Abstract
Ribosomal protein (rp) S5 belongs to a family of ribosomal proteins that includes bacterial rpS7. rpS5 forms part of the exit (E) site on the 40S ribosomal subunit and is essential for yeast viability. Human rpS5 is 67% identical and 79% similar to Saccharomyces cerevisiae rpS5 but lacks a negatively charged (pI ∼3.27) 21 amino acid long N-terminal extension that is present in fungi. Here we report that replacement of yeast rpS5 with its human homolog yielded a viable yeast strain with a 20%–25% decrease in growth rate. This replacement also resulted in a moderate increase in the heavy polyribosomal components in the mutant strain, suggesting either translation elongation or termination defects, and in a reduction in the polyribosomal association of the elongation factors eEF3 and eEF1A. In addition, the mutant strain was characterized by moderate increases in +1 and −1 programmed frameshifting and hyperaccurate recognition of the UAA stop codon. The activities of the cricket paralysis virus (CrPV) IRES and two mammalian cellular IRESs (CAT-1 and SNAT-2) were also increased in the mutant strain. Consistently, the rpS5 replacement led to enhanced direct interaction between the CrPV IRES and the mutant yeast ribosomes. Taken together, these data indicate that rpS5 plays an important role in maintaining the accuracy of translation in eukaryotes and suggest that the negatively charged N-terminal extension of yeast rpS5 might affect the ribosomal recruitment of specific mRNAs.
Keywords: ribosomal protein S5(S7), E-site, eEF3, translation accuracy, IRES
INTRODUCTION
Significant progress has been recently made in understanding ribosomal function. X-ray and cryo-electron microscopy (cryo-EM) analyses have provided a wealth of information regarding the structure of the ribosome peptidyl transferase center and interactions of the ribosome with mRNA, tRNAs, and translation factors (Yonath and Bashan 2004; Nilsson and Nissen 2005; Noller 2005; Ogle and Ramakrishnan 2005; Yonath 2005; Mitra and Frank 2006; Beringer and Rodnina 2007). Although it became clear that a primary role in ribosomal function belongs to rRNA (Beringer and Rodnina 2007), ribosomal proteins are nevertheless essential (Brodersen and Nissen 2005; Wilson and Nierhaus 2005; Dresios et al. 2006). Thus, bacterial ribosomal protein (rp) S1 was shown to be involved in mRNA binding, rpS12 was implicated in tRNA decoding at the A-site, rpL9 was shown to affect the stability of binding of the tRNA at the P-site, and rpL2 was suggested to participate in the peptidyl transferase activity of the ribosome (for reviews, see Brodersen and Nissen 2005; Wilson and Nierhaus 2005). Mutations in ribosomal proteins have been shown to affect both the efficiency and the fidelity of the translation process, processing of ribosomal RNA, and ribosome assembly (Brodersen and Nissen 2005; Wilson and Nierhaus 2005; Dresios et al. 2006). However, the specific functions of most bacterial ribosomal proteins have yet to be discovered and even less is known about the functions of eukaryotic ribosomal proteins. The functions of eukaryotic ribosomal proteins have recently attracted significant interest not only in connection with their roles in the basic mechanism of protein synthesis, but also in connection with their involvement in some key cellular regulatory processes such as translational control of inflammation (Mazumder et al. 2006; Kapasi et al. 2007; Ray et al. 2007), translation initiation on a subset of IRESs (Odreman-Macchioli et al. 2000; Fukushi et al. 2001; Otto et al. 2002; Sarnow et al. 2005; Laletina et al. 2006; Pfingsten et al. 2006; Schuler et al. 2006; Nishiyama et al. 2007), and the development and progression of diseases such as Diamond-Blackfan anemia (Sylvester et al. 2004 Gazda and Sieff 2006; Morimoto et al. 2006).
Eukaryotic ribosomal protein S5 belongs to a family of ribosomal proteins that includes bacterial rpS7. rpS5/rpS7 forms part of the exit (E) site on the small ribosomal subunit (Fig. 1A; Yusupov et al. 2001; Spahn et al. 2004) and cross-links to the E-site tRNA (Wower et al. 1993; Doring et al. 1994). rpS7 also contributes to the formation of the so-called mRNA exit channel and interacts with rpS11, which is located on the platform of the 30S subunit (Robert and Brakier-Gingras 2003). This interaction was suggested to contribute to the structural rearrangements of the head of the 30S subunit during translation (Robert and Brakier-Gingras 2003). Mutations that disrupt this interaction affect translational fidelity in Escherichia coli, leading to an increased capacity for frameshifting and readthrough (Robert and Brakier-Gingras 2003). In addition, E. coli rpS7 initiates assembly of the 30S subunit by binding to 16S rRNA (Nowotny and Nierhaus 1988; Fredrick et al. 2000; Grondek and Culver 2004). In contrast, very little is known about the function of rpS7’s eukaryotic rpS5 counterpart. In Saccharomyces cerevisiae, rpS5 is represented by a single gene copy and is essential for cell viability (Ignatovich et al. 1995). The bacterial rpS7/rpS11 interaction is not conserved in eukaryotes (rpS5/rpS14) (Robert and Brakier-Gingras 2003) and the mRNA exit channel formed by the rpS5/rpS14 interaction is open in isolated yeast 40S subunits (Passmore et al. 2007). Interestingly, eukaryotic rpS5 interacts with the cricket paralysis virus (CrPV) IRES (Pfingsten et al. 2006; Schuler et al. 2006) and was suggested to be one of the determinants that might facilitate recruitment of this IRES to the 40S subunit.
