and up to six times if there was a discrepancy in the first three amplifications, until obtaining at least three concordant results. Such discrepancies occurred on the average in 25% cases of total amplifications for each microsatellite-based marker, but were resolved in further three amplifications.
Data analysis
The expected heterozygosity (HE) of each locus was calculated according to the formula HE=n(1-Σpi2)/(n-1), where pi is the frequency of the i-th allele and n is the number of gene copies in the sample for the given locus (16).
Expected and observed heterozygosities were calculated considering that specimens with only one amplified product for the given locus were homozygotes for that locus. Hence, heterozygosities reported here might have been underestimated in cases when null alleles occurred.
Results
Genotyping analysis was performed on the total of 44 different olive samples, and the total of 163 amplified polymorphic products were obtained after applying 16 previously developed primer pairs used for amplification of microsatellite-based markers (Table 1). The absence of any amplified product occurred only once, ie in genotyping one cultivar applying marker UDO99-028 (Table 2).
Table 1.
Composition, size range, and number of alleles detected in 44 olive specimens; observed (Ho) and expected heterozygosity (HE)
| Locus |
Repeat motif |
Size range (bp) |
No. of polymorphic alleles |
Ho |
HE |
|---|---|---|---|---|---|
| (GA)n repeats: | |||||
| ssrOeUA-DCA3 | (HB)19 | (229-252) | 8 | 0.932 | 0.836 |
| ssrOeUA-DCA8 | (HB)18 | (124-154) | 9 | 0.932 | 0.811 |
| ssrOeUA-DCA9 | (HB)23 | (152-209) | 10 | 0.886 | 0.829 |
| ssrOeUA-DCA10 | (UB)14(HB)17 | (153-241) | 20 | 0.750 | 0.910 |
| EMO2 | (AG)5-G-(GA)10 | (184-244) | 14 | ND* | ND* |
| (CA)n repeats: | |||||
| UDO99-008 | (AC)13 | (155-166) | 7 | 0.273 | 0.792 |
| UDO99-012 | (GT)10 | (155-166) | 5 | 0.659 | 0.610 |
| UDO99-031 | (TG)21 (TATG)6 | (107-151) | 9 | 0.455 | 0.583 |
| UDO99-043 | (GT)12 | (171-219) | 13 | 0.886 | 0.746 |
| ssrOeUA-DCA14 | (DB)18B6(UBB)7 | (145-191) | 11 | ND* | ND* |
| EMO3 | (CA)7 | (203-214) | 9 | 0.841 | 0.841 |
| (GA)n and (CA)n compound repeats: | |||||
| ssrOeUA-DCA16 | (HU)13(HB)29 | (124-182) | 12 | 0.886 | 0.815 |
| (CA)n or (GT)n and (TA)n compound repeats: | |||||
| UDO99-019 | (GT)20(AT)5 | ( 99-163) | 6 | 0.636 | 0.499 |
| UDO99-024 | (CA)11(TA)2(CA)4 | (166-195) | 9 | 0.682 | 0.747 |
| UDO99-028 | (CA)23(TA)3 | (126-176; 0) | 10 | 0.886 | 0.831 |
| UDO99-039 | (AT)5(GT)11 | (106-186) | 12 | ND* | ND* |
| Total | 164 | ||||
| Average | 10.25 | 0.747 | 0.758 | ||
*Ho and HE were not determined since some specimens yielded three different amplified products.
Table 2.
