Abstract
Many RNA-binding proteins help RNAs to fold via their RNA chaperone activity. This term has been used widely without accounting for the diversity of the observed reactions, which include complex events like restructuring of misfolded catalytic RNAs, promoting the assembly of RNA-protein complexes, and mediating RNA–RNA interactions. Proteins display very diverse activities depending on the assays used to measure RNA chaperone activity. To classify proteins with this activity, we compared three exemplary proteins from E. coli, host factor Hfq, ribosomal protein S1, and the histone-like protein StpA for their abilities to promote two simple reactions, RNA annealing and strand displacement. The results of a FRET-based assay show that S1 promotes only RNA strand displacement while Hfq solely enhances RNA annealing. StpA, in contrast, is active in both reactions. To test whether the two activities can be assigned to different domains of the bipartite-structured StpA, we assayed the purified N- and C- terminal domains separately. While both domains are unable to promote RNA annealing, we can attribute the RNA strand displacement activity of StpA to the C-terminal domain. Correlating with their RNA annealing activities, only Hfq and full-length StpA display simultaneous binding of two RNAs, suggesting a matchmaker-like model for this activity. For StpA, this “RNA crowding” requires protein–protein interactions, since a dimerization-deficient StpA mutant lost the ability to bind and anneal two RNAs. These results underline the difference between the two reaction types, making it necessary to distinguish and classify proteins according to their specific RNA chaperone activities.
Keywords: Hfq, S1, StpA dimerization, FRET, RNA annealing, strand displacement
INTRODUCTION
RNA molecules have to reach a defined native conformation in order to assemble with other RNAs and proteins and to fulfill their multiple roles in the cell. Despite their potential to easily misfold in vitro (Zuker 1989; Uhlenbeck 1995), RNAs appear to fold efficiently within cells. It has been suggested that the interaction with proteins accounts for efficient folding in vivo (Herschlag 1995; Schroeder et al. 2004). Proteins with RNA chaperone activity help RNAs to find their native conformations either by preventing kinetic traps or by resolving them. These proteins are very heterogeneous, and their number is increasing constantly (Herschlag 1995; Woodson 2000; Cristofari and Darlix 2002; Schroeder et al. 2004). Assays for RNA chaperone activity monitor a variety of reactions, ranging from simple reactions like the annealing of two complementary RNAs or the dissociation of RNA duplexes to not very well defined systems such as the rescuing of misfolded complex tertiary structures of catalytic RNAs in vitro and in vivo (Cristofari and Darlix 2002; Rajkowitsch et al. 2005).
Here, we compare the RNA annealing and strand displacement activities of three exemplary Escherichia coli RNA chaperones, Hfq, S1, and StpA. Hfq is a highly abundant protein that was isolated originally as part of the phage Qβ replicase enzyme (Franze de Fernandez et al. 1968; Blumenthal and Carmichael 1979). It was shown to function as a pleiotropic post-transcriptional regulator that enhances the interaction between small, noncoding RNAs (sRNAs) with their target mRNAs (Zhang et al. 1998, 2002; Moller et al. 2002). Hfq forms a homo-hexameric ring structure that contains at least two distinct RNA-binding surfaces, offering an explanation for how the protein facilitates RNA–RNA interactions (Schumacher et al. 2002; Sauter et al. 2003). Interestingly, Hfq has been repeatedly copurified with ribosomal protein S1 as part of the bacteriophage Qβ replication complex (Inouye et al. 1974; Wahba et al. 1974), both proteins are present in stoichiometric amounts in preparations of RNA polymerase (Sukhodolets and Garges 2003), and Hfq, S1, and RNA polymerase subunits are found as binding partners of small RNAs (N. Windbichler and R. Schroeder, unpubl.). S1 is an abundant cellular protein that is involved in mRNA binding to the small ribosomal subunit; it disrupts RNA secondary structures in vitro and unwinds mRNAs during translation initiation in vivo (Bear et al. 1976; Kolb et al. 1977; Subramanian 1983; Tedin et al. 1997).
The third protein tested is StpA, a small, basic protein that was identified originally as suppressor of a splicing-defective mutant of the phage T4 thymidylate-synthase gene, and it has been reported to function as a molecular back-up of its intraspecies homolog (paralog), the E. coli nucleoid-structuring protein H-NS (Zhang and Belfort 1992; Zhang et al. 1995, 1996; Dorman 2004). StpA is a known RNA chaperone that rescues splicing of a misfolded td pre-mRNA in vitro and in vivo and is active in both RNA annealing and strand displacement (Zhang et al. 1995; Mayer et al. 2002; Rajkowitsch et al. 2005). Recently, we showed that StpA can bind two RNA 21mers simultaneously, thereby supporting an “RNA crowding” mechanism for the nonspecific RNA annealing activity of StpA (Mayer et al. 2007). StpA and H-NS are composed of two domains that are joined by a linker region with the N-terminal domain reported to be involved in protein–protein interactions, while the C-terminal domain mediates nucleic acid binding (Cusick and Belfort 1998; Dorman et al. 1999; Dorman 2004). Mapping a discrete function to the two domains has previously been hampered by the emerging picture that they appear to work cooperatively. For example, the N-terminal domain of H-NS actively contributes to DNA binding, and a residue in the C-terminal domain is crucial for oligomerization and to some extent also for dimerization (Spurio et al. 1997; Bloch et al. 2003; Stella et al. 2005).