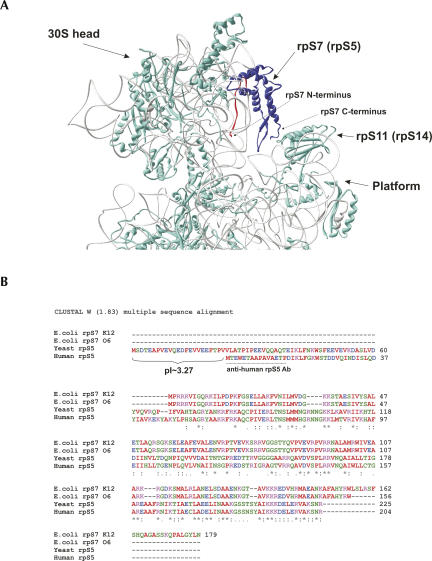
FIGURE 1.
Structure, ribosomal location, and sequence alignments of ribosomal protein S7 (rpS5). (A) Location of rpS7 within the head of the E. coli 30S ribosomal subunit. PDB file 1VS7 was used for modeling. Image was produced using Deep View 3.7 software. The rpS7 protein is in blue, and its 20 amino-terminal amino acids are in red. (B) Sequence alignments of E. coli strain K12 rpS7 (GenBank accession no. NP_417800), E. coli strain O6:H1/CFT073 (NP_755978), S. cerevisiae rpS5 (NP_012657), and H. sapiens rpS5 (NP_001000). The pI value of the N-terminal extension of yeast rpS5 is shown, and the peptide used to elicit anti-human rpS5 antibodies is underlined.
Alignment of rpS5/rpS7 from metazoans (Homo sapiens), fungi (S. cerevisiae), and bacteria (E. coli) shows that the proteins contain a conserved central/C-terminal core and possess variable N-terminal regions (Fig. 1B). The very C-terminal regions also vary even between different E. coli strains. Thus, rpS7 in the K12 strain is 23 amino acids longer than in the O6 and B+ strains (Fig. 1B). Although human rpS5 is 67% identical and 79% similar to S. cerevisiae rpS5, it lacks a negatively charged (pI ∼3.27) 21-amino acid long N-terminal extension that is present in fungi. To investigate the function of rpS5 and in particular the role of the negatively charged N-terminal extension of the yeast protein, we obtained and characterized a yeast strain in which yeast rpS5 was replaced by its human homolog. Our data suggest that rpS5 plays important roles in ensuring the efficiency of elongation, in maintaining the reading frame for translation, and in stop codon recognition and that the negatively charged N-terminal extension of yeast rpS5 might affect the ribosomal recruitment of specific mRNAs.
RESULTS
Human rpS5 substitutes for its yeast homolog in vivo
To determine whether human rpS5 (hrpS5) can substitute for yeast rpS5 (yrpS5) in vivo, we used an experimental approach that would ensure that human rpS5 in the mutant strain was expressed at a level equal to that of yeast rpS5 in the wild-type (WT) strain. In eukaryotic cells and particularly in yeast, balanced expression of ribosomal proteins is mainly achieved at the post-transcriptional level and, more specifically, through the regulated turnover of ribosomal proteins (elBaradi et al. 1986; Maicas et al. 1988; Tsay et al. 1988). Thus, individual ribosomal proteins that are present in excess in the cell are rapidly degraded until they reach levels identical to other ribosomal proteins (elBaradi et al. 1986; Maicas et al. 1988; Tsay et al. 1988). The hrpS5 coding sequence was therefore cloned into the high copy number 2μ vector to ensure high level hrpS5 expression; the assumption that putative excess hrpS5 would be degraded to maintain hrpS5 at the same level in the mutant strain as yrpS5 in the WT strain was confirmed by Western blotting (see below). hrpS5 was placed under the control of the constitutive (TEF) promoter (derived from the elongation factor 1A gene), which was chosen because the TEF gene belongs to the cluster of ribosomal protein genes and its expression is tightly co-regulated with the expression of ribosomal proteins in yeast (Ihmels et al. 2002).
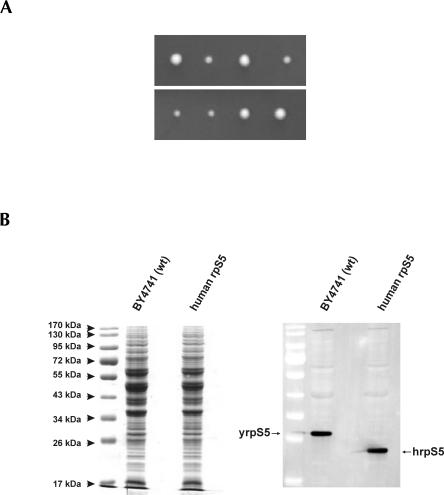
The resulting pTEF_humanS5 (2μ, LEU2) plasmid was transformed into a diploid heterozygous yeast strain with a single disrupted copy of the RPS5 gene. Transformants were allowed to sporulate and tetrads were dissected. Tetrad dissection analysis revealed that all four spores were viable, however two of them gave rise to colonies growing at reduced rates (Fig. 2A). PCR analysis using chromosomal DNA isolated from all four clones showed that clones growing at reduced rates contained a disrupted copy of the yeast RPS5 gene (not shown). Thus, hrpS5 was the sole source of rpS5 in these two strains, and we therefore conclude that human rpS5 can substitute for its yeast homolog in vivo. The BY4743 strain transformed with the empty p425TEF plasmid (2μ, LEU2) gave rise to only two viable colonies after sporulation and tetrad dissection, confirming that rpS5 is essential for yeast viability (not shown). A haploid strain (BY47hS5) with the genotype (MATa his3–1, leu2–0, ura3–0, rps5∷kanMX, <hrps5; LEU2, 2μ>) was used for further detailed characterization. This strain displayed slightly reduced (∼20%–25%) growth rates in comparison with the WT strain BY4741 transformed with the empty p425TEF LEU2 when grown on either rich YEPD medium or minimal SD medium. The doubling time for this strain at 30°C in liquid YEPD glucose medium was found to be ∼2 h, while for the WT BY4741 strain it is about 1.5 h. The expression levels of yrpS5 and hrpS5 in the WT and the mutant yeast strain were determined by Western blotting using an antibody directed against the AIKKKDELERVAKSNRC C-terminally conserved rpS5 peptide (provided by Shuetsu Fukushi, BioMedical Laboratories, Saitama, Japan) that is able to recognize the yeast and the human protein. This analysis showed equal levels of expression of the rpS5 proteins in the two strains (Fig. 2B), validating comparisons of other aspects of translation in the two strains.