The lengths of DNA sequences (bp) encompassing 16 microsatellites as a results of genotyping 44 different Croatian olive specimens*
| Markers |
| Cultivars | U-008 | U-012 | U-019 | U-024 | U-028 | U-031 | U-039 | U-043 | DCA3 | DCA14 | DCA16 | DCA8 | DCA9 | DCA10 | EMO2 | EMO3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Resulja | 165 | 155/157 | 129/143 | 172/186 | 134/151 | 108/137 | 177/180 | 175/177 | 236/250 | 170/178 | 152 | 136/154 | 164/188 | 208/217 | 184/210/225 | 203/212 |
| Grozdulja | 164 | 155/160 | 129/154 | 179/184 | 172 | 108/146 | 156/177 | 181 | 229/246 | 187 | 152 | 124/134 | 205/207 | 153/194 | 219/225/230 | 210 |
| Pačica | 165 | 155 | 129 | 172/186 | 151/172 | 148 | 180 | 175/177 | 236/246 | 170/187 | 157/161 | 136/138 | 205/207 | 163/213 | 219/225/230 | 211/212 |
| Pontoguza | 160/165 | 155/164 | 129/143 | 172/186 | 151/172 | 108 | 177/180 | 177/215 | 229/246 | 187 | 127/157 | 134/136 | 164/207 | 215/239 | 220/225 | 211/212 |
| Paštrica | 160/165 | 155/164 | 129/143 | 172/186 | 151/172 | 108 | 177/180 | 177/215 | 229/246 | 187 | 127/157 | 134/136 | 164/207 | 215/239 | 219/225 | 208/212 |
| Sitnica 1 | 160 | 155/164 | 129/143 | 172/186 | 134/172 | 108 | 178 | 179/215 | 229/236 | 184/187 | 152/157 | 124/134 | 205/207 | 159/239 | 219/225 | 203/212 |
| Sitnica 2 | 162 | 155/164 | 129/143 | 172/186 | 134/172 | 108 | 178 | 179/215 | 229/236 | 187 | 152/157 | 124/134 | 207/209 | 159/239 | 219/225 | 203/212 |
| Grozdača | 166 | 155 | 129/143 | 184/186 | 134/151 | 110 | 179/184 | 177/179 | 229/246 | 178/187 | 152/179 | 134/136 | 164/205 | 155/239 | 225 | 203/210 |
| Dužica | 166 | 155 | 129 | 166/186 | NA | 108 | 106 | 177/179 | 246/250 | 178 | 157/182 | 136 | 164/184 | 217 | 224/225 | 208 |
| Bjelica 1 | 163/165 | 155/164 | 129 | 184 | 132/153 | 108/151 | 106/177 | 177/179 | 229/240 | 153/175/187 | 152/157 | 130/134 | 205/207 | 153/237 | 219/223/230 | 210/211 |
| Bjelica 2 | 163/165 | 155/164 | 129 | 184 | 132/153 | 108/151 | 106/179 | 179 | 229/240 | 153/175/187 | 152/157 | 130/134 | 205/207 | 153/237 | 219/225/230 | 211 |
| Bjelica 3 | 163/165 | 155/164 | 129 | 184 | 132/153 | 108/151 | 106/177 | 179/181 | 229/240 | 153/175/187 | 152/157 | 130/134 | 197/207 | 153/237 | 220/225/230 | 210 |
| Mezanica 1 | 160/166 | 155/166 | 129 | 186 | 134/151 | 110 | 177/180 | 177/179 | 240/250 | 178/191 | 124/126 | 136/138 | 174/196 | 155/237 | 202/225/230 | 208/211 |
| Mezanica 2 | 160/166 | 155/166 | 129 | 186 | 134/151 | 110 | 177/180 | 177/179 | 240/250 | 178/187 | 124/126 | 136/138 | 174/196 | 155/239 | 202/224/228 | 208/211 |
| Lastovka | 155/164 | 157/164 | 99/129 | 184/188 | 134 | 108 | 106/179 | 171/219 | 229 | 187 | 152/157 | 130/138 | 188/196 | 153/195 | 211/219/244 | 210/213 |
| Uljarica 1 | 164 | 155/164 | 129/163 | 184 | 134/141 | 108/151 | 106/177 | 179/217 | 229/240 | 180/187 | 152/157 | 126/134 | 174 | 239 | 202/210/223 | 211 |