We find that Hfq displays only RNA annealing activity and that S1 exclusively promotes RNA strand displacement, whereas StpA has both activities. These observations suggest that it is necessary to distinguish between these activities, which would require a more defined classification of proteins according to their specific RNA chaperone activities. We further addressed the question whether the RNA annealing and strand displacement activities of StpA could be allocated to distinct regions of the protein. For this purpose, the N- and C-terminal domains were separately assessed for their activities. We find that the C-terminal domain retains the ability to promote strand displacement. In contrast, the RNA annealing activity seems to require the full-length protein, with the dimerization capacity provided by the N-terminal domain and the RNA-binding function by the C-terminal domain.
RESULTS AND DISCUSSION
Comparing the RNA annealing and strand displacement activities of Hfq, S1, and StpA
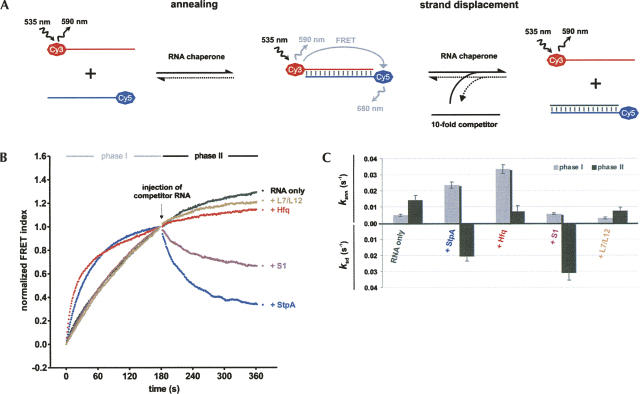
To compare the RNA annealing and strand displacement activities of the E. coli proteins Hfq, S1, and StpA, we employed a recently developed assay that combines monitoring of the two activities in a single set-up and detects double-stranded RNA by FRET (fluorescence resonance energy transfer) (Rajkowitsch and Schroeder 2007). In the first phase of the assay, two fluorophore-labeled complementary RNA 21mers are annealed in the absence or presence of the protein tested, and injection of an excess of nonlabeled competitor RNA starts the second phase, which monitors strand displacement (Fig. 1A). This pair of RNA oligonucleotides has been employed before to monitor RNA annealing by StpA (Zhang et al. 1995; Rajkowitsch et al. 2005; Rajkowitsch and Schroeder 2007), and we observed similar kinetics and protein effects with a number of RNA 21mers differing in their G/C content from 5% to 71% (S. Stampfl, pers. comm.). As the short RNAs used here do not form stable intramolecular secondary structures, they can anneal by themselves with an observed annealing rate constant k ann,1 of 0.005 sec−1 (Fig. 1B,C). In contrast, strand displacement does not take place in the absence of proteins, because the RNA duplex is stable at the assay temperature of 37°C. Annealing continues in the second phase with a higher rate (k ann,2 of 0.012 sec−1) due to an increase in the concentration of one reaction partner (Table 1).
FIGURE 1.
RNA chaperone activities in RNA annealing and strand displacement can be analyzed in a fluorescence-based assay. (A) Annealing of two fluorophore-labeled RNA 21mers yields a FRET-signal that is reduced upon RNA chaperone-facilitated strand displacement with a competitor RNA. (B) In phase I, 5 nM each of two fully complementary RNAs (Cy5–21R+, Cy3–21R−) were annealed in a microplate reader in the absence or presence of 1 μM of protein. The donor (Cy3) and acceptor (Cy5) fluorescence emissions were quantified every second; the FRET index was calculated as FCy5/FCy3 and normalized at t180s. Hfq and StpA accelerated this reaction. Phase II was initiated by the injection of an excess of nonlabeled competitor RNA, and either RNA annealing continued (L7/L12 and Hfq) or the tested protein induced strand displacement (S1 and StpA). Representative curves are shown. (C) Comparison of the observed reaction constants for RNA annealing in phase I and II (k ann,1 and k ann,2) and strand displacement (k SD).
TABLE 1.
Reaction constants of RNA annealing and strand displacement
As previously reported, StpA promotes both reactions: It enhances the RNA annealing rate fivefold, and it strongly induces strand displacement in phase II (Fig. 1B,C; Zhang et al. 1995; Rajkowitsch et al. 2005; Rajkowitsch and Schroeder 2007). In contrast, Hfq is only active in one of the reactions: While Hfq is not able to facilitate strand displacement, its presence accelerates annealing of the RNAs sevenfold, which is in good agreement with manifold reports of its matchmaker role (Valentin-Hansen et al. 2004). Annealing is almost completed in phase I, and therefore, the curve fitting of phase II shows only residual annealing (Fig. 1B,C; Table 1). Recently, an ATPase activity of Hfq was reported, which could provide external energy for reactions such as strand displacement (Sukhodolets and Garges 2003). Therefore, we also performed our assay with ATP added up to a concentration of 1 mM, but we found Hfq's role in RNA annealing and strand displacement to be independent from ATP (data not shown).