FIGURE 2.
Tetrad analysis and expression levels of the rpS5 in the WT and the mutant yeast strains. (A) Tetrad dissection analysis. Two representative dissections are shown. (B) SDS-PAGE and Coomassie staining (left panel) and Western blot analysis of the crude cell extracts done using anti-rpS5 antibody derived against the AIKKKDELERVAKSNRC C-terminally conserved S5 peptide capable of recognizing both yeast and the human protein (right panel). Arrows point to the yeast (yrpS5) and the human hrpS5 correspondingly.
Polyribosome analysis of the mutant hrpS5 yeast strain
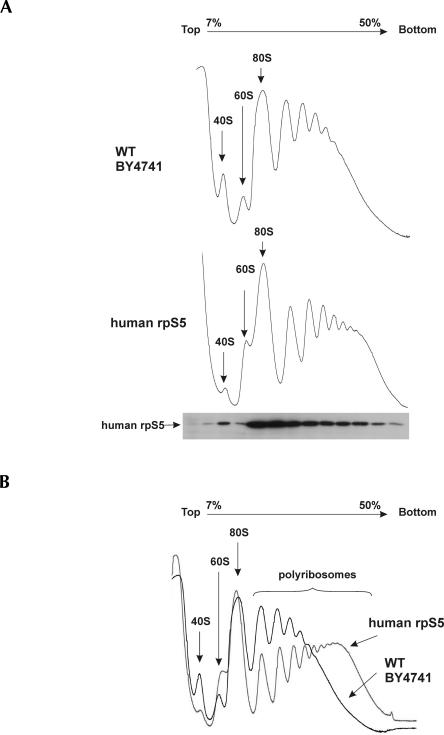
Sucrose density-gradient centrifugation analysis of cytoplasmic extracts prepared from the WT or BY47hS5 strains showed similar amounts of 80S ribosomes in both strains, but reduced levels of 40S subunits and increased levels of 60S subunits in the mutant strain compared to the WT strain (Fig. 3A,B). Interestingly, rpS5 substitution also led to a moderate, but reproducible, increase in the heavy polyribosomal components in the mutant yeast strain compared to the WT strain (Fig. 3A,B). Western blotting analysis done using antibodies directed against a specific N-terminal MTEWETAAPAVAETPD peptide of hrpS5 showed that hrpS5 was present in fractions containing 40S subunits, 80S ribosomes, and polyribosomes (Fig. 3A), thus confirming that it is a stable component of hybrid ribosomes that are actively involved in protein synthesis.
FIGURE 3.
Ribosome profiles of the WT and mutant yeast strain containing human rpS5 protein and Western blot analysis of the mutant yeast strain. (A) Extracts from WT (upper panel) and human rpS5 mutant (lower panel) strains were resolved by velocity sedimentation on 7%–50% sucrose gradients. Fractions were collected while scanning at A254. The positions of different ribosomal species are indicated. The distribution of human rpS5 in different ribosomal fractions from the human rpS5 mutant strain was assessed by Western blot analysis of sucrose gradient fractions, collected, and resolved by SDS-PAGE followed by immunoblot analysis using antibodies against human rpS5. (B) Ribosome profiles of WT and human rpS5 strains superimposed for better visualization of the profile differences. To underline the difference, ribosome profile showing the highest level of heavy polysome accumulation for the human rpS5 strain is presented in this panel.
Interaction of eEF3 with ribosomes is reduced in the mutant hrpS5 yeast strain
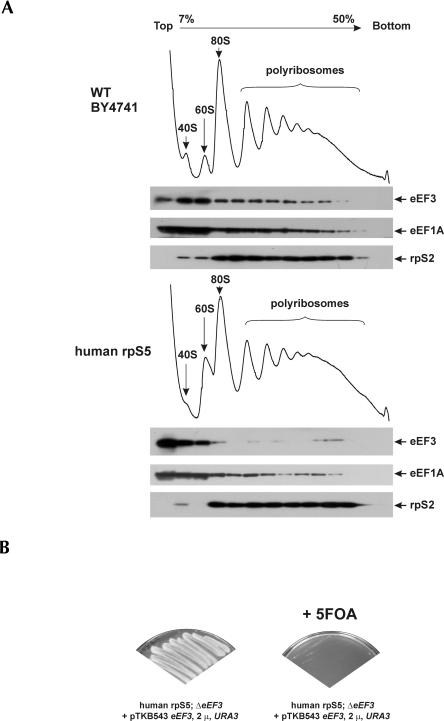
Increased accumulation of the heavy polyribosomal components in the mutant yeast strain suggests that either elongation or termination is compromised in the mutant strain. Yeast-specific elongation factor eEF3 is suggested to facilitate release of deacylated tRNA from the E-site in an ATP-dependent manner, thereby allowing binding of the next aminoacyl-tRNA to the A-site (Chakraburtty 2001; Andersen et al. 2006). Recent cryo-EM visualization of eEF3 bound to 80S ribosomes showed that this unique fungal elongation factor uses an entirely novel factor-binding site near the E-site (Andersen et al. 2006). Although the resolution was not sufficient to assign all the eEF3 interacting partners on the ribosome surface, the unassigned density (rpSX2) (Andersen et al. 2006) was suggested to correspond to the N-terminal part of rpS5. Thus, modification of rpS5 and specifically of its N-terminal region could potentially compromise ribosomal binding of eEF3, leading to reduced elongation rates and accumulation of the heavy polyribosomal components. To verify whether this was indeed the case, the distribution of eEF3 across the gradient was determined. Consistent with our hypothesis, Western blotting showed that polysomal association of eEF3 had decreased in the mutant strain (Fig. 4A). eEF3 interacts with eEF1A through its C-terminal end, and it has been suggested that this interaction facilitates the recruitment of eEF1A to yeast ribosomes (Anand et al. 2006). Consistently, polyribosomal association of eEF1A in the mutant strain was also reduced (Fig. 4A). Replacement of yrpS5 with hrpS5 could potentially change the interaction of deacylated tRNA with the E-site and increase its spontaneous dissociation, which in turn could influence the requirement for eEF3. We therefore determined whether eEF3 was still required for viability of the mutant yeast strain. Disruption of the YEF3 gene (encoding the essential copy of eEF3) by homologous recombination resulted in a nonviable strain (Fig. 4B), thus indicating that eEF3 was still essential for the viability of the mutant hrpS5 yeast strain.