| Uljarica 2 | 164 | 155/164 | 129/163 | 184 | 134/141 | 108/151 | 106/177 | 179/217 | 229/240 | 180/187 | 152/157 | 126/134 | 174 | 239 | 201/210/223 | 211/212 |
| Uljarica 3 | 164 | 155/164 | 129/163 | 184 | 134/141 | 108/151 | 106/177 | 179/217 | 229/240 | 178/187 | 152/157 | 126/134 | 152/174 | 158/239 | 202/210/223 | 211/212 |
| Vel. Lastovka | 165 | 155/166 | 129/143 | 172/186 | 126/134 | 108 | 178/180 | 177/215 | 229/250 | 187/189 | 157/177 | 134/136 | 164/205 | 157/239 | 202/225/230 | 208/212 |
| Želudarica | 165 | 155 | 129 | 166/186 | 126/153 | 108 | 106 | 177/179 | 242/250 | 178/187 | 124/127 | 130/136 | 164/205 | 155 | 225/230 | 210/211 |
| Murgulja | 165 | 155/164 | 129/143 | 186 | 126/172 | 108 | 106/177 | 179/213 | 236/246 | 178/187 | 152 | 124/136 | 164/207 | 159/217 | 220/225 | 203/208 |
| Trsteno Olive | 163 | 157/166 | 99/129 | 181/184 | 176 | 108/137 | 175/177 | 189/219 | 242/252 | 145/153/175 | 124/163 | 130/140 | 188/207 | 153/175 | 210/225/230 | 212/214 |
| Zuzorka | 160 | 155/164 | 129/143 | 172/186 | 134/172 | 108 | 178 | 179/215 | 229/236 | 187 | 152/157 | 124/134 | 205/207 | 159/241 | 219/225/230 | 203/211 |
| Žabarka | 165 | 155 | 129 | 186/195 | 134/172 | 108 | 144/177 | 175/179 | 229/250 | 153/176/187 | 177 | 134/140 | 164/188 | 220 | 220/225/240 | 208 |
| Levantinka | 166 | 155 | 129 | 186 | 151/153 | 110/151 | 177/186 | 179/183 | 236 | 178/187 | 152/176 | 124/134 | 164/207 | 177/215 | 225/230 | 208/211 |
| Jeruzalemka | 165 | 155 | 99/129 | 172/186 | 172 | 108/137 | 123/138 | 175/179 | 229/246 | 145/178 | 152/157 | 136/140 | 184/207 | 159/217 | 220 | 208/212 |
| Crnica | 166 | 155/164 | 129/143 | 186 | 151/172 | 108 | 106/179 | 177/179 | 236/246 | 187 | 152/176 | 124/136 | 196/207 | 155/239 | 202/219/230 | 203/208 |
| Lumbardeška 1 | 165 | 155 | 129/143 | 172/186 | 134/151 | 108 | 177/180 | 177/179 | 236/250 | 178/187 | 152/177 | 134/136 | 164/205 | 217 | 225/230 | 208/212 |
| Lumbardeška 2 | 165 | 155 | 129/143 | 171/186 | 134/151 | 108 | 177/180 | 177/179 | 236/250 | 178/187 | 152/177 | 134/136 | 164/205 | 217 | 225 | 208/212 |
| Konavoska | 166 | 155/164 | 129/143 | 186 | 134/151 | 108 | 106/177 | 177/179 | 236/250 | 187 | 157/177 | 136 | 196/205 | 155/159 | 202/225/230 | 210 |
| Dubravka | 165 | 155 | 129 | 166/186 | 126/153 | 108 | 106 | 177/179 | 242/250 | 178/187 | 124/127 | 130/136 | 164/205 | 155 | 225/230 | 210/211 |
| Krvavica | 166 | 155/164 | 129/143 | 186 | 126/172 | 108 | 106/177 | 179/211 | 236/248 | 178/187 | 127/152 | 124/136 | 164/207 | 159/215 | 220/225 | 203/208 |
| Polituša | 166 | 155 | 113/129 | 172/186 | 151/172 | 107/148 | 180 | 175/177 | 236/246 | 170/187 | 157/161 | 136/138 | 205/207 | 163/215 | 220/225/230 | 211/212 |
| Puizica 1 | 164 | 157/166 | 129 | 166/186 | 134/162 | 124/151 | 177 | 179 | 240/252 | 170/187 | 146/152 | 134/148 | 174 | 169/237 | 202/224 | 206/212 |
| Puizica 2 | 164 | 157/166 | 129 | 166/186 | 134/162 | 124/151 | 177 | 179 | 240/252 | 170/187 | 152 | 134/148 | 174 | 169/237 | 202/223 | 206/212 |
| Šljivača | 166 | 155 | 113/129 | 172/186 | 151/172 | 107/148 | 180 | 179 | 236/246 | 170/187 | 157/161 | 136/138 | 205/207 | 163/215 | 219/225/230 | 210/211 |
| Piculja 1 | 155/164 | 157/166 | 129 | 166/181 | 153/172 | 108/144 | 106/123/138 | 179/185 | 229/242 | 170/187 | 124/146 | 124/130 | 174/205 | 153/212 | 202/225/230 | 206/209 |
| Piculja 2 | 155/164 | 157/166 | 129 | 166/181 | 153/172 | 108/151 | 106/123/138 | 179/185 | 229/242 | 170/187 | 124/146 | 124/130 | 174/205 | 153/215 | 202/225/230 | 206/209 |
| Piculja 3 | 155/164 | 157/166 | 129 | 166/181 | 153/172 | 108/151 | 106/123/138 | 179/185 | 229/242 | 170/187 | 124/146 | 124/130 | 174/205 | 153/212 | 202/225/230 | 206/209 |
| Piculja 4 | 155/164 | 157/166 | 113/129 | 166/181 | 153/172 | 108/151 | 106/123/138 | 179/185 | 229/242 | 170/187 | 124/146 | 124/130 | 174/197 | 153/212 | 202/225/230 | 206/209 |
| Šipan Mezanica | 164 | 155/164 | 129/163 | 184 | 134/141 | 108/151 | 106/177 | 179/217 | 229/240 | 180/187 | 152/157 | 126/134 | 174 | 158/239 | 202/210/223 | 211/212 |
| Oblica 1 | 166 | 155 | 129/143 | 172/186 | 134/151 | 108 | 177/180 | 177/179 | 236/250 | 178/187 | 152/176 | 134/136 | 164/205 | 217 | 225/230 | 208/211 |
| Oblica 2 | 166 | 155 | 129/143 | 172/186 | 134/151 | 108 | 177/180 | 177/179 | 250 | 178/187 | 152/176 | 134/136 | 164/205 | 217 | 225 | 208/212 |
| Oblica 3 | 166 | 155 | 129/143 | 172/186 | 134/151 | 108 | 177/180 | 177/179 | 236/250 | 178/187 | 152/176 | 134 | 164/205 | 217 | 225/230 | 208/212 |
*Abbreviations: U-No. refer to UDO99-No.; DCANo. refer to ssrOeUA-DCANo.; NA – no amplified product.
The allele size ranges and the total number of alleles, as well as the expected and observed heterozygosities for each locus are presented in Table 1. The average expected heterozygosity amounted to 0.758.
An electropherogram of microsatellite-based olive DNA, serving as an illustration of the amplified products obtained in this study, is demonstrated in the Figure 2.
Figure 2.
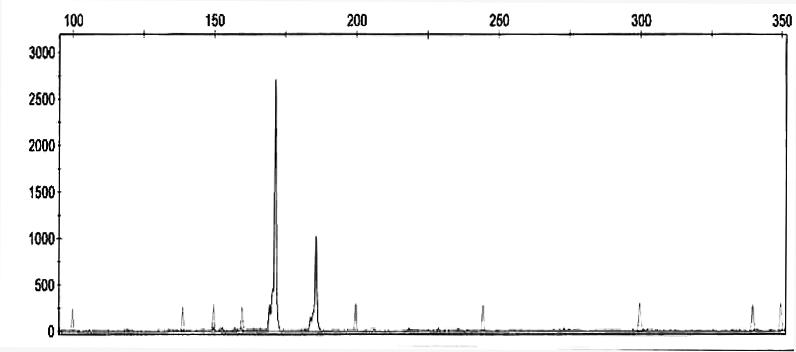
The electropherogram of olive DNA obtained in this study by applying UDO99-024 microsatellite-based marker (internal size standard – LIZ 500). X-axis indicates fragment lenght (bp), while Y-axis indicates relative flourescent units (RFU) that are proportional to the amount of PCR product.
Three microsatellite-based primer pairs (UDO99-039, ssrOeUA-DCA14, and EMO2) amplified three different products for some cultivars (UDO99-039 for all four Piculja varieties, ssrOeUA-DCA14 for all three Bjelica varieties, as well as for two cultivars – Trsteno Olive and Žabarka, and EMO2 for 25 out of 44 specimens), thus increasing their power of discrimination of genotyped olive specimens.
Discussion
By genotyping more than one different specimen of the same olive cultivar denominations we have demonstrated that it is possible to distinguish among different intracultivar varieties by means of a set of 16 microsatellite-based markers.
In our study, 164 polymorphic alleles were characterized using 16 microsatellite-based markers over 44 different specimens and 30 denominations defined previously by agronomic and morphological means (17,18). In comparison, in the study employing 14 microsatellite-based markers over 130 specimens comprising 67 different denominations, 135 alleles were detected (19), while in the study employing 12 microsatellite-based markers over 50 specimens comprising 34 different cultivars, 119 alleles were detected (20). Furthermore, in the study applying 15 microsatellite-based markers over 47 cultivars, 124 alleles were detected (9), while in the study applying 7 microsatellite-based markers over 23 cultivars, 45 polymorphic alleles were detected (12). Finally, in our study the average expected heterozygosity amounted to 0.758, while in other studies it amounted to 0.679 (19), 0.760 (20), 0.693 (9), 0.648 (12), and 0.681 (13). Hence, comparable levels of genetic variability were observed in other studies as well. In addition, we showed that the applied set of microsatellite-based markers efficiently resolved all the cases of intracultivar variability, which points to a high potential of 16 chosen microsatellite-based markers in revealing olive genetic diversity.
The mechanism of the occurrence of three different microsatellite-based alleles amplified by one primer pair is not elucidated, but might be ascribed to chromosome rearrangements (21), genome fusions (22,23), or “chimerism” (24).
Microsatellite-based olive genotyping application is justified in solving criminal and civil cases by its ability to assign olive DNA to an individual tree and at the same time by its capability of yielding the same molecular profile for the same tree due to the olive microsatellite somatic stability.
General reproducibility of microsatellite-based genotyping data among laboratories should result in a comprehensive olive genotyping database that might be searched in case of a need to assign the origin of the found plant material connected to crime or accident or suicide scene.
In conclusion, we demonstrated that Croatian olive cultivar genotyping using 16 microsatellite-based markers may provide the possibility of olive specimen identification up to the individual tree and may open the possibility of their successful application in forensic investigations.
Acknowledgements
This research was supported by a grant from the Croatian Ministry of Science, Education, and Sports (grant No. 141-2160800-0333). We gratefully acknowledge the provision of samples by Franko Marinović and Antun Kotlar from the Croatian Agricultural Extension Institute (CAEI) of Dubrovačko-neretvanska County. Equipment was provided by the Laboratory of Forensic and Clinical Genetics, headed by Prof Šimun Anđelinović, Split university Hospital.
Disclaimer
Snježana Štambuk is an employee of SMS – Food Development Center, Klis, Croatia. SMS - Food Development Center participated to the financing of the experimental work, organized sampling and contributed to designing the work.
References
- 1.Miller Coyle H, Ladd C, Palmbach T, Lee HC. The Green Revolution: botanical contributions to forensics and drug enforcement. Croat Med J. 2001;42:340–5. [PubMed] [Google Scholar]
- 2.Yoon CK. Forensic science. Botanical witness for the prosecution. Science. 1993;260:894–5. doi: 10.1126/science.8493521. [DOI] [PubMed] [Google Scholar]
- 3.Korpelainen H, Virtanen V. DNA fingerprinting of mosses. J Forensic Sci. 2003;48:804–7. [PubMed] [Google Scholar]
- 4.Craft KJ, Owens JD, Ashley MV. Application of plant DNA markers in forensic botany: genetic comparison of Quercus evidence leaves to crime scene trees using microsatellites. Forensic Sci Int. 2007;165:64–70. doi: 10.1016/j.forsciint.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Miller Coyle H, Lee CL, Lin WY, Lee HC, Palmbach TM. Forensic botany: using plant evidence to aid in forensic death investigation. Croat Med J. 2005;46:606–12. [PubMed] [Google Scholar]
- 6.Jobling MA, Gill P. Encoded evidence: DNA in forensic analysis. Nat Rev Genet. 2004;5:739–51. doi: 10.1038/nrg1455. [DOI] [PubMed] [Google Scholar]
- 7.Bartolini G, Prevost G, Messeri C, Carignani C. Olive germplasm: cultivars and world-wide collections. Rome: Seed and Plant Genetic Resources Service, FAO; 1998. Available from: http://www.fao.org/waicent/faoinfo/agricult/agp/agps/seed/oliv.htm. Accessed: July 4, 2007.
- 8.Rallo P, Dorado G, Martin A. Development of simple sequence repeats (SSRs) in olive tree (Olea europaea L.). Theor Appl Genet. 2000;101:984–9. [Google Scholar]
- 9.Sefc KM, Lopes MS, Mendonça D, Dos Santos MR, Da Câmara Machado ML, Da Câmara Machado A. Identification of microsatellite loci in olive (Olea europaea) and their characterization in Italian and Iberian olive trees. Mol Ecol. 2000;9:1171–3. doi: 10.1046/j.1365-294x.2000.00954.x. [DOI] [PubMed] [Google Scholar]
- 10.Carriero F, Fontanazza G, Cellini F, Giorio G. Identification of simple sequence repeats (SSRs) in olive (Olea europaea L.). Theor Appl Genet. 2002;104:301–7. doi: 10.1007/s001220100691. [DOI] [PubMed] [Google Scholar]
- 11.Cipriani G, Marrazzo MT, Marconi R, Cimato A, Testolin R. Microsatellite markers isolated in olive (Olea europaea L.) are suitable for individual fingerprinting and reveal polymorphism within ancient cultivars. Theor Appl Genet. 2002;104:223–8. doi: 10.1007/s001220100685. [DOI] [PubMed] [Google Scholar]
- 12.de la Rosa R, James CM, Tobutt KR. Isolation and characterization of polymorphic microsatellites in olive (Olea europaea L.) and their transferability to other genera in the Oleaceae. Mol Ecol Notes. 2002;2:265–7. [Google Scholar]
- 13.Sabino Gil F, Busconi M, Da Câmara Machado A, Fogher C. Development and characterization of microsatellite loci from Olea europaea. Mol Ecol Notes. 2006;6:1275–7. [Google Scholar]
- 14.Diaz A, De la Rosa R, Martin A, Rallo P. Development, characterization and inheritance of new microsatellites in olive (Olea europaea L.) and evaluation of their usefulness in cultivar identification and genetic relationship studies. Tree Genet Genomes. 2006;2:165–75. [Google Scholar]
- 15.Busconi M, Foroni C, Corradi M, Bongiorni C, Cattapan F, Fogher C. DNA extraction from olive oil and its use in the identification of the production cultivar. Food Chem. 2003;83:127–34. [Google Scholar]
- 16.Nei M, Roychoudhury AK. Sampling variances of heterozygosity and genetic distance. Genetics. 1974;76:379–90. doi: 10.1093/genetics/76.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakarić P. Olive species in the Dubrovnik littoral [in Croatian]. Dubrovnik: Bakarić; 2002. [Google Scholar]
- 18.Bakarić P. Autochthonous olive species on the Elafiti islands [in Croatian]. Dubrovnik: Bakarić; 2007. [Google Scholar]
- 19.Lopes MS, Mendonça D, Sefc KM, Gil FS, Da Câmara Machado A. Genetic evidence of intra-cultivar variability within Iberian olive cultivars. HortScience. 2004;39:1562–5. [Google Scholar]
- 20.La Mantia M, Lain O, Caruso T, Testolin R. SSR-based DNA fingerprints reveal the genetic diversity of Sicilian olive (Olea europaea L.) germplasm. J Hortic Sci Biotechnol. 2005;80:628–32. [Google Scholar]
- 21.Minelli S, Maggini F, Gelati MT, Angiolillo A, Cionini PG. The chromosome complement of Olea europaea L.: characterization by differential staining of the chromatin and in-situ hybridization of highly repeated DNA sequences. Chromosome Res. 2000;8:615–9. doi: 10.1023/a:1009286008467. [DOI] [PubMed] [Google Scholar]
- 22.Buteler MI, Jarret RL, LaBonte DR. Sequence characterization of microsatellites in diploid and polyploidy Ipomoea. Theor Appl Genet. 1999;99:123–32. [Google Scholar]
- 23.Belaj A, Satovic Z, Cipriani G, Baldoni L, Testolin R, Rallo L, et al. Comparative study of the discriminating capacity of RAPD, AFLP and SSR markers and of their effectiveness in establishing genetic relationships in olive. Theor Appl Genet. 2003;107:736–44. doi: 10.1007/s00122-003-1301-5. [DOI] [PubMed] [Google Scholar]
- 24.Franks T, Botta R, Thomas MR, Franks J. Chimerism in grapevines: implications for cultivar identity, ancestry and genetic improvement. Theor Appl Genet. 2002;104:192–9. doi: 10.1007/s001220100683. [DOI] [PubMed] [Google Scholar]