In contrast to Hfq, ribosomal protein S1 does not accelerate RNA annealing but effectively promotes strand displacement. This activity matches S1’s proposed role in resolving secondary structures in translation initiation. As a control for an RNA-binding protein without RNA chaperone activity, we used the E. coli ribosomal protein L7/L12 (Semrad et al. 2004). L7/L12 was inactive in both reaction types (Fig. 1B,C). These results show that proteins that can function as RNA chaperones have different activities in RNA annealing and strand displacement, and hence, their performance in these reactions can be useful to classify them accordingly and to study the reaction mechanisms of the two activities in more detail.
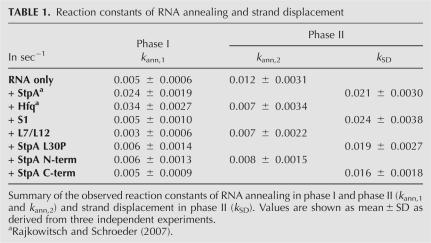
The strand displacement activity of StpA can be attributed to the C-terminal domain
We further wanted to know whether the RNA annealing and strand displacement activities of StpA can be separated and allocated to distinct domains. StpA is composed of two domains, the N-terminal domain, which is essential for dimerization of the protein, and the C-terminal domain, which has nucleic acid binding properties (Fig. 2A; Dorman et al. 1999). The two domains were assessed separately for their activities. As shown in Figure 2B, neither domain can by itself accelerate the annealing of the two short RNAs, whereas the C-terminal domain but not the N-terminal domain retains the ability of the full-length protein to promote strand displacement.
FIGURE 2.
Only dimerization-competent StpA enhances RNA annealing, whereas the ability to facilitate strand displacement resides in the C-terminal domain. (A) The N- and C-terminal domains, as well as the position of the L30P mutation, are indicated in the protein sequence of E. coli StpA. (B) Wild-type StpA (wt), StpA L30P, the N-terminal, and the C-terminal fragment were assayed for their RNA annealing and strand displacement activity as described in the Materials and Methods. (C) Kinetic evaluation of the reactions. Only StpA wt accelerates annealing; StpA wt, StpA L30P, and StpA C-term facilitate strand displacement in phase II. (D) Coomassie blue–stained SDS-polyacrylamide gels showing the analysis of protein–protein cross-link reactions. X-link indicates the incubation with cross-link agents EDC/NHS. The calculated molecular weights of the monomers are as follows: StpA, 15 kDa; StpA L30P, 15 kDa; N-terminal domain, 9 kDa; and C-terminal domain, 5 kDa.
The two domains have been tested for their RNA chaperone activity before: A recent study from our group showed that both termini display cis-splicing activity in vitro; i.e., they can promote the folding and concomitant splicing of a misfolded group I intron forming aberrant base-pairs (Mayer et al. 2007). While the C-terminal domain evidently is active in both strand displacement and cis-splicing, the N-terminal domain is only active in the latter assay, indicating that these two RNA chaperone activities require different protein properties. In another study, similar but not identical terminal fragments have been tested for their activities in annealing and trans-splicing (Cusick and Belfort 1998). Both reactions could be promoted by an extended C-terminal domain (comprising 11 residues from the adjacent linker region) but not by an N-terminal fragment including four linker amino acids. The finding that this C-terminal fragment promotes RNA annealing contrasts results gained in this study with a C terminus defined by the domain boundaries (Fig. 2) and with a dimerization-deficient full-length StpA mutant with a single amino acid exchange in the N-terminal domain (see below).
Promotion of RNA annealing depends on dimerization of the full-length StpA protein
We have previously shown that StpA is able to bind two RNAs simultaneously (Mayer et al. 2007). To test whether this ability is provided by the dimerization of StpA and whether dimerization is essential for RNA annealing activity, we constructed an StpA mutant with a leucine to proline change at amino acid position 30 (Fig. 2A). We designed this mutant in consideration of the homology of StpA to H-NS. A study searching for dominant-negative mutants of H-NS, which fail to repress the transcription of one of its target promoters proVWX, yielded only one mutant (Ueguchi et al. 1997). This mutant H-NS L30P lost the ability to dimerize, most likely due to the shortening of a coiled-coil region in the N-terminal domain that is important for protein–protein interactions (Dorman et al. 1999; Bloch et al. 2003; Cerdan et al. 2003). Similarly, for the L30P mutant of StpA, the EMBnet program Coils (http://www.ch.embnet.org/software/coils_form.html) predicts a loss of coiled-coils at the N terminus (Lupas et al. 1991).
We purified the StpA L30P protein and tested it for its dimerization ability in a chemical cross-linking assay (Fig. 2D). Indeed, no band corresponding to a protein dimer was detected when compared to the wild type. In parallel, the two domains of StpA were assayed for dimerization. As expected, the N-terminal domain cross-links efficiently, showing dimeric and tetrameric forms on a denaturing SDS-polyacrylamide gel. Notably, dimerization of the non-cross-linked sample cannot be resolved by the denaturing conditions applied. The C-terminal domain cannot be cross-linked and migrates as a monomer.