FIGURE 4.
Reduced association of eEF1A and eEF3 with mutant yeast ribosomes containing human rpS5. (A) Analysis of polysome profiles of WT and human rpS5 mutant yeast strains. Cell extracts were resolved by velocity sedimentation on 7%–50% sucrose gradients. Fractions were collected while scanning at A254. The positions of different ribosomal species are indicated. Western blot analysis of sucrose gradient fractions, resolved by SDS-PAGE and analyzed by immunoblotting using antibodies against the proteins indicated on the right. (B) eEF3 is required for growth of the human rpS5 mutant yeast strain. This strain was transformed with a (CEN, URA3) vector harboring a WT copy of the eEF3 gene under its own promoter (Anand et al. 2006). The chromosomal copy of the eEF3 gene was disrupted in this strain by homologous recombination using the HIS3 cassette (as described in Materials and Methods). This strain was unable to grow on the selective media containing 5-Fluoroorotic Acid (5-FOA), which counter-selects cells containing a functional copy of the URA3 gene. A YNB minimal medium, with appropriate amino acids, containing 2% glucose and 5-FOA (2 mg/mL) was used (right panel).
Fidelity of translation is compromised in the mutant hrpS5 yeast strain
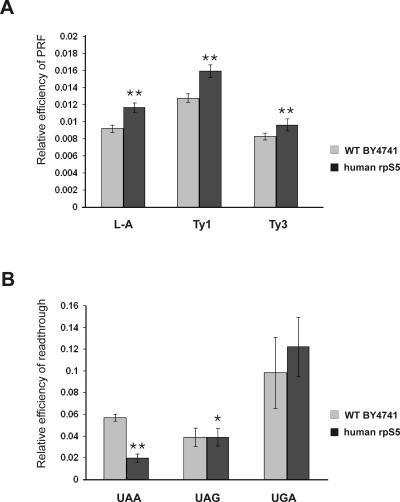
The ribosomal E-site plays a significant role in maintaining the accuracy of translation, and modifications of the E-site have been shown to result in increased frameshifting and readthrough of nonsense codons (Nierhaus 2006; Wilson and Nierhaus 2006). Thus, mutations in prokaryotic rpS7 yielded more error-prone ribosomes (Robert and Brakier-Gingras 2003). We therefore determined whether translation fidelity was also compromised in the mutant hrpS5 yeast strain. The yeast L-A virus system provides a highly sensitive assay to detect programmed −1 ribosomal frameshifting (Harger and Dinman 2003, 2004). The genome of L-A virus contains two overlapping ORFs (encoding Gag and pol proteins), which are joined by a programmed −1 ribosomal frameshift (PRF) signal, and a −1 PRF event is needed for the synthesis of the fusion protein (Gag–pol) (Dinman 1995). The S. cerevisiae Ty1 and Ty3 endogenous retrotransposons provide an assay system to study programmed +1 ribosomal frameshifting (Dinman 1995). To monitor translation fidelity, we used a bicistronic dual-luciferase reporter system, in which the frameshift signals L-A, Ty1, or Ty3 were inserted between the Renilla and firefly luciferase genes so that firefly luciferase can only be produced in the event of a frameshift (Harger and Dinman 2003, 2004). Substitution of yrpS5 with its human homolog resulted in a moderate increase in programmed frameshifting (∼1.2–1.3-fold) for all three signals (Fig. 5A). To investigate whether nonsense suppression is also affected in the mutant strain, we employed similar dual-luciferase reporters each containing one of the stop codons (UAA, UAG, or UGA) inserted into the firefly luciferase gene so that firefly luciferase can only be produced as a result of a nonsense suppression (Harger and Dinman 2003, 2004). We have found that the S5 substitution affected the ability of the mutant strain to suppress stop codons in a codon-dependent manner. Thus, the mutant strain exhibited hyperaccurate recognition of the UAA codon (approximately twofold enhancement), and no significant change in the recognition of the UAG or UGA codons (Fig. 5B). Interestingly, the UAA codon is the most frequently used stop codon in yeast and is recognized more efficiently than the other stop codons (Bertram et al. 2000; Baxter-Roshek et al. 2007). Our data, together with the recent findings that changes in the ribosomal structure caused by undermodification of rRNA differentially affect recognition of termination codons in yeast leading in particular to hyperaccurate recognition of the UAA codon (Baxter-Roshek et al. 2007), support the suggestion that the efficiency of recognition of different stop codons might depend on the ribosomal structure (Baxter-Roshek et al. 2007).
FIGURE 5.
Altered translation fidelity in the human rpS5 mutant yeast strain expressing human rpS5 protein. (A) Programmed frameshifting efficiency. WT and human rpS5 strain were transformed with the control, L-A, Ty1, and Ty3 frameshift reporter plasmids. Dual-luciferase assays were performed and programmed frameshifting (PRF) efficiencies were calculated as described in Materials and Methods. Mean efficiencies determined from at least six independent experiments are plotted and bars represent the corresponding standard error. (B) Termination codon readthrough efficiency in WT and human rpS5 mutant yeast strains. The WT and human rpS5 mutant yeast strains expressing human rpS5 were transformed with control, UAA, UAG, and UGA reporter plasmids and readthrough efficiencies at each terminator were determined as described in Materials and Methods. Mean efficiencies determined from at least six independent experiments are plotted and bars represent the corresponding standard error. The significance of differences in signals between mutant and WT strain is indicated: *P <0.05, **P <0.001.
IRES-mediated initiation in the mutant hrpS5 yeast strain
The CrPV IRES binds directly to 40S subunits in the absence of initiation factors (Wilson et al. 2000), and its interaction with rpS5 has been proposed to play a critical role in ribosomal recruitment of this IRES (Pfingsten et al. 2006; Schuler et al. 2006). We therefore investigated the influence of substitution of yrpS5 with its human homolog on the ability of yeast cells to support translation mediated by the CrPV IRES. In addition, the effect on translation directed by cellular IRESs including yeast URE2 and mammalian CAT-1 and SNAT-2 IRESs was assessed.
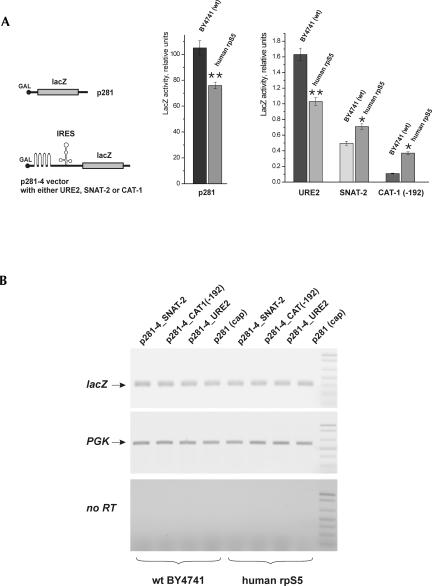
To investigate cap-dependent translation, we used a construct (p281), in which the lacZ gene is under control of the GAL1 promoter (Mueller et al. 1987). LacZ expression in the mutant strain was reduced by ∼20%–25% compared to the WT strain (Fig. 6A). To study the effect of the yrpS5 substitution on yeast URE2 IRES-mediated expression, the previously described Ure2p–lacZ reporter system was employed, in which the URE2 IRES was inserted in frame in front of the lacZ reporter gene but behind a stable hairpin structure (ΔG >−30 kcal/mol), which nearly abolished cap-dependent lacZ expression (Komar et al. 2003, 2005). Expression from the URE2 IRES in the mutant strain was 25%–30% lower than in the WT strain (Fig. 6A), similar to the reduction observed for cap-dependent lacZ expression. These data indicate that the yrpS5 substitution did not specifically affect URE2 IRES-mediated expression. To investigate the activities of two mammalian IRESs in yeast, the SNAT-2 IRES (Gaccioli et al. 2006) and the CAT-1 IRES (Fernandez et al. 2001; Yaman et al. 2003), these IRESs were cloned into the p281–4 vector behind the stable hairpin structure described above and fused to the lacZ reporter gene. The SNAT-2 and the CAT-1(-192) IRESs (Yaman et al. 2003) were active in the WT yeast strain (Fig. 6A), and the activities of both SNAT-2 and CAT-1 IRESs were enhanced in the mutant yeast strain (Fig. 6A), suggesting that substitution of yrpS5 with its human homolog likely facilitated the recruitment of these IRESs to the yeast translational apparatus. No apparent differences in lacZ mRNA levels were observed between the WT and the mutant yeast strains (Fig. 6B).
FIGURE 6.
The activities of mammalian SNAT-2 and CAT-1 IRESs are enhanced in the human rpS5 mutant yeast strain. (A) Expression of reporter constructs (schematic of the constructs is shown in the left panel) under the control of the GAL1/10 promoter was transformed into BY4741 (WT) and human rpS5 (mutant) yeast strains. β-Galactosidase activity (relative units) for URE2, SNAT-2, and CAT-1(-192) IRES constructs and for the p281 monocistronic construct (absolute units) was determined as described in Materials and Methods after a 20-h induction of BY4741 (WT) or human rpS5 (mutant) yeast cells, as indicated, in 2% galactose at 30°C. The significance of differences in signals between mutant and WT strain is indicated: *P <0.05, **P <0.01. (B) Semi-quantitative RT-PCR analysis of the expression of the lacZ reporter mRNA in comparison with the expression of the endogenous phosphoglycerate kinase (PGK) mRNA.
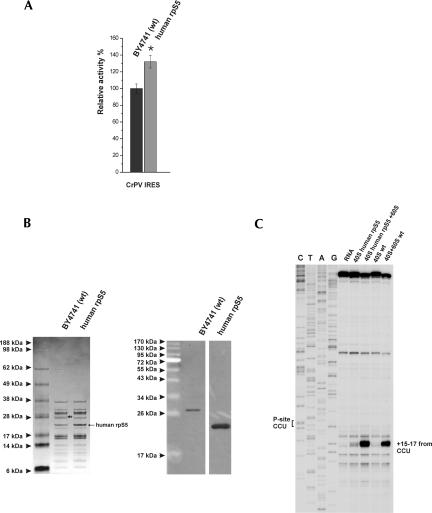
Although the CrPV IRES is active in yeast, under normal growth conditions its activity is very low but can be strongly increased by reducing the concentration of eIF2•GTP•Met-tRNAMet i ternary complex (Thompson et al. 2001). To test whether the yrpS5 substitution might affect CrPV IRES activity, the CrPV IRES was cloned into the p281–4 plasmid behind the stable hairpin structure and fused to the lacZ reporter gene. We were unable to detect significant CrPV IRES activity under normal growth conditions in either WT or mutant yeast strains (not shown). However, in agreement with previously reported data (Thompson et al. 2001), reduction in the concentration of eIF2•GTP•Met-tRNAMet i ternary complex due to overexpression of constitutively active GCN2 kinase resulted in stimulation of CrPV IRES activity in both WT and mutant strains. Under such conditions, the activity of the CrPV IRES in the mutant strain was ∼1.3-fold higher than in the WT strain (Fig. 7A). To verify that the increase in the activity of the CrPV IRES in the mutant strain was due to enhanced affinity of the IRES to hrpS5-containing hybrid yeast ribosomes, the direct interaction between the CrPV IRES and the WT and mutant ribosomes was assayed by toeprinting. Gel electrophoresis and Western blotting confirmed that hrpS5 was a stable constituent of purified mutant 40S ribosomal subunits (Fig. 7A,B). Toeprinting analysis of a reaction mixture that contained the CrPV IRES and WT 40S subunits did not yield any detectable characteristic toeprint +15–17 nucleotides (nt) downstream from the P-site CCU codon (Fig. 7C), which indicated that the binding of CrPV IRES to WT yeast 40S subunits was either very inefficient or extremely unstable. In contrast, hrpS5-containing hybrid yeast 40S ribosomes yielded a weak characteristic stop at the +15–17 position (Fig. 7C), suggesting that the presence of hrpS5 enhanced the affinity of yeast 40S subunits to the CrPV IRES. However, very prominent toeprints at +15–17 nt downstream from the P-site CCU codon were observed for both WT and hybrid 80S ribosomes (Fig. 7C), but, again, the signal obtained with hybrid ribosomes was stronger. Comparison of the band intensities (normalized to the background) by the use of the ImageJ program (Wayne Rasband, NIH) showed that the toeprint from the hybrid 80S ribosomes is ∼1.33-fold stronger than that of the WT 80S. This correlates strongly with the in vivo ∼1.3-fold increased activity of the CrPV IRES in the mutant yeast strain (Fig. 7A). However, in contrast to higher eukaryotes, the CrPV IRES was found to bind stably only to yeast 80S ribosomes but not to yeast 40S subunits. Our data therefore suggest that the 21 amino acid long N-terminal extension of yrpS5 has a negative effect on the interaction of the CrPV IRES RNA with the yeast WT 40S subunits and 80S ribosomes.
FIGURE 7.
Expression of human rpS5 in human rpS5 (mutant) yeast cells and its presence in mutant ribosomes enhance the activity of the CrPV IRES and its interaction with yeast ribosomes. (A) Expression of reporter constructs under the control of the GAL1/10 promoter transformed into BY4741 (WT) and human rpS5 (mutant) yeast strains, which were also cotransformed with a constitutive allele of the GCN2 kinase GCN2 c-515 (GCN2 c-E601K-E1606G). IRES activities were normalized to the level of cap-dependent initiation in the same strains. The significance of differences in signals between mutant and WT strain is indicated: *P <0.05. (B) Human rpS5 is an integral component of isolated 40S ribosomal subunits, as shown by SDS-PAGE and Coomassie staining (left panel) and Western blot analysis done using anti-yeast and anti-human rpS5 antibodies (right panel). The short black arrow indicates the position of the yeast rpS5; the position of the human rpS5 is as indicated. (C) Toeprint analysis of 40S and 80S ribosomal complexes assembled on CrPV IRES RNA. The CCU triplet present in the ribosomal P-site is indicated on the left, and the positions of toeprints relative to nucleotide +1 of this triplet are indicated to the right of the panel. Lanes C, T, A, and G depict the CrPV sequence.
DISCUSSION
To gain insight into the function of S. cerevisiae rpS5 and its N-terminal extension, which is absent in human rpS5, we replaced S. cerevisiae rpS5 with its human homolog. Although human rpS5 is 21 amino acids shorter than yeast rpS5, this replacement did not result in any significant negative growth phenotypes. The mutant yeast strain was viable and its growth rate was only moderately (20%–25%) reduced. This suggests that the 21 amino acids N-terminal extension of yeast rpS5 may play only a modulatory role in the function of the yeast ribosome. To date there are only a few reports that a yeast ribosomal protein can be replaced with a mammalian (or insect) homolog. Yeast rpL25 can be replaced by rat and Drosophila melanogaster rpL23A (Jeeninga et al. 1996; Ross et al. 2007) and mouse ribosomal protein rpL27′ can substitute for the yeast rpL29 (Fleming et al. 1989). These replacements all involved the large (60S) ribosomal subunit. Thus, the replacement of the yeast rpS5 with its human homolog is the first example of such a substitution to be made in the small ribosomal subunit. This result also shows the conservation of function between the yeast and the human rpS5 proteins. However, certain ribosomal activities were perturbed. The increase in heavy polyribosomes in the mutant was suggestive of a mild defect in elongation or termination. The former hypothesis is consistent with the decreased association of eEF1A and the unique fungal elongation factor eEF3 with the mutant polyribosomes. The decreased ribosomal association of eEF3 could be due either to a decreased affinity to the mutant ribosomes or because the complex formed by binding of eEF3 to mutant ribosomes is less stable and therefore short-lived. Recent cryo-EM reconstructions of eEF3 bound to the yeast ribosome (Andersen et al. 2006) have identified several contacts between eEF3 and both 40S and 60S ribosomal subunits. Two regions in eEF3 interact with the small ribosomal subunit: the eEF3 HEAT domain interacts with helix 39 of 18S rRNA, an unknown adjacent ribosomal protein and rpS18, whereas the chromodomain/ABC2 region interacts with rpS18 and an as-yet unassigned density (rpSX2) that likely corresponds to the N-terminal extension of rpS5 (Andersen et al. 2006). Our data corroborate the suggestion that the positively charged C terminus of eEF3 might interact with the negatively charged N-terminal extension of rpS5. This interaction might be necessary to adjust the position of eEF3 on the yeast ribosome following its proposed initial weak binding. Interestingly, deletion of the eEF3 C-terminal 64 amino acids (980–1044), containing 40% basic residues, also yields a strain that is characterized by slightly decreased growth rates (Anand et al. 2006), similarly to what we observed for the mutant yeast strain expressing human rpS5. Further experiments are required to elucidate the exact role of the N-terminal extension of yeast rpS5 in positioning eEF3 on yeast ribosomes.
The translational fidelity of the strain expressing the hybrid ribosomes was altered relative to the wild-type strain, displaying moderately increased levels of frameshifting as well as an altered ability to recognize the UAA stop codon. It has been proposed that the E-site is allosterically coupled to the A-site and that this coupling controls the ability of the ribosomes to discriminate between cognate and noncognate tRNAs at the A-site (Nierhaus 2006). The observation that the translational fidelity of mutant ribosomes with a modified E-site was altered is consistent with this hypothesis. The reduced binding of eEF3 to mutant ribosomes, which favors binding of cognate aminoacyl-tRNA at the A-site (Uritani and Miyazaki 1988) may also contribute to altered translational fidelity. A similar effect has been previously observed in the case of mutations in E. coli rpS7 (the bacterial rpS5 homolog), which disrupted its interaction with rpS11 (Robert and Brakier-Gingras 2003). The authors likewise suggested that mutations in rpS7 might have impaired the coupling between the E- and the A-site and that this contributed to the reduced translation fidelity (Robert and Brakier-Gingras 2003). Allosteric coupling between A- and E-sites has also been suggested to influence translation termination events (Nierhaus 2006). Our experiments revealed the hyperaccurate (approximately twofold enhanced) recognition of a UAA stop codon in the mutant strain relative to the WT strain, whereas there was no significant change in the recognition of a UAG or UGA codon. The reason for the hyperaccurate recognition of UAA codons but not of other termination codons in the mutant strain is not clear. It might be that the relative positioning of UAA, UAG, and UGA codons in the ribosomal A-site differs slightly and that their recognition by eRF1 could be influenced by an aspect of ribosomal architecture, presumably altered in the mutant strain. Similar hyperaccurate recognition of the UAA codon has been recently reported in a mutant yeast strain containing undermodified rRNA bases (Baxter-Roshek et al. 2007). It was proposed that the efficiency of recognition of the three stop codons might be critically dependent on the ribosome structure that can be affected by modifications of rRNA (Baxter-Roshek et al. 2007). Modifications of rpS5 might have a similar effect.
We also investigated whether modifications to the E-site caused by heterologous replacement of rpS5 affected translation in an mRNA-specific manner. We focused on IRES-containing mRNAs, utilizing the CrPV IRES that functions in yeast (Thompson et al. 2001) because it interacts primarily with the ribosomal E-site and because it has been suggested that rpS5 may play a critical role in recruiting this IRES to the 40S subunit (Pfingsten et al. 2006; Schuler et al. 2006). Indeed, toeprinting experiments indicated that purified mutant yeast ribosomes containing human rpS5 had an increased affinity to the CrPV IRES in vitro. This observation was supported by in vivo experiments showing increased levels of CrPV IRES activity in the mutant yeast strain under permissive conditions when eIF2•GTP•Met-tRNAMet i levels were reduced by constitutive overexpression of the GCN2 kinase. In sharp contrast to mammalian 40S subunits, the CrPV IRES does not bind stably to yeast 40S subunits, even to those containing human rpS5, but does bind to WT 80S yeast ribosomes and somewhat more strongly to mutant 80S ribosomes (Fig. 7C). This binding may account for the enhancement in translation mediated by this IRES in conditions that favor accumulation of 80S ribosomes. Direct and stable binding of the IRES to mammalian 80S ribosomes has been reported previously (Pestova et al. 2004). Interestingly, two mammalian cellular IRESs (the SNAT-2 IRES and the CAT-1(-192) IRES) were active in yeast and mediated a higher level of translation in the mutant yeast strain than in the wild-type strain. The factors required for initiation on the SNAT-2 and the CAT-1 IRESs and the mechanism(s) of their recruitment to the ribosome are not known. In light of the importance of the E-site for the activity of other viral IRESs (e.g., Spahn et al. 2001b), it is possible that the architecture of the E-site might also be critical for the activity of these cellular IRESs and that the presence of the homologous mammalian rpS5 might facilitate their recruitment to the yeast ribosome. However, the precise location of the 21 amino acids C-terminal extension of yeast rpS5 has not been determined, so that details of how it might affect IRES binding are not known. The resolution of cryo-EM reconstructions of yeast ribosomes (Spahn et al. 2001a, 2004) is not sufficient to visualize the rpS5 N-terminal extension. Interestingly, the N terminus of the homologous bacterial rpS7 points to the mRNA channel between the head and the platform (Fig. 1A; Yusupov et al. 2001). It is tempting to speculate that the additional 50 N-terminal amino acids present in human rpS5 (and ∼70 amino acids present in yeast rpS5) might protrude into the mRNA channel and thus interfere with binding of some mRNAs. Taken together the data presented here indicate that eukaryotic rpS5 plays a role in a variety of ribosomal activities, including maintenance of the translation reading frame, recognition of stop codons as well as recruitment of specific mRNAs to the translational machinery.
MATERIALS AND METHODS
Yeast strains and growth methods
Yeast strains used in this study are: Research Genetics wild-type haploid BY4741 (MATa, his3–1, leu2–0, met15–0, ura3–0), diploid BY4743 ([4741/4742] MATa/MATα, his3–1/his3–1, leu2–0/leu2–0, lys2–0/+, met15–0/+, ura3–0/ura3–0, RPS5/rps5∷kanMX). The human rpS5 strain BY47hS5 Mata his3–1, leu2–0, ura3–0, rps5∷kanMX, <hrps5; LEU2, 2μ> and strain BY47hS51 MATα his3–1, leu2–0, lys2–0, ura3–0, rps5∷kanMX, <hrps5; LEU2, 2μ> were obtained as follows: cDNA of human ribosomal protein S5 was amplified by PCR using 5′-AACGCGGATCCGCTCAGGCTGTGTTCTCAG-3′ and 5′-AAAAAAAAGTCGACGGCTGGGACTGCCCCAAAG-3′ primers and pCMV-SPORT6 plasmid containing cDNA for human rpS5 (MGC-21949) as a template. The PCR fragment was further digested with BamHI and SalI and cloned into p425TEF (2μ, LEU2) (Mumberg et al. 1995) digested with the same enzymes. The resultant pTEF_hS5 plasmid was transformed into the heterozygous diploid. Transformants were allowed to sporulate (using standard protocols; Rose et al. 1990) and tetrads were dissected. Haploid clones able to grow on YPED medium, resistant to G418 sulfate, and expressing human rpS5 were selected. An rps5∷kanMX genotype of these clones was further verified by PCR using 5′-CATAATACCAAGAAAAGAGACTAGAAATAACCG-3′ and 5′-GAAAAACATATGTAATATTGAAAATCTTTCACTTTTTTTAG-3′ primers.
Yeast cultures were grown as indicated using either synthetic media containing 0.67% Difco yeast nitrogen base, 1% ammonium sulfate, 2% glucose (or galactose), and supplemented with the appropriate amino acids or YEPD medium (Rose et al. 1990). Transformation was done using the lithium acetate method (Ito et al. 1983). For polysome analysis, yeast cells were grown in YEPD medium with 2% glucose or galactose.
Plasmids
The p281 plasmid containing lacZ under GAL1/10 promoter has been described previously (Mueller et al. 1987). The p281–4-URE2 and p281–4-URE2_CTT vectors have also been previously described (Komar et al. 2003, 2005). The p281–4-CAT-1(-192), p281–4-SNAT-2, and p281–4-CrPV plasmids were made by subcloning the respective IRES elements as XhoI-EcoRI or SacII-EcoRI fragments into the p281–4 plasmid (Altmann et al. 1993). The CAT-1(-192) IRES (Yaman et al. 2003) was amplified by PCR using 5′-AAAAACTCGAGCCTTGCAGGGGCGTGAAGCTACT-3′ and 5′-AAAAAGAATTCTCATCGCGCTGAGCAAATCTGTCTG-3′ primers. The SNAT-2 IRES (Gaccioli et al. 2006) was amplified using 5′-AAAAACTCGAGCGACGCCGCCGCCTTAGAAC-3′ and 5′-AAAAAGAATTCTCATGCTAAGCACTGGGAGGAATCGG-3′ primers. PCR fragments were digested with XhoI and EcoRI and were cloned into the p281–4 vector. The CrPV-IGR IRES (Wilson et al. 2000) was amplified by PCR using 5′-AAAAACCGCGGAAAAATGTGATCTTGCTTGTAAATACAATTTTGAG-3′ and 5′-AAAAAGAATTCTAGCAGGTAAATTTCTTAGGTTTTTCGACTACCA-3′ primers, digested with SacII and EcoRI and cloned into the p281–4 vector. The GCN2c-515 (GCN2c-E601K-E1606G) allele of GCN2 kinase (Ramirez et al. 1992) in the p1054 plasmid was kindly provided by Dr. Tom Dever (NIH). This allele was cloned into pRS313 (CEN, HIS3) (Sikorski and Hieter 1989) as a SalI-NotI fragment. Programmed –1 and +1 frameshifting test reporters containing L-A, Ty1, or Ty3 frameshift signals, respectively, between the Renilla and firefly luciferase genes and nonsense suppression test reporters containing three different stop codons (UAA, UAG, or UGA) in the firefly luciferase gene (Harger and Dinman 2003, 2004) were provided by Dr. Jonathan D. Dinman (University of Maryland). The monocistronic CrPV IGR IRES-containing transcription vector has been described (Wilson et al. 2000). All luciferase reporter plasmids were transformed into WT and BY47hS5 strains and grown on the minimal YNB medium.
Semiquantitative RT-PCR analysis of RNA expression levels
The levels of IRES-containing RNA expressed from the respective p281–4 plasmids were assessed by RT-PCR analysis using the SuperScript system from Invitrogen. Total RNAs were extracted after glass bead yeast cell disruption using the TRIzol Reagent (Invitrogen). Prior to RT-PCR reactions the RNAs were additionally treated with DNAse I (Ambion) to remove possible DNA admixtures. The following oligonucleotide primers were used: 5′-GCGTGGCAGCATCAGGGG-3′ and 5′-CGTGCAGCAGATGGCGATGGC-3′ to amplify the lacZ containing mRNAs and 5′-GGACTTGAAGGACAAGCGTGTCTTC-3′ and 5′-CCACCTAAGATGGCCAAGAATGGT-3′ to amplify the phosphoglycerate kinase (PGK) mRNA for use as an internal control. No more than 25 amplification cycles were routinely used in the second PCR step.
Fractionation of polyribosomes and isolation of 40S and 60S subunits
Fractionation of polyribosomes was done essentially as described (Komar et al. 2005). All procedures were performed at 4°C except where indicated. Yeast cells from 50 mL of log phase culture were pelleted, treated for 1 min with 10 μg/mL cycloheximide, and repelleted. Lysates were made by glass bead cell disruption (3–5 cycles of 1 min each), with intermittent cooling on ice, in buffer which contained 100 mM KCl, 2.5 mM magnesium acetate, 20 mM HEPES•KOH, pH 7.4, 14.4 mM β-mercaptoethanol, 100 μg/mL cycloheximide. Cell debris was removed by centrifugation at 7000 rpm for 8 min. Polyribosomes, ribosomes, and subunits were resolved in either 7%–25% (20,000 rpm, 18 