Abstract
Aim
To provide a screening tool to reduce time and sample consumption when attempting mtDNA haplogroup typing.
Methods
A single base primer extension assay was developed to enable typing, in a single reaction, of twelve mtDNA haplogroup specific polymorphisms. For validation purposes a total of 147 samples were tested including 73 samples successfully haplogroup typed using mtDNA control region (CR) sequence data, 21 samples inconclusively haplogroup typed by CR data, 20 samples previously haplogroup typed using restriction fragment length polymorphism (RFLP) analysis, and 31 samples of known ancestral origin without previous haplogroup typing. Additionally, two highly degraded human bones embalmed and buried in the early 1950s were analyzed using the single nucleotide polymorphisms (SNP) multiplex.
Results
When the SNP multiplex was used to type the 96 previously CR sequenced specimens, an increase in haplogroup or macrohaplogroup assignment relative to conventional CR sequence analysis was observed. The single base extension assay was also successfully used to assign a haplogroup to decades-old, embalmed skeletal remains dating to World War II.
Conclusion
The SNP multiplex was successfully used to obtain haplogroup status of highly degraded human bones, and demonstrated the ability to eliminate possible contributors. The SNP multiplex provides a low-cost, high throughput method for typing of mtDNA haplogroups A, B, C, D, E, F, G, H, L1/L2, L3, M, and N that could be useful for screening purposes for human identification efforts and anthropological studies.
Mitochondrial DNA (mtDNA) is a 16 569 bp circular molecule present, on average, in 500 copies per cell (1). MtDNA analysis is utilized in several areas of science including, but not limited to, anthropology, evolutionary studies, and forensic science (2-5). The high copy number, and possibly the cellular location and molecular features of mtDNA, allow for increased recovery, thus providing a distinct advantage over nuclear DNA when working with highly compromised samples (6). The maternal inheritance and high mutation rate are characteristics extremely useful for evolutionary studies (7,8); in fact, mtDNA has been used to resolve evolutionary questions related to extinct species and to human migrations throughout the continents (9-12). The field of forensic science also relies upon mtDNA to identify missing persons, locate maternal relatives, identify victims in mass disasters, and, in some situations, include an individual at a crime scene (13-19).
Early studies of the mtDNA genome revealed patterns of variation that were linked to geographic regions. Individuals with the same sequence variations were clustered into haplogroups defined by mutations at particular nucleotide positions (20-27). A closer examination of the mtDNA genomes of various populations led to the following assumptions: 1) several of the mtDNA mutations are highly correlated with the ethnic and geographic origin of the individual, 2) all mutations originated from a single mtDNA tree, and 3) the greatest variation and deepest root of the mtDNA tree is present in the African population. Furthermore, a calculation of the variation between mtDNA haplogroups demonstrated that 35% of the mutations were continent-specific, and therefore useful indicators of geographic origin (24,25,28-31).
Before the advent of modern sequencing methods, the primary approach to identifying polymorphic sites throughout the mtDNA coding region was restriction fragment length polymorphism (RFLP) analysis. While this methodology is still utilized in certain contexts, direct sequencing of the mtDNA molecule is rapidly gaining acceptance as the method of choice for haplogroup typing (20,23,30,32-35). Single base primer extension, also known as minisequencing, is an example of a direct sequencing technique that is currently utilized for mtDNA haplogroup typing (36-42). This methodology, described in detail by Fiorentino et al (43), offers several advantages to the investigator over RFLP and conventional sequence analysis methods including the use of small amplicons (<150 bp), increased sensitivity and robustness, and multiplexing capability. Multiplexing capability is particularly important, especially in regard to forensic DNA analysis, as it reduces sample consumption while increasing throughput of sample processing and data analysis. Increased sensitivity allows for improved amplification success with DNA samples that contain limited starting template. Additionally, the possibility for high throughput processing can aid in population screening studies in situations where numerous samples need to be typed (44). This is particularly true in mass disaster or other mass screening situations, where a simple and rapid population screening tool that consumes little extract could effectively direct subsequent identification testing. In these situations, coding region sequencing would be expensive and time-consuming, and the subsequent data analysis a lengthy, burdensome, and potentially error-prone process (42,45-48). Furthermore, the possibility of obtaining interpretable results from poor quality polymerase chain reaction (PCR) products while simultaneously typing several polymorphisms throughout the mtDNA genome make it a more feasible method than conventional PCR fragment sequence analysis, especially in forensic cases and anthropological studies involving highly degraded or otherwise compromised human remains (16,36-38,49,50).
In this article, we describe the development of a multiplex assay designed to simultaneously type twelve polymorphic positions located throughout the coding region of the mtDNA genome for the identification of haplogroups A, B, C, D, E, F, G, H, I, L1/L2, L3, M, N, and X. The intended utility of this assay is haplogroup typing of highly degraded human remains for either forensic casework or as a low cost, high throughput alternative for screening anthropological specimens. Validation of this assay included the analysis of 20 samples previously haplogroup typed by RFLP, and 94 samples for which haplogroup had been inferred based on control region (CR) sequence data. Additionally, and to evaluate the potential application of the assay for population screening, 31 samples were tested for which only the population of origin, and not the mtDNA haplogroup, was known. Finally, to verify robustness and sensitivity of the assay, we also tested two highly degraded human bones embalmed and buried in the early 1950s.
Methods
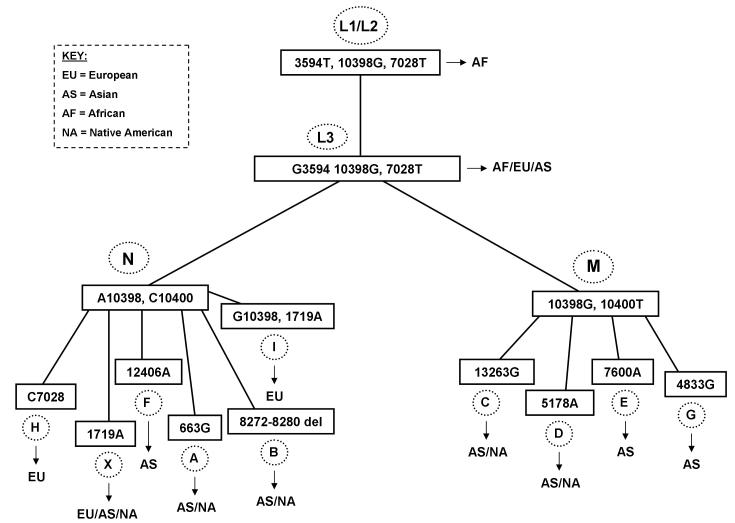
A review of the relevant literature (20-27,33,34), which collectively utilized 2500 fully sequenced mtDNA coding regions to generate haplogroup phylogenies, was the foundation for the selection of the twelve single nucleotide polymorphisms (SNP) included in this assay. The intention was to maximize the number of haplogroups that could be identified using a specific set of polymorphisms (Figure 1). Table 1 summarizes the expected results from each haplogroup typed by the assay. The SNPs were also selected for their ability to discriminate among the major ancestral lineages (Europeans, Africans, Asians, and Native Americans) by examining only the coding region of the mitochondrial genome (24,25,28-31).
Figure 1.
Visual representation of the diagnostic polymorphisms used to identify the haplogroup and ancestry of an individual using the SNP assay. The presence of the polymorphism(s) displayed in boxes leads to the identification of the circled haplogroup. Inferred ancestry into at least one of the four lineages is established from the haplogroup status.
Table 1.
Expected single nucleotide polymorphisms typing results and inferred ancestries for each haplogroup represented by the assay
| Nucleotide position and base* |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplogroup | 8272-8280 del | 13263 | 1719 | 5178 | 663 | 10398 | 10400r† | 3594r | 7028r | 12406 | 4833 | 7600 | Inferred ancestry |
| A | C | A | G | C | B | A | C | C | T | G | A | G | Asian/Native American |
| B | G | A | G | C | A | A | C | C | T | G | A | G | Asian/Native American |
| C | C | G | G | C | A | G | T | C | T | G | A | G | Asian/Native American |
| D | C | A | G | A | A | G | T | C | T | G | A | G | Asian/Native American |
| E | C | A | G | C | A | G | T | C | T | G | A | A | Asian |
| F | C | A | G | C | A | A | C | C | T | A | A | G | Asian |
| G | C | A | G | C | A | G | T | C | T | G | G | G | Asian |
| H | C | A | G | C | A | A | C | C | C | G | A | G | European |
| I | C | A | A | C | A | G | C | C | T | G | A | G | European |
| L1/L2 | C | A | G | C | A | G | C | T | T | G | A | G | African |
| L3 | C | A | G | C | A | G | C | C | T | G | A | G | African/European/Asian |
| M | C | A | G | C | A | G | T | C | T | G | A | G | Asian/Native American |
| N | C | A | G | C | A | A | C | C | T | G | A | G | European/Asian/Native American |
| X | C | A | A | C | A | A | C | C | T | G | A | G | European/Asian/Native American |
*Bases in bold are different than the Cambridge reference sequence (haplogroup H) and are used to differentiate haplogroup status.
†r – the single base extension primer is in the reverse orientation.
Primer design
Amplification and minisequencing primers were designed using the Primer Express Version 2.0 software. Primers with comparable Tm and GC content properties and similar primer lengths were selected. Primer specificity was tested using the National Center for Biotechnology Information (NCBI) nucleotide-BLAST search to eliminate the possibility of non-specific products during PCR. The PCR primers were designed to satisfy three criteria – 1) must flank the desired SNP site, 2) must produce an amplicon no greater than 110 bp in length, and 3) the amplicon must retain the minisequencing primer annealing site for proper single base extension reaction (Table 2). The single base extension primers were designed one base contiguous to the polymorphic site of interest in either the forward or reverse orientation. Additionally, variable length polymeric-T tails were added to the 5′ end of the primer in order to separate the products during electrophoresis (Table 3).
Table 2.
Nucleotide position of the polymorphism, oligonucleotide sequence (5′ → 3′), primer orientation, primer length (bp), amplicon length (bp), Tm (°C), GC content (%), and final concentration (nM) for each primer used during polymerase chain reaction amplification.
| Nucleotide position | Primer sequence (5′ → 3′) | Orientation | Primer length (bp) | Amplicon length (bp) | Tm (°C) | GC content (%) | Final (nM) |
|---|---|---|---|---|---|---|---|
| 8272-8280 del | TAAAAATCTTTGAAATAGGGCCC | F | 23 | 89 (del) 80 | 51.4 | 34.7 | 200 |
| GTTAATGCTAAGTTAGCTTTACAGTGG | R | 27 | 54.2 | 37 | |||
| 13263 | CAAAAAAATCGTAGCCTTCTCC | F | 22 | 67 | 52.2 | 40.9 | 150 |
| GTTGATGCCGATTGTAACTATTATG | R | 25 | 52.3 | 36 | |||
| 1719 | CCCACTCCACCTTACTACCAGA | F | 22 | 84 | 57.7 | 54.5 | 500 |
| TGCGCCAGGTTTCAATTT | R | 18 | 53.1 | 44.4 | |||
| 5178 | TAAACTCCAGCACCACGACC | F | 20 | 79 | 57.2 | 55 | 200 |
| GTGGATGGAATTAAGGGTGTTAG | R | 23 | 53.3 | 43.4 | |||
| 663 | ACATCACCCCATAAACAAATAGG | F | 23 | 108 | 52.9 | 39.1 | 200 |
| TGGTGATTTAGAGGGTGAACTCA | R | 23 | 55.6 | 43.4 | |||
| 10398/10400* | AGTCTGGCCTATGAGTGACTAC | F | 22 | 86 | 55.5 | 50 | 500 |
| AATGAGTCGAAATCATTCGTTT | R | 22 | 50.7 | 31.8 | |||
| 3594 | CTTAGCTCTCACCATCGCTCT | F | 21 | 90 | 56.2 | 52.3 | 300 |
| AGAATAAATAGGAGGCCTAGGTTG | R | 24 | 53.9 | 41.6 | |||
| 7028 | TATTAGCAAACTCATCACTAGACA TCGT | F | 28 | 96 | 55.7 | 35.7 | 200 |
| TGGCAAATACAGCTCCTATTGA | R | 22 | 54 | 40.9 | |||
| 12406 | AATTCCCCCCATCCTTACC | F | 19 | 78 | 54.2 | 52.6 | 300 |
| GCGACAATGGATTTTACATAATG | R | 23 | 50.6 | 34.7 | |||
| 4833 | AATAGCCCCCTTTCACTTCTG | F | 21 | 72 | 54.7 | 47.6 | 400 |
| AGAAGAAGCAGGCCGGA | R | 17 | 56.4 | 58.8 | |||
| 7600 | GGCTAAATCCTATATATCTTAATGGCA | F | 27 | 64 | 52.6 | 33.3 | 100 |
| GGGAAGTAGCGTCTTGTAGACC | R | 22 | 59.9 | 54.5 |
*The 10398 and 10400 polymorphisms are located on the same amplicon.
Table 3.
Nucleotide position of the polymorphism, sequence of the single base extension primer including polymeric T-stretch (5′ → 3′), orientation of the primer, primer length (bp), Tm of primer (°C), GC content of primer (%), final concentration (nM) of each minisequencing primer, and the expected base substitution observed at the polymorphic site*
| Nucleotide position |
Primer sequence |
Orientation |
Primer length (bp) |
Tm (°C) |
GC content (%) |
Final (nM) |
Base substitution |
|---|---|---|---|---|---|---|---|
| (5′ → 3′) | |||||||
| 8272-8280 del | CCCTATAGCACCCCCTCTA | F | 19 | 54.9 | 57.8 | 84 | C>G |
| 13263 | (3-poly-T tail)-TAGCCTTCTCCACTTCAAGTCA | F | 25 | 56.3 | 40.0 | 83 | A>G |
| 1719 | (9-poly-T tail)-CACTCCACCTTACTACCAGACAAC | F | 33 | 59.2 | 36.3 | 292 | G>A |
| 5178 | (13-poly-T tail)-CTACTATCTCGCACCTGAAACAAG | F | 37 | 58.5 | 29.7 | 167 | C>A |
| 663 | (19-poly-T tail)-CCATAAACAAATAGGTTTGGTCCT | F | 43 | 58.8 | 20.9 | 84 | A>G |
| 10398 | (21-poly-T tail)-GAGTGACTACAAAAAGGATTAGACTGA | F | 48 | 59.7 | 20.8 | 292 | A>G |
| 10400 | (24-poly-T tail)-TTCGTTTTGTTTAAACTATATACCAATTC | R | 53 | 58.4 | 13.2 | 292 | C>T |
| 3594 | (29-poly-T tail)-TAGGAGGCCTAGGTTGAGGTT | R | 58 | 62.2 | 20.6 | 167 | C>T |
| 7028 | (33-poly-T tail)-CCTATTGATAGGACATAGTGGAAGTG | R | 63 | 62.6 | 20.6 | 84 | C>T |
| 12406 | (50-poly-T tail)-CCCATCCTTACCACCCTC | F | 68 | 63.5 | 16.1 | 167 | G>A |
| 4833 | (54-poly-T tail)-CCAGAGGTTACCCAAGGC | F | 73 | 64.7 | 16.4 | 292 | A>G |
| 7600 | (51-poly-T tail)-TATCTTAATGGCACATGCAGC | F | 78 | 64.1 | 12.8 | 166 | G>A |
*For the single base extension primers designed in the reverse orientation the recorded base substitution remains in the forward orientation to facilitate interpretation
Multiplex PCR
The simultaneous amplification of the twelve amplicons containing the polymorphic sites of interest was carried out in a total volume of 50 μL. The reaction was conducted using the following reagents and concentrations: 0.05 U/μL of AmpliTaq Gold® DNA polymerase (Applied Biosystems, Foster City, CA, USA), 1X GeneAmp® PCR Gold Buffer (Applied Biosystems), 2 mM MgCl2 (Applied Biosystems), 200 μM each dNTP (Roche, Mannheim, Germany), 2 μL DNA extract, and 7.6 μL of DNA grade dH2O. The genomic DNA concentration of the samples varied between 0.2 to 1 ng/μL. The final concentrations of the PCR primers are listed in Table 2.
Thermocycling conditions followed a “reverse touch down” program adapted for single base extension assays by Vallone et al (35). The conditions were as follows: 95°C for 11 minutes, 3 cycles of 95°C for 30 seconds, 50°C for 55 seconds, 72°C for 30 seconds, 19 cycles of 95°C for 30 seconds, 50°C for 55 seconds +0.2°C per cycle, 72°C for 30 seconds, 11 cycles of 95°C for 30 seconds, 55°C for 55 seconds, 72°C for 30 seconds, 72°C for 7 minutes, and storage at 4°C. PCR amplification was carried out using GeneAmp® PCR System 9700 thermocyclers (Applied Biosystems), followed by agarose gel electrophoresis of the PCR product to verify that proper amplification occurred. Purification of 5 μL of PCR product was performed using 10 U of Exonuclease I (EXO) (USB, Cleveland, OH, USA) and 1 U of Shrimp Alkaline Phosphatase (SAP) (Roche Diagnostics Corporation, Indianapolis, IN, USA) enzymes in a final volume of 7 μL. The thermocycling conditions for EXO-SAP purification were as follows: 70 minutes at 37°C followed by 20 minutes at 72°C.
Single base extension and detection of the twelve polymorphic sites
Single base primer extension was carried out using 2 μL of the SNaPshot® Ready Reaction Mix (Applied Biosystems), 2 μL purified PCR product, and 10 µL minisequencing primer mix in a 12 μL final volume. The final concentrations of the single base extension primers are listed in Table 3. The thermocycling conditions were as follows: 25 cycles of 95°C for 10 seconds, 55°C for 5 seconds, 60°C for 30 seconds, and storage at 4°C. Samples were then purified with 1 unit of SAP for 70 minutes at 37°C, and 20 minutes at 72°C.
The SNP multiplex panel was developed in the laboratories of the Department of Forensic Sciences at The George Washington University using the ABI Prism® 310 Genetic Analyzer. Purified minisequencing products were prepared for capillary electrophoresis on the ABI Prism® 310 Genetic Analyzer by adding 1µL of product to a 0.5 mL tube containing 11.85 µL of HIDI Formamide (Applied Biosystems) and 0.15 µL of Genescan-120 LIZ Size Standard (Applied Biosystems). The samples were denatured at 95°C for 3 minutes and cooled at 4°C. The samples were then loaded on the ABI Prism® 310 Genetic Analyzer using a 47 cm × 50 μM capillary filled with denaturing performance optimized polymer POP-4 (Applied Biosystems). The run parameters on ABI Prism® 310 Collection version 2.5 5-Dye Chemistry Software were as follows: 5-dye chemistry set up with module GS STR POP4 (1mL) E5, 5-second injection time, 15 kV injection and run voltage, 60°C run temperature, and a 15-minute run time. The data was then analyzed using the Genescan Version 3.7 software (Applied Biosystems).
Additional testing conducted at the Armed Forces DNA Identification Laboratory (AFDIL) was carried out using the ABI Prism® 3130 Genetic Analyzer utilizing the run parameters described in Vallone et al (35). Analysis was conducted using the Genemapper software (Applied Biosystems) with a custom panel and bin set to facilitate results interpretation. In addition, AFDIL results were analyzed with a custom-designed Excel macro that enabled automatic haplogroup typing from GeneMapper export files.
DNA samples
In all, 147 samples were analyzed to evaluate the ability of the assay to properly assign haplogroups. The set of DNA samples tested included 20 extracts from individuals previously typed using RFLP analysis (data from Dr Theodore Schurr, Pennsylvania State University), 73 samples for which the haplogroup was inferred based on the control region (CR) sequence data (Armed Forces DNA Identification Laboratory, Rockville, MD, USA, unpublished data), 21 AFDIL samples for which the CR-inferred haplogroup was inconclusive, and 31 extracts with no haplogroup assigned but of known ancestral origin (11 Europeans, 9 Africans, and 11 Asians). To evaluate the ability of the assay to obtain results at low concentrations, progressive dilutions, ranging from 1 to 0.007 ng of genomic DNA, were tested with the multiplex SNP panel. Additionally, to evaluate the assay’s potential application in the forensic field, analysis of two extracts obtained from World War II era skeletal remains was conducted at the AFDIL. Sample 1 was a human femur whose potential origin was an American soldier killed in 1945 in a plane crash on Negros Island, Philippines (Figure 2). Sample 2 was a human femur from one of four men lost in a 1943 plane crash in New Guinea. Both samples yielded no genomic DNA when quantitated with real time PCR.
Figure 2.
Human femur (sample 1) allegedly collected near a 1945 plane crash site in the Philippine Islands.
Results
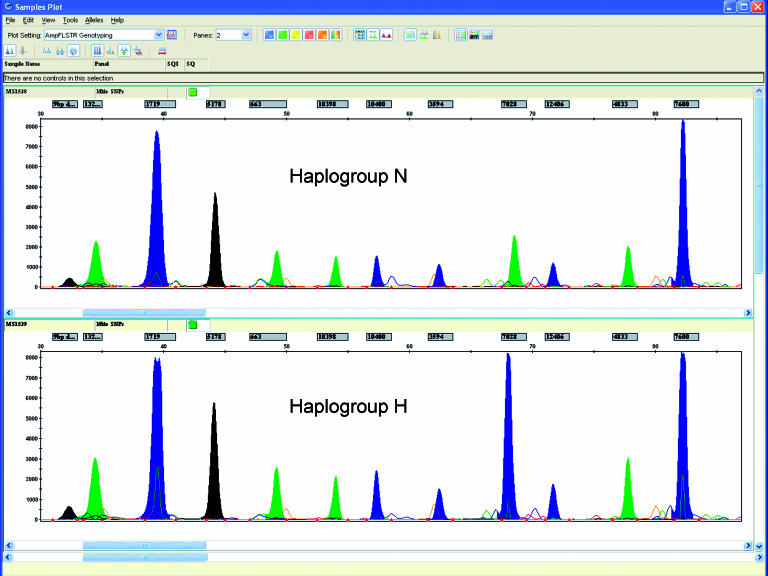
The order of migration of the primers and possible nucleotides incorporated is as follows (“F” and “R” represent the forward or reverse orientation of the primers, respectively): 9 bp deletion (C→G) F, 13263 (A→G) F, 1719 (G→A) F, 5178 (C→A) F, 663 (A→G) F, 10398 (A→G) F, 10400 (G→A) R, 3594 (G→A) R, 7028 (G→A) R, 12406 (G→A) F, 4833 (A→G) F, and 7600 (G→A) F (Figure 3). The primers designed in the reverse orientation incorporate the reverse complement nucleotide; therefore, the complementary base pair of the nucleotide incorporated during the reaction represents the nucleotide compared to the Cambridge reference sequence (CRS). As a result of the variable polymeric-T tails, the primers exhibited sufficient separation during electrophoresis to allow for simple identification and interpretation of the peaks.
Figure 3.
Electropherogram representing macrohaplogroup N and Cambridge Reference Sequence haplogroup H are included to demonstrate the ability to detect polymorphic sequences. The x-axis represents the size (bp) of the primer with the incorporated nucleotide, while the y-axis corresponds to the relative fluorescent unit (RFU) of the peak. Each fluorescent dye corresponds to a different nucleotide where blue represents G, green represents A, yellow (depicted here as black for better visual contrast) represents C, and red represents T.
The ideal genomic DNA concentration range for the initial PCR reaction is between 0.2 and 0.5 ng/µL. Sensitivity tests indicated though that sufficient signal strength (300 relative fluorescent units and greater) could be obtained down to 0.007 ng of input genomic DNA. Adding more than 1 ng of genomic DNA to the reaction frequently resulted in off-scale peaks; however these generally did not compromise data interpretation. In the few cases in which the signal strength was high enough to complicate analysis, a reduced injection time was sufficient for successful troubleshooting.
The 20 samples with known ancestral origin and haplogroup designation revealed consistent results, with a single exception. One sample previously typed as haplogroup D (5178A) using the RFLP system exhibited an additional mutation (1719A) found among individuals belonging to haplogroup X and I (data not shown).
Of the 73 AFDIL samples for which a haplogroup was inferred based on CR sequence data, 59 samples gave identical results with the SNP typing. Of the 14 samples that did not give identical results, 3 samples were SNP typed as the same macrohaplogroup rather than the more specific haplogroup assignment based on the CR data, 5 samples gave an inconclusive haplogroup based on the SNP typing, and 6 samples were SNP typed as a different haplogroup than that determined from the CR data. Of the 5 samples that gave an inconclusive haplogroup based on the SNP typing, 2 samples were SNP typed as the correct haplogroup but also had the 9 bp deletion found among haplogroup B individuals, 1 sample was heteroplasmic at position 10398, and 2 samples displayed mutations at both np 4833 and np 7600. Of the 6 samples that were SNP typed as a different haplogroup than that inferred from the CR data, 3 samples were SNP typed as haplogroup X or I based on a mutation at np 1719, 1 sample was SNP typed as L3 rather than the CR typed haplogroup D, and 2 samples were SNP typed as haplogroup H rather than the CR typed haplogroup U. These results are summarized in Table 4.
Table 4.
Single nucleotide polymorphisms (SNP) typing results for the Armed Forces DNA Identification Laboratory samples with a control region (CR) haplogroup assignment
| No. of samples (n = 73) | CR haplogroup | SNP haplogroup |
|---|---|---|
| 9 | A | A |
| 4 | B | B |
| 5 | C | C |
| 1 | C | M* |
| 7 | D | D |
| 1 | D | L3‡ |
| 1 | F | F |
| 2 | F | N* |
| 2 | G | E or G† |
| 3 | H | H |
| 1 | HV | N |
| 1 | I | I |
| 1 | I | I or X† |
| 1 | I | I + 9bp deletion† |
| 5 | L | L |
| 1 | L | L + 9bp deletion† |
| 11 | L3 | L3 |
| 1 | L3 | I‡ |
| 7 | M | M |
| 2 | N | X‡ |
| 3 | U | N |
| 2 | U | H‡ |
| 2 | X | X |
*SNP typing identified the correct macrohaplogroup but failed to determine the more specific haplogroup assigned based on the CR data (3 samples).
†SNP typing did not result in a conclusive haplogroup (5 samples).
‡CR and SNP haplogroup typing results did not agree (6 samples).
All 21 AFDIL samples for which a single haplogroup could not be inferred from CR sequence data were successfully SNP typed. Of these, one sample is known to have been incorrectly SNP typed as haplogroup I. The haplogroup I SNP type was based on the presence of the 1719A mutation, however the CR sequence is missing numerous mutations characteristic of haplogroup I (A16129G, C16223T, G16391A, T199C, T204C, T250C) (51). The results of the SNP typing of these 21 samples, including inconclusive haplogroup assignments (where available) based on the CR data, are summarized in Table 5.
Table 5.
Single nucleotide polymorphism (SNP) typing results for the Armed Forces DNA Identification Laboratory samples without a conclusive control region (CR) haplogroup assignment*
| No. of samples (n = 21) | CR haplogroup | SNP haplogroup |
|---|---|---|
| 10 | D or G | D |
| 1 | D or G | G |
| 1 | ? | H |
| 1 | ? | I* |
| 4 | ? | N |
| 4 | ? | L3 |
*One sample is known to have been incorrectly SNP typed.
The 31 samples of known ancestral origin but which had not been previously haplogroup typed were successfully SNP typed. These results are summarized in Table 6.
Table 6.
Results from the analysis of 31 samples of known self-defined ancestral origin
| Known ancestral origin | Haplogroup designation | Inferred ancestry* |
|---|---|---|
| EU | N | EU/AS/NA |
| EU | I | EU |
| EU | H | EU |
| EU | N | EU/AS/NA |
| EU | H | EU |
| EU | N | EU/AS/NA |
| EU | N | EU/AS/NA |
| EU | L3 | EU/AF/AS |
| EU | B | AS/NA |
| EU | H | EU |
| EU | N | EU/AS/NA |
| AF | L1/L2 | AF |
| AF | L1/L2 | AF |
| AF | L1/L2 | AF |
| AF | L1/L2 | AF |
| AF | L3 | EU/AF/AS |
| AF | L1/L2 | AF |
| AF | N | EU/AS/NA |
| AF | N | EU/AS/NA |
| AF | L1/L2 | AF |
| AS | D | AS/NA |
| AS | N | EU/AS/NA |
| AS | D | AS/NA |
| AS | L3 | EU/AF/AS |
| AS | M | AS/NA |
| AS | B | AS/NA |
| AS | M | AS/NA |
| AS | N | EU/AS/NA |
| AS | L3 | EU/AF/AS |
| AS | F | AS |
| AS | N | EU/AS/NA |
*Abbreviations: EU – European; AF – African; AS – Asian; NA – Native American.
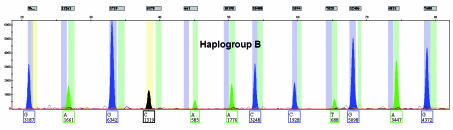
The ancient skeletal remains also produced successful SNP typing results: samples 1 and 2, both extracted from World War II-era bones, SNP typed as haplogroup B (Figure 4) and macrohaplogroup N (data not shown), respectively.
Figure 4.
Electropherogram obtained from the mtDNA extracted from the femur recovered from a WWII plane crash in the Philippine Islands shown in Figure 2. The first peak is blue (G) which represents a C to G base change caused by the 8272-8280 9 bp deletion that defines Haplogroup B.
All 12 SNPs tested were clearly typed in most of the tested samples, however all haplogroup A samples and five of the AFDIL haplogroup D samples exhibited drop out (null allele) of the peak corresponding to np 4833 and np 10398, respectively. A null allele can be caused by a failure of the initial amplification of the fragment encompassing the polymorphism or by a failed single base primer extension reaction. The null allele in the haplogroup A samples was most likely caused by an A to G transition (np 4824) in the 4833 extension primer annealing region (30), however all samples had the expected base substitutions 663A and 7028T. The five AFDIL samples that exhibited drop out of the 10398 SNP originated from the same region (Hong Kong), and are the only haplogroup D samples that exhibited this null allele. Coding region sequencing could be performed to determine the cause of the null allele, but such sequence data was not generated for these samples.
Discussion
The assay characterizes 12 mtDNA coding region polymorphisms selected to identify the European (haplogroup H and I), African (haplogroup L1/L2), Asian (haplogroups E, F and G), and Asian/Native American (haplogroups A, B, C, and D) lineages. The assay is also able to identify macrohaplogroup L3, which includes several haplogroups in and out of Africa (52).
When the SNP assay was applied to the 73 AFDIL samples previously haplogroup typed using CR sequence data, the correct haplogroup or macrohaplogroup was unambiguously identified ~ 85% of the time. Among the 94 AFDIL samples SNP typed, a presumably correct haplogroup or macrohaplogroup was unambiguously assigned ~ 87% of the time. As a first step screening tool for haplogroup assignment, the SNP typing compares quite favorably to the considerably more time consuming, expensive, and labor-intensive alternative method of CR sequencing that correctly inferred a specific haplogroup slightly less frequently.
Of the AFDIL samples that did not result in a conclusive SNP haplogroup (5 samples, or ~ 7%), four instances resulted from one of two mutations (7600A or the 9 bp deletion) identified in a sample of a different haplogroup, and one instance occurred as the result of heteroplasmy at np 10398. However, in each of these cases, the correct haplogroup was one of the two assigned. The 9 bp deletion that defines haplogroup B has occasionally been found in populations from different geographic origins, suggesting that this mutation has independently occurred more than once during human evolution (29,53,54). This explains why, in a small number of samples, the 9 bp deletion could be observed concurrent with a mutation specific for a different haplogroup. Overall, the data here demonstrate that inconclusive haplogroup assignment could be expected to occur less than 10% of the time. In practice, these few instances of inconclusive haplogroup assignment could be corrected by sequencing the CR or the coding region in the area of another haplogroup specific polymorphic site.
Among the 7 AFDIL samples for which the SNP haplogroup disagreed with the CR-inferred haplogroup, more than half of the disagreements (4 samples) were the result of a mutation at np 1719. Thus, one limitation of this SNP assay is the correct identification of haplogroups X and I, as the mutation 1719A, initially chosen because found in both haplogroups, has also been observed in other haplogroups (10,26,30,38). It has been suggested that np 1719A is rather mutable, thus accounting for the appearance of the base substitution in several geographic lineages (55). Consequently, in order to specifically identify haplogroup X, the 1719 SNP should be replaced with a more informative polymorphism (eg, 6371) that can distinguish haplogroup X from the root of macrohaplogroup N (29,33,55).
Of the remaining 3 AFDIL samples for which the SNP and CR haplogroups disagreed, it is difficult to determine the correct haplogroup from the data available. In one case, the sample was tentatively designated as haplogroup D based on a specific CR mutation (16362C), while the SNP typing resulted in a L3 assignment. Like np 1719, the 16362 mutation has been observed on multiple branches of the mtDNA tree and may mutate frequently (32-34). As such, the CR haplogroup assignment may be incorrect for this sample. In the second and third case, the samples were designated as haplogroup U based on a specific mutation in the CR, but the SNP typing identified these samples as haplogroup H due to the lack of mutation at np 7028. Sequencing relevant portions of the coding region, or SNP typing of additional haplogroup specific positions of interest, would be the best (and likely only) way to resolve these haplogroup disagreements.
One area in which the SNP haplogroup typing proved to have a significant advantage over the CR haplogroup typing was in the capability to distinguish between haplogroup D and haplogroup G. More than half of the samples for which a single haplogroup could not be inferred on the basis of the CR sequence data were believed to belong to either haplogroup D or haplogroup G. In each of these cases, the SNP typing confidently assigned the samples to one of these two haplogroups (Table 5).
World War II-era Sample 1 was a human femur whose potential origin was an American soldier killed in 1945 in a plane crash in the Philippine Islands. In late 1946, American personnel visited the crash site and recovered wreckage but no human remains. In July of 1950, a Filipino native contacted the US Army and provided the femur, identifying the crash site as its source. However, in order to identify the femur as that of the missing American soldier, the mtDNA data would need to be consistent with the H haplogroup of the soldier’s maternal relatives. The SNP typing of Sample 1 identified the sample as haplogroup B, thus excluding the American soldier as the source of the sample.
Sample 2 was a human femur found in the wreckage of a B-25D-1 Mitchell bomber that crashed in New Guinea in 1943 after being attacked by Japanese aircraft. Human remains from the crash were initially recovered and buried in New Guinea in the late 1940s, and later exhumed in 1947 for transfer to the Philippine Islands for storage. Before re-interment in 1950 at the Manila American Military Cemetery, the remains were treated with preservatives, and then finally exhumed again in 2004 for identification purposes.
Four American soldiers were aboard the B-25D-1 Mitchell bomber at the time of the crash. The mtDNA haplogroups of each were determined using maternal references: 2 soldiers belonged to haplogroup H, the third soldier to haplogroup T, and the last soldier to haplogroup K. SNP typing of the human femur submitted to AFDIL for analysis identified the sample as haplogroup N, thus excluding the two haplogroup H soldiers.
In these two cases, the SNP typing of the ancient skeletal remains confirmed the haplogroup obtained by CR sequencing. Though amplification and sequencing of the CR was successful when amplicons smaller than 150 bp were targeted, obtaining CR sequence data is generally time consuming, expensive, labor intensive, and can consume large quantities of limited extract. In both cases, the SNP typing confirmed the haplogroups identified by CR sequence data and resolved questions relating to the source of the femurs with a quick and easy assay that required only a single multiplexed amplification.
Based upon the specific SNPs included in this assay, only limited information was obtained from certain samples. For example, 7028T excludes a sample from haplogroup H, however the sample can still be placed within macrohaplogroup N. This macrohaplogroup contains mtDNA types from the European, Asian, and Native American lineages, thus providing inconclusive information on ancestral origin. The inconclusive ancestral determination is currently being addressed in our laboratories with the development of a European haplogroup assay (using the same minisequencing technology) designed to simultaneously type additional SNPs that allow identification of haplogroups H, H1, I, J, K, T, U, V, W, and X. The multiplex assay is currently being tested in our laboratories.
In conclusion, the minisequencing method utilized for this panel of SNPs demonstrated the ability to rapidly screen for haplogroups A, B, C, D, E, F, G, H, L1/L2, L3, M, and N when working either pristine or highly degraded DNA samples, therefore making it a potentially useful screening tool for molecular anthropology studies. Furthermore, the minisequencing strategy allowed for simple multiplexing, the design of amplicons with minimal length, and the ability to target multiple nucleotide positions located throughout the mtDNA coding region. These properties reduce sample consumption and enable simplistic interpretation of multiple SNPs simultaneously, which are desirable qualities for forensic DNA analysis.
Acknowledgments
Financial support for this project was provided by The George Washington University Department of Forensic Sciences. The authors thank Theodore G. Schurr for providing research samples, Mariel Candelario for the valuable research she conducted while at The George Washington University, Kimberly Sturk for assistance with GeneMapper and AFDIL sample processing, Jessica Saunier for the creation of an excel macro for automatic haplogroup typing, Alec Christensen for case background and material, and Jodi Irwin for CR haplogroup assignment discussion. Additionally, the authors thank Mike Coble, Tom Holland, Jim Canik, Jim Ross, and Lou Finelli for logistical and administrative support, and Kim Kempton and Pat Kavanaugh for manuscript review.
Official disclaimer
Certain commercial equipment, instruments, and materials are identified in order to specify experimental procedures as completely as possible. In no case does such identification imply a recommendation or endorsement by the United States Department of Defense or the United States Department of the Army, nor does it imply that any of the materials or equipment identified are necessarily the best available for the purpose. The opinions and assertions contained herein are solely those of the authors and are not to be construed as official or as views of the United States Department of Defense or the United States Department of the Army.
References
- 1.Robin ED, Wong R. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J Cell Physiol. 1988;136:507–13. doi: 10.1002/jcp.1041360316. [DOI] [PubMed] [Google Scholar]
- 2.Paabo S. Ancient DNA: extraction, characterization, molecular cloning, and enzymatic amplification. Proc Natl Acad Sci U S A. 1989;86:1939–43. doi: 10.1073/pnas.86.6.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ingman M, Kaessmann H, Paabo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–13. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- 4.Budowle B, Allard MW, Wilson MR, Chakraborty R. Forensics and mitochondrial DNA: applications, debates, and foundations. Annu Rev Genomics Hum Genet. 2003;4:119–41. doi: 10.1146/annurev.genom.4.070802.110352. [DOI] [PubMed] [Google Scholar]
- 5.Hagelberg E, Clegg JB. Isolation and characterization of DNA from archaeological bone. Proc Biol Sci. 1991;244:45–50. doi: 10.1098/rspb.1991.0049. [DOI] [PubMed] [Google Scholar]
- 6.Foran DR. Relative degradation of nuclear and mitochondrial DNA: an experimental approach. J Forensic Sci. 2006;51:766–70. doi: 10.1111/j.1556-4029.2006.00176.x. [DOI] [PubMed] [Google Scholar]
- 7.Mishmar D, Ruiz-Pesini E, Golik P, Macaulay V, Clark AG, Hosseini S, et al. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci U S A. 2003;100:171–6. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303:223–6. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- 9.Kivisild T, Shen P, Wall DP, Do B, Sung R, Davis K, et al. The role of selection in the evolution of human mitochondrial genomes. Genetics. 2006;172:373–87. doi: 10.1534/genetics.105.043901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maca-Meyer N, Gonzalez AM, Larruga JM, Flores C, Cabrera VM. Major genomic mitochondrial lineages delineate early human expansions. BMC Genet. 2001;2:13. doi: 10.1186/1471-2156-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, et al. Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet. 2000;67:1251–76. [PMC free article] [PubMed] [Google Scholar]
- 12.Macaulay V, Hill C, Achilli A, Rengo C, Clarke D, Meehan W, et al. Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science. 2005;308:1034–6. doi: 10.1126/science.1109792. [DOI] [PubMed] [Google Scholar]
- 13.Holland MM, Parsons TJ. Mitochondrial DNA sequence analysis – validation and use of forensic casework. Forensic Science Review. 1999;11:21–50. [PubMed] [Google Scholar]
- 14.Holland MM, Fisher DL, Michell LG, Rodriguez WC, Canik JJ, Merril CR, et al. Mitochondrial DNA sequence analysis of human skeletal remains: identification of remains from the Vietnam War. J Forensic Sci. 1993;38:542–53. [PubMed] [Google Scholar]
- 15.Parsons TJ, Coble MD. Increasing the forensic discrimination of mitochondrial DNA testing through analysis of the entire mitochondrial DNA genome. Croat Med J. 2001;42:304–9. [PubMed] [Google Scholar]
- 16.Just RS, Irwin JA, O’Callaghan JE, Saunier JL, Coble MD, Vallone PM, et al. Toward increased utility of mtDNA in forensic identifications. Forensic Sci Int. 2004;146(Suppl):S147–9. doi: 10.1016/j.forsciint.2004.09.045. [DOI] [PubMed] [Google Scholar]
- 17.Irwin JA, Edson SM, Loreille O, Just RS, Barritt SM, Lee DA, et al. DNA identification of “Earthquake McGoon” 50 years postmortem. J Forensic SciForthcoming2007 [DOI] [PubMed] [Google Scholar]
- 18.Andelinovic Š, Sutlovic D, Erceg Ivkosic I, Skaro V, Ivkosic A, Paic F, et al. Twelve-year experience in identification of skeletal remains from mass graves. Croat Med J. 2005;46:530–9. [PubMed] [Google Scholar]
- 19.Melton T, Nelson K. Forensic mitochondrial DNA analysis: two years of commercial casework experience in the United States. Croat Med J. 2001;42:298–303. [PubMed] [Google Scholar]
- 20.Kong QP, Bandelt HJ, Sun C, Yao YG, Salas A, Achilli A, et al. Updating the East Asian mtDNA phylogeny: a prerequisite for the identification of pathogenic mutations. Hum Mol Genet. 2006;15:2076–86. doi: 10.1093/hmg/ddl130. [DOI] [PubMed] [Google Scholar]
- 21.Roostalu U, Kutuev I, Loogvali EL, Metspalu E, Tambets K, Reidla M, et al. Origin and expansion of haplogroup H, the dominant human mitochondrial DNA lineage in West Eurasia: the Near Eastern and Caucasian perspective. Mol Biol Evol. 2007;24:436–48. doi: 10.1093/molbev/msl173. [DOI] [PubMed] [Google Scholar]
- 22.Palanichamy MG, Sun C, Agrawal S, Bandelt H, Kong QP, Khan F, et al. Phylogeny of mitochondrial DNA macrohaplogroup N in India, based on complete sequencing: implications for the peopling of South Asia. Am J Hum Genet. 2004;75:966–78. doi: 10.1086/425871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka M, Cabrera VM, Gonzalez AM, Larruga JM, Takeyasu T, Fuku N, et al. Mitochondrial genome variation in eastern Asia and the peopling of Japan. Genome Res. 2004;14:1832–50. doi: 10.1101/gr.2286304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wallace DC. 1994 William Allan Award Address. Mitochondrial DNA variation in human evolution, degenerative disease, and aging. Am J Hum Genet. 1995;57:201–23. [PMC free article] [PubMed] [Google Scholar]
- 25.Wallace DC, Brown MD, Lott MT. Mitochondrial DNA variation in human evolution and disease. Gene. 1999;238:211–30. doi: 10.1016/s0378-1119(99)00295-4. [DOI] [PubMed] [Google Scholar]
- 26.Finnila S, Lehtonen MS, Majamaa K. Phylogenetic network for European mtDNA. Am J Hum Genet. 2001;68:1475–84. doi: 10.1086/320591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith DG, Malhi RS, Eshleman J, Lorenz JG, Kaestle FA. Distribution of mtDNA haplogroup X among Native North Americans. Am J Phys Anthropol. 1999;110:271–84. doi: 10.1002/(SICI)1096-8644(199911)110:3<271::AID-AJPA2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 28.Bonatto SL, Salzano FM. A single and early migration for the peopling of the Americas supported by mitochondrial DNA sequence data. Proc Natl Acad Sci U S A. 1997;94:1866–71. doi: 10.1073/pnas.94.5.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen YS, Torroni A, Excoffier L, Santachiara-Benerecetti AS, Wallace DC. Analysis of mtDNA variation in African populations reveals the most ancient of all human continent-specific haplogroups. Am J Hum Genet. 1995;57:133–49. [PMC free article] [PubMed] [Google Scholar]
- 30.Herrnstadt C, Elson JL, Fahy E, Preston G, Turnbull DM, Anderson C, et al. Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet. 2002;70:1152–71. doi: 10.1086/339933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schurr TG, Ballinger SW, Gan YY, Hodge JA, Merriwether DA, Lawrence DN, et al. Amerindian mitochondrial DNAs have rare Asian mutations at high frequencies, suggesting they derived from four primary maternal lineages. Am J Hum Genet. 1990;46:613–23. [PMC free article] [PubMed] [Google Scholar]
- 32.Bandelt HJ, Herrnstadt C, Yao YG, Kong QP, Kivisild T, Rengo C, et al. Identification of Native American founder mtDNAs through the analysis of complete mtDNA sequences: some caveats. Ann Hum Genet. 2003;67:512–24. doi: 10.1046/j.1469-1809.2003.00049.x. [DOI] [PubMed] [Google Scholar]
- 33.Kivisild T, Tolk HV, Parik J, Wang Y, Papiha SS, Bandelt HJ, et al. The emerging limbs and twigs of the East Asian mtDNA tree. Mol Biol Evol. 2002;19:1737–51. doi: 10.1093/oxfordjournals.molbev.a003996. [DOI] [PubMed] [Google Scholar]
- 34.Kong QP, Yao YG, Sun C, Bandelt HJ, Zhu CL, Zhang YP. Phylogeny of east Asian mitochondrial DNA lineages inferred from complete sequences. Am J Hum Genet. 2003;73:671–6. doi: 10.1086/377718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vallone PM, Just RS, Coble MD, Butler JM, Parsons TJ. A multiplex allele-specific primer extension assay for forensically informative SNPs distributed throughout the mitochondrial genome. Int J Legal Med. 2004;118:147–57. doi: 10.1007/s00414-004-0428-5. [DOI] [PubMed] [Google Scholar]
- 36.Wiesbauer M, Meierhofer D, Mayr JA, Sperl W, Paulweber B, Kofler B. Multiplex primer extension analysis for rapid detection of major European mitochondrial haplogroups. Electrophoresis. 2006;27:3864–8. doi: 10.1002/elps.200600086. [DOI] [PubMed] [Google Scholar]
- 37.Brandstatter A, Parsons TJ, Parson W. Rapid screening of mtDNA coding region SNPs for the identification of west European Caucasian haplogroups. Int J Legal Med. 2003;117:291–8. doi: 10.1007/s00414-003-0395-2. [DOI] [PubMed] [Google Scholar]
- 38.Alvarez-Iglesias V, Jaime JC, Carracedo A, Salas A. Coding region mitochondrial DNA SNPs: targeting East Asian and Native American haplogroups. Forensic Science International: Genetics. 2007;1:44–55. doi: 10.1016/j.fsigen.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 39.Quintans B, Alvarez-Iglesias V, Salas A, Phillips C, Lareu MV, Carracedo A. Typing of mitochondrial DNA coding region SNPs of forensic and anthropological interest using SNaPshot minisequencing. Forensic Sci Int. 2004;140:251–7. doi: 10.1016/j.forsciint.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 40.Grignani P, Peloso G, Achilli A, Turchi C, Tagliabracci A, Alu M, et al. Subtyping mtDNA haplogroup H by SNaPshot minisequencing and its application in forensic individual identification. Int J Legal Med. 2006;120:151–6. doi: 10.1007/s00414-005-0059-5. [DOI] [PubMed] [Google Scholar]
- 41.Sigurdsson S, Hedman M, Sistonen P, Sajantila A, Syvänen AC. A microarray system for genotyping 150 single nucleotide polymorphisms in the coding region of human mitochondrial DNA. Genomics. 2006;87:534–42. doi: 10.1016/j.ygeno.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 42.Salas A, Quintans B, Alvarez-Iglesias V. SNaPshot typing of mitochondrial DNA coding region variants. Methods Mol Biol. 2005;297:197–208. doi: 10.1385/1-59259-867-6:197. [DOI] [PubMed] [Google Scholar]
- 43.Fiorentino F, Magli MC, Podini D, Ferraretti AP, Nuccitelli A, Vitale N, et al. The minisequencing method: an alternative strategy for preimplantation genetic diagnosis of single gene disorders. Mol Hum Reprod. 2003;9:399–410. doi: 10.1093/molehr/gag046. [DOI] [PubMed] [Google Scholar]
- 44.Szibor R, Plate I, Schmitter H, Wittig H, Krause D. Forensic mass screening using mtDNA. Int J Legal Med. 2006;120:372–6. doi: 10.1007/s00414-006-0085-y. [DOI] [PubMed] [Google Scholar]
- 45.Bandelt HJ, Quintana-Murci L, Salas A, Macaulay V. The fingerprint of phantom mutations in mitochondrial DNA data. Am J Hum Genet. 2002;71:1150–60. doi: 10.1086/344397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bandelt HJ, Salas A, Lutz-Bonengel S. Artificial recombination in forensic mtDNA population databases. Int J Legal Med. 2004;118:267–73. doi: 10.1007/s00414-004-0455-2. [DOI] [PubMed] [Google Scholar]
- 47.Yao YG, Bravi CM, Bandelt HJ. A call for mtDNA data quality control in forensic science. Forensic Sci Int. 2004;141:1–6. doi: 10.1016/j.forsciint.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 48.Salas A, Carracedo A, Macaulay V, Richards M, Bandelt HJ. A practical guide to mitochondrial DNA error prevention in clinical, forensic, and population genetics. Biochem Biophys Res Commun. 2005;335:891–9. doi: 10.1016/j.bbrc.2005.07.161. [DOI] [PubMed] [Google Scholar]
- 49.Haak W, Forster P, Bramanti B, Matsumura S, Brandt G, Tanzer M, et al. Ancient DNA from the first European farmers in 7500-year-old Neolithic sites. Science. 2005;310:1016–8. doi: 10.1126/science.1118725. [DOI] [PubMed] [Google Scholar]
- 50.Yao YG, Kong QP, Man XY, Bandelt HJ, Zhang YP. Reconstructing the evolutionary history of China: a caveat about inferences drawn from ancient DNA. Mol Biol Evol. 2003;20:214–9. doi: 10.1093/molbev/msg026. [DOI] [PubMed] [Google Scholar]
- 51.Malyarchuk BA, Grzybowski T, Derenko MV, Czarny J, Wozniak M, Miscicka-Sliwka D. Mitochondrial DNA variability in Poles and Russians. Ann Hum Genet. 2002;66:261–83. doi: 10.1017/S0003480002001161. [DOI] [PubMed] [Google Scholar]
- 52.Gonder MK, Mortensen HM, Reed FA, de Sousa A, Tishkoff SA. Whole-mtDNA genome sequence analysis of ancient African lineages. Mol Biol Evol. 2007;24:757–68. doi: 10.1093/molbev/msl209. [DOI] [PubMed] [Google Scholar]
- 53.Ballinger SW, Schurr TG, Torroni A, Gan YY, Hodge JA, Hassan K, et al. Southeast Asian mitochondrial DNA analysis reveals genetic continuity of ancient mongoloid migrations. Genetics. 1992;130:139–52. doi: 10.1093/genetics/130.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Torroni A, Sukernik RI, Schurr TG, Starikorskaya YB, Cabell MF, Crawford MH, et al. mtDNA variation of aboriginal Siberians reveals distinct genetic affinities with Native Americans. Am J Hum Genet. 1993;53:591–608. [PMC free article] [PubMed] [Google Scholar]
- 55.Reidla M, Kivisild T, Metspalu E, Kaldma K, Tambets K, Tolk HV, et al. Origin and diffusion of mtDNA haplogroup X. Am J Hum Genet. 2003;73:1178–90. doi: 10.1086/379380. [DOI] [PMC free article] [PubMed] [Google Scholar]