When we tested the StpA L30P mutant in the combined FRET assay, it displayed a loss of the ability to accelerate annealing, but it retained the RNA strand displacement activity of the wild-type protein (Fig. 2B). These results suggest that dimerization of StpA is essential for promoting RNA annealing but not for strand displacement, and this is in good agreement with the fact that the C-terminal domain by itself can promote RNA strand displacement.
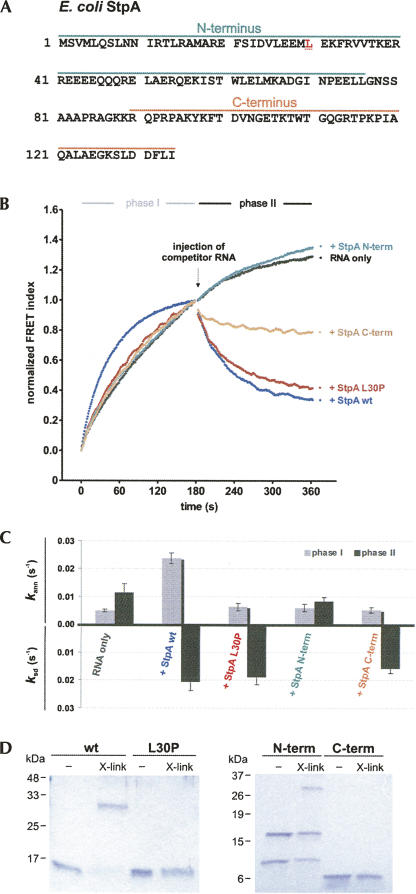
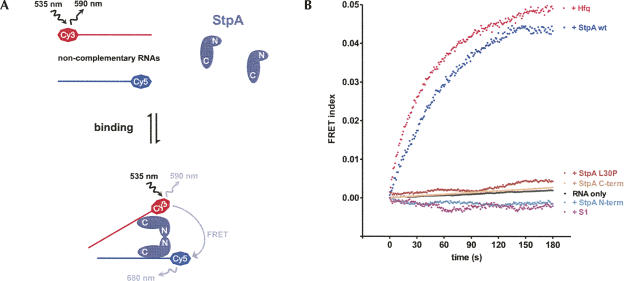
The annealing-active proteins StpA and Hfq can bind two RNAs simultaneously
Wild-type StpA can bind two 21mers simultaneously, indicating that RNA annealing is promoted by a local increase in RNA concentration (Mayer et al. 2007). The corresponding assay monitors the occurrence of FRET between noncomplementary single-stranded RNAs, which requires mediation by a protein binding partner. We employed this assay to determine whether the dimerization and annealing properties of StpA and its variants are mirrored in their ability for dual RNA binding (Fig. 3A). Indeed, only the full-length protein, but neither StpA L30P nor the two domains, gives rise to a FRET signal (Fig. 3B). This indicates that only the dimerization-competent wild-type StpA can bind at least two differently labeled RNAs, thereby bringing the two fluorophores in close proximity. A similar signal increase was observed for Hfq but not for S1, corresponding to their activities in RNA annealing. We therefore conclude that the stimulation of this reaction is caused by the properties of Hfq and wild-type StpA to bind two RNAs simultaneously, suggesting that RNA annealing depends on a matchmaker activity of these proteins.
FIGURE 3.
Two noncomplementary short RNAs bind simultaneously to Hfq or dimerization-competent wild-type StpA. (A) Monitoring “RNA crowding.” Two noncomplementary fluorophore-labeled RNAs give a FRET signal when they are in close proximity because of simultaneous binding to a protein. In this model, dual RNA binding is mediated by two StpA proteins that dimerize via their N-terminal domain. (B) In a microplate reader, the noncomplementary RNAs Cy5–21R+ and Cy3-Duplex− were injected into buffer containing the respective proteins. The FRET index was measured and calculated as described in the Materials and Methods but not normalized. Only incubation with StpA and Hfq yielded quantifiable reaction curves; the observed rate constants for dual binding k db are 0.028±0.002 sec−1 and 0.027±0.002 sec−1.
Conclusions
In this study, we employed a fluorescence-based assay to assess the RNA annealing and strand displacement activities of exemplary RNA chaperones. We find that Hfq and StpA enhance the annealing of two RNA 21mers significantly and that they both can bring two noncomplementary RNAs in close proximity to yield a FRET signal. This supports the theory of a “molecular crowding” mechanism as a basis for the RNA annealing activity of these proteins. In this model, the RNA–protein and protein–protein interactions create a microenvironment that facilitates annealing of complementary RNAs (Cristofari and Darlix 2002). The proteins function hereby as “matchmakers” by binding to the RNA and presenting it in an annealing-ready state (e.g., by decreasing the electrostatic repulsion). This prolongs the lifetime of the RNA–RNA complex and thereby increases the likelihood of annealing (Portman and Dreyfuss 1994). The short RNAs used in this study do not form significant intramolecular base-pairings. Native and longer RNA will form structures that have to be resolved prior to annealing. Therefore, protein-facilitated RNA annealing of natural substrates is likely to also require a nucleic acid melting activity. This is currently discussed for Hfq, whose annealing activity is described to occur both with and without concomitant unfolding of target RNAs (Lease et al. 1998; Brescia et al. 2003; Moll et al. 2003; Arluison et al. 2007). Additional factors such as the ribosomal protein S1, which copurifies with Hfq, could be responsible for the opening of base-pairings in vivo, resulting in a complex that could first disrupt RNA secondary structures or duplexes and then bring in another hybridization partner. The StpA L30P mutant allows the dissection of these two activities: While still able to separate double-stranded RNA, it lost the capability for protein–protein interactions and the associated matchmaker activity.
The finding that Hfq promotes RNA annealing but not strand displacement whereas S1 is active vice versa indicates that strand displacement is fundamentally different from RNA annealing. This is also supported by the observation that StpA wild type, the C-terminal domain, and StpA L30P displace RNA strands with similar rates (Table 1) suggesting that this reaction is independent from an enhancement of RNA annealing shown e.g., by StpA wild type. Some members of the DEAD-box RNA helicase family show both activities, but in an ATP-dependent manner (Cordin et al. 2006). They unwind RNA duplexes in the presence of ATP but also promote annealing in the absence of ATP (Yang and Jankowsky 2005; Uhlmann-Schiffler et al. 2006; Halls et al. 2007).
We propose that RNA annealing and strand displacement are the simplest basic properties of proteins with RNA chaperone activity, which can have either or both of them. We recently established a Web site for proteins with RNA chaperone activity in order to be able to better compare all the proteins that have been reported to promote RNA folding (http://www.projects.mfpl.ac.at/rnachaperones). It will be of significant interest to analyze these proteins for these two basic activities in order to understand their mode of action.
We also find proteins that help RNAs to fold by binding to and stabilizing RNAs specifically can have nucleic acid melting activity. While RNA chaperones such as StpA allow RNA to refold by opening up RNA secondary structures, RNA-binding proteins conversely stabilize the structure of their target RNA. This is the case for CYT-18, a Neurospora crassa mitochondrial tyrosyl-tRNA synthetase, that binds to and specifically stabilizes the catalytically competent form of the phage T4 td group I intron (Waldsich et al. 2002a,b). We find that in our assay, CYT-18 was also capable of inducing RNA strand displacement (Rajkowitsch and Schroeder 2007). This supports the model of a “preassociation binding pathway,” in which the nonspecific RNA chaperone activity of a protein allows for a “conformational search” of the target RNA whose correct fold can then be bound specifically (Herschlag 1995). This idea is further backed by a recent, very elegant single molecule FRET study showing that CBP2, a protein with specific binding activity for the bI5 group I intron, induces conformational movements in the RNA before achieving strong binding (Bokinsky et al. 2006). Furthermore, the human proteins La and hnRNP I can promote in vitro cis-splicing, but they lose this ability upon specific binding to Y RNA (Belisova et al. 2005).
In conclusion, the term RNA chaperone activity is being used for reactions as heterogeneous as the proteins found to exhibit this activity. Annealing, matchmaker, and RNA chaperone activity are hard to discern in some cases, and RNA helicases utilizing energy derived from ATP hydrolysis are also referred to as RNA chaperones, thereby extending a proposed definition by Daniel Herschlag (1995). These ambiguities promoted a categorization of proteins with RNA chaperone activity according to their membership with known protein families such as nucleoid structuring (H-NS, StpA), Sm-like (Hfq), or OB-fold containing proteins (S1). The emerging data now enable us to classify these proteins because of their activities in distinct reactions. In this article, we approach this kind of classification by testing proteins in well-defined RNA annealing and strand displacement assays, which we hope will eventually be of triple benefit: A discrete functional characterization will enable us to align proteins according to their activities regardless of consensus sequences or motifs, proteins can be dissected for their functional domains, and the knowledge about diverse proteins with similar RNA chaperone activities can provide insights into the mechanisms involved.
MATERIALS AND METHODS
Combined FRET assay for RNA annealing and strand displacement
This method is described in detail by Rajkowitsch and Schroeder (2007). In brief, two fluorophore-tagged RNA 21mers (Cy5–5′-AUGUGGAAAAUCUCUAGCAGU-3′ and Cy3–5′-ACUGCUAGAGAUUUUCCACAU-3′, VBC-Biotech, Austria) were annealed in a microplate reader (Tecan GENios Pro) at 37°C in a buffer containing 50 mM Tris-HCl p(H 7.5), 3 mM MgCl2, and 1 mM DTT. Annealing was started by injection of 20 μL of 10 nM Cy5–21R+ into a well (96-well black microtiter plate, half-area, medium binding, Greiner Bio-One) containing an equal volume of 10 nM Cy3–21R− and, where applicable, 1 μM (final concentration) of the protein. The molarity of Hfq refers to its hexameric form Hfq6. The reaction was allowed to proceed for 180 sec, and with Cy3 excited, donor and acceptor dye fluorescence emissions were measured once every second. Then, 5 μL of 400 nM nonlabeled competitor RNA (21R−) were injected to yield a 10-fold molar excess over the labeled strands, the mixture was shaken vigorously for 2 sec, and readings were taken for another 180 sec. The time-resolved ratio of the fluorescence emissions (FRET index FCy5/FCy3) was normalized to 1 at t180s and least-square fitted with Prism 4.03 (GraphPad Software Inc.). For phase I, the second-order reaction equation for equimolar initial reactant concentrations was used: y=A[1−1/(k ann,1t+1)], where k ann,1 is the observed annealing reaction constant and A is the maximum reaction amplitude. Phase II was assessed to be either describing continuing RNA annealing (FRET index increasing) or strand displacement (FRET index decreasing) and fitted accordingly with a single-exponential function for signal increase y=y0+A[1−exp(−k ann,2t)] or signal decay y=y0+A exp(−k SDt). Values are shown as mean ± SD as derived from three independent experiments.
FRET assay for dual RNA binding
The setup was almost identical to the one of the combined FRET assay except that the noncomplementary RNAs Cy5–21R+ and Cy3-Duplex- (Cy3–5′-CUUUCAUUGGUCGGUCUCUCC-3′) were used. After injection of both RNAs into the protein-containing well, the FRET signal was monitored for 180 sec. The reaction curves of StpA and Hfq were fitted with y=A [1−1/(k dbt+1)], where k db is the observed double binding reaction constant.
Plasmid construction and protein purification
For StpA expression and cloning, the plasmid pTWIN1 of the New England Biolabs IMPACT-TWIN system was used. The StpA L30P mutant was created by site-directed mutagenesis amplifying the complete template plasmid pTWIN1-StpA-intein with primers StpA-L30P+ (5′-CTTGAAGAAATGCCCGAAAAATTCAGGGTTG-3′) and StpA-L30P- (5′-CCCTGAATTTTTCGGGCATTTCTTCAAGAACG-3′). Wild-type StpA and StpA L30P proteins were purified according to the manufacturer's protocol as described before (Grossberger et al. 2005).
Protein–protein cross-linking
StpA cross-linking was performed with 2 μg of protein in 8 μL of reaction buffer (50 mM Tris-HCl at pH 7.5, 3 mM MgCl2, 1 mM DTT) (Williams et al. 1996). To the protein sample, 2 μL of a freshly prepared mixture containing the zero-length cross-linker 1-ethyl-3-(3′-dimethylaminopropyl) carbodiimide (EDC, Fluka) and the catalyst N-hydroxy-succinimide (NHS, Fluka) were added to yield a final concentration of 50 mM EDC and 200 mM NHS (Grabarek and Gergely 1990). The control reaction was mixed with 2 μL of deionized water instead. The samples were incubated at room temperature for 45 min before the reaction was stopped by adding 5 μL of 3× SDS-loading buffer (β-mercaptoethanol final concentration 150 mM) and subsequent denaturing for 5 min at 95°C. The ice-chilled, complete sample volume was loaded on a denaturing 15% SDS-polyacrylamide gel and separated at 10 V/cm. The gel was stained with Coomassie brilliant blue and destained with 20% methanol/10% acetic acid solution before being scanned.
ACKNOWLEDGMENTS
We thank members of the Max F. Perutz Laboratories (University of Vienna) for the generous gift of proteins used in this study: Mads Beich-Frandsen and Udo Bläsi for Hfq, Oliver Mayer for the StpA domains, Nikolai Windbichler for S1, and Katharina Semrad for L7/L12. We thank members of the Schroeder laboratory for providing helpful input and for constant discussion. This project was funded by the Austrian Science Fund FWF grant SFB1703.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.671807.
REFERENCES
- Arluison, V., Hohng, S., Roy, R., Pellegrini, O., Regnier, P., Ha, T. Spectroscopic observation of RNA chaperone activities of Hfq in post-transcriptional regulation by a small noncoding RNA. Nucleic Acids Res. 2007;35:999–1006. doi: 10.1093/nar/gkl1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear, D.G., Ng, R., Van Derveer, D., Johnson, N.P., Thomas, G., Schleich, T., Noller, H.F. Alteration of polynucleotide secondary structure by ribosomal protein S1. Proc. Natl. Acad. Sci. 1976;73:1824–1828. doi: 10.1073/pnas.73.6.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belisova, A., Semrad, K., Mayer, O., Kocian, G., Waigmann, E., Schroeder, R., Steiner, G. RNA chaperone activity of the protein components of human Ro RNPs. RNA. 2005;11:1084–1094. doi: 10.1261/rna.7263905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch, V., Yang, Y., Margeat, E., Chavanieu, A., Auge, M.T., Robert, B., Arold, S., Rimsky, S., Kochoyan, M. The H-NS dimerization domain defines a new fold contributing to DNA recognition. Nat. Struct. Biol. 2003;10:212–218. doi: 10.1038/nsb904. [DOI] [PubMed] [Google Scholar]
- Blumenthal, T., Carmichael, G.G. RNA replication: Function and structure of Qβ-replicase. Annu. Rev. Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- Bokinsky, G., Nivon, L.G., Liu, S., Chai, G., Hong, M., Weeks, K.M., Zhuang, X. Two distinct binding modes of a protein cofactor with its target RNA. J. Mol. Biol. 2006;361:771–784. doi: 10.1016/j.jmb.2006.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brescia, C.C., Mikulecky, P.J., Feig, A.L., Sledjeski, D.D. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA. 2003;9:33–43. doi: 10.1261/rna.2570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdan, R., Bloch, V., Yang, Y., Bertin, P., Dumas, C., Rimsky, S., Kochoyan, M., Arold, S.T. Crystal structure of the N-terminal dimerisation domain of VicH, the H-NS-like protein of Vibrio cholerae . J. Mol. Biol. 2003;334:179–185. doi: 10.1016/j.jmb.2003.09.051. [DOI] [PubMed] [Google Scholar]
- Cordin, O., Banroques, J., Tanner, N.K., Linder, P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Cristofari, G., Darlix, J.L. The ubiquitous nature of RNA chaperone proteins. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:223–268. doi: 10.1016/s0079-6603(02)72071-0. [DOI] [PubMed] [Google Scholar]
- Cusick, M.E., Belfort, M. Domain structure and RNA annealing activity of the Escherichia coli regulatory protein StpA. Mol. Microbiol. 1998;28:847–857. doi: 10.1046/j.1365-2958.1998.00848.x. [DOI] [PubMed] [Google Scholar]
- Dorman, C.J. H-NS: A universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2004;2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- Dorman, C.J., Hinton, J.C., Free, A. Domain organization and oligomerization among H-NS-like nucleoid-associated proteins in bacteria. Trends Microbiol. 1999;7:124–128. doi: 10.1016/s0966-842x(99)01455-9. [DOI] [PubMed] [Google Scholar]
- Franze de Fernandez, M.T., Eoyang, L., August, J.T. Factor fraction required for the synthesis of bacteriophage Qβ-RNA. Nature. 1968;219:588–590. doi: 10.1038/219588a0. [DOI] [PubMed] [Google Scholar]
- Grabarek, Z., Gergely, J. Zero-length crosslinking procedure with the use of active esters. Anal. Biochem. 1990;185:131–135. doi: 10.1016/0003-2697(90)90267-d. [DOI] [PubMed] [Google Scholar]
- Grossberger, R., Mayer, O., Waldsich, C., Semrad, K., Urschitz, S., Schroeder, R. Influence of RNA structural stability on the RNA chaperone activity of the Escherichia coli protein StpA. Nucleic Acids Res. 2005;33:2280–2289. doi: 10.1093/nar/gki515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halls, C., Mohr, S., Del Campo, M., Yang, Q., Jankowsky, E., Lambowitz, A.M. Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J. Mol. Biol. 2007;365:835–855. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag, D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- Inouye, H., Pollack, Y., Petre, J. Physical and functional homology between ribosomal protein S1 and interference factor i. Eur. J. Biochem. 1974;45:109–117. doi: 10.1111/j.1432-1033.1974.tb03535.x. [DOI] [PubMed] [Google Scholar]
- Kolb, A., Hermoso, J.M., Thomas, J.O., Szer, W. Nucleic acid helix-unwinding properties of ribosomal protein S1 and the role of S1 in mRNA binding to ribosomes. Proc. Natl. Acad. Sci. 1977;74:2379–2383. doi: 10.1073/pnas.74.6.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease, R.A., Cusick, M.E., Belfort, M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl. Acad. Sci. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas, A., Van Dyke, M., Stock, J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Mayer, O., Waldsich, C., Grossberger, R., Schroeder, R. Folding of the td pre-RNA with the help of the RNA chaperone StpA. Biochem. Soc. Trans. 2002;30:1175–1180. doi: 10.1042/bst0301175. [DOI] [PubMed] [Google Scholar]
- Mayer, O., Rajkowitsch, L., Lorenz, C., Konrat, R., Schroeder, R. RNA chaperone activity and RNA-binding properties of the E. coli protein StpA. Nucleic Acids Res. 2007;35:1257–1269. doi: 10.1093/nar/gkl1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll, I., Leitsch, D., Steinhauser, T., Blasi, U. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep. 2003;4:284–289. doi: 10.1038/sj.embor.embor772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller, T., Franch, T., Hojrup, P., Keene, D.R., Bachinger, H.P., Brennan, R.G., Valentin-Hansen, P. Hfq. A bacterial Sm-like protein that mediates RNA–RNA interaction. Mol. Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- Portman, D.S., Dreyfuss, G. RNA annealing activities in HeLa nuclei. EMBO J. 1994;13:213–221. doi: 10.1002/j.1460-2075.1994.tb06251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowitsch, L., Schroeder, R. Coupling RNA annealing and strand displacement: A FRET-based microplate reader assay for RNA chaperone activity. Biotechniques. 2007;43:304–310. doi: 10.2144/000112530. [DOI] [PubMed] [Google Scholar]
- Rajkowitsch, L., Semrad, K., Mayer, O., Schroeder, R. Assays for the RNA chaperone activity of proteins. Biochem. Soc. Trans. 2005;33:450–455. doi: 10.1042/BST0330450. [DOI] [PubMed] [Google Scholar]
- Sauter, C., Basquin, J., Suck, D. Sm-like proteins in Eubacteria: The crystal structure of the Hfq protein from Escherichia coli . Nucleic Acids Res. 2003;31:4091–4098. doi: 10.1093/nar/gkg480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, R., Barta, A., Semrad, K. Strategies for RNA folding and assembly. Nat. Rev. Mol. Cell Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- Schumacher, M.A., Pearson, R.F., Moller, T., Valentin-Hansen, P., Brennan, R.G. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: A bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semrad, K., Green, R., Schroeder, R. RNA chaperone activity of large ribosomal subunit proteins from Escherichia coli . RNA. 2004;10:1855–1860. doi: 10.1261/rna.7121704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurio, R., Falconi, M., Brandi, A., Pon, C.L., Gualerzi, C.O. The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA bending. EMBO J. 1997;16:1795–1805. doi: 10.1093/emboj/16.7.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella, S., Spurio, R., Falconi, M., Pon, C.L., Gualerzi, C.O. Nature and mechanism of the in vivo oligomerization of nucleoid protein H-NS. EMBO J. 2005;24:2896–2905. doi: 10.1038/sj.emboj.7600754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, A.R. Structure and functions of ribosomal protein S1. Prog. Nucleic Acid Res. Mol. Biol. 1983;28:101–142. doi: 10.1016/s0079-6603(08)60085-9. [DOI] [PubMed] [Google Scholar]
- Sukhodolets, M.V., Garges, S. Interaction of Escherichia coli RNA polymerase with the ribosomal protein S1 and the Sm-like ATPase Hfq. Biochemistry. 2003;42:8022–8034. doi: 10.1021/bi020638i. [DOI] [PubMed] [Google Scholar]
- Tedin, K., Resch, A., Blasi, U. Requirements for ribosomal protein S1 for translation initiation of mRNAs with and without a 5′ leader sequence. Mol. Microbiol. 1997;25:189–199. doi: 10.1046/j.1365-2958.1997.4421810.x. [DOI] [PubMed] [Google Scholar]
- Ueguchi, C., Seto, C., Suzuki, T., Mizuno, T. Clarification of the dimerization domain and its functional significance for the Escherichia coli nucleoid protein H-NS. J. Mol. Biol. 1997;274:145–151. doi: 10.1006/jmbi.1997.1381. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck, O.C. Keeping RNA happy. RNA. 1995;1:4–6. [PMC free article] [PubMed] [Google Scholar]
- Uhlmann-Schiffler, H., Jalal, C., Stahl, H. Ddx42p—A human DEAD box protein with RNA chaperone activities. Nucleic Acids Res. 2006;34:10–22. doi: 10.1093/nar/gkj403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Hansen, P., Eriksen, M., Udesen, C. The bacterial Sm-like protein Hfq: A key player in RNA transactions. Mol. Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- Wahba, A.J., Miller, M.J., Niveleau, A., Landers, T.A., Carmichael, G.G., Weber, K., Hawley, D.A., Slobin, L.I. Subunit I of G β replicase and 30 S ribosomal protein S1 of Escherichia coli. Evidence for the identity of the two proteins. J. Biol. Chem. 1974;249:3314–3316. [PubMed] [Google Scholar]
- Waldsich, C., Grossberger, R., Schroeder, R. RNA chaperone StpA loosens interactions of the tertiary structure in the td group I intron in vivo. Genes & Dev. 2002a;16:2300–2312. doi: 10.1101/gad.231302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldsich, C., Masquida, B., Westhof, E., Schroeder, R. Monitoring intermediate folding states of the td group I intron in vivo. EMBO J. 2002b;19:5281–5291. doi: 10.1093/emboj/cdf504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, R.M., Rimsky, S., Buc, H. Probing the structure, function, and interactions of the Escherichia coli H-NS and StpA proteins by using dominant negative derivatives. J. Bacteriol. 1996;178:4335–4343. doi: 10.1128/jb.178.15.4335-4343.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson, S.A. Recent insights on RNA folding mechanisms from catalytic RNA. Cell. Mol. Life Sci. 2000;57:796–808. doi: 10.1007/s000180050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Q., Jankowsky, E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–13601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- Zhang, A., Belfort, M. Nucleotide sequence of a newly-identified Escherichia coli gene, stpA, encoding an H-NS-like protein. Nucleic Acids Res. 1992;20:6735. doi: 10.1093/nar/20.24.6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, A., Derbyshire, V., Salvo, J.L., Belfort, M. Escherichia coli protein StpA stimulates self-splicing by promoting RNA assembly in vitro. RNA. 1995;1:783–793. [PMC free article] [PubMed] [Google Scholar]
- Zhang, A., Rimsky, S., Reaban, M.E., Buc, H., Belfort, M. Escherichia coli protein analogs StpA and H-NS: Regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J. 1996;15:1340–1349. [PMC free article] [PubMed] [Google Scholar]
- Zhang, A., Altuvia, S., Tiwari, A., Argaman, L., Hengge-Aronis, R., Storz, G. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, A., Wassarman, K.M., Ortega, J., Steven, A.C., Storz, G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- Zuker, M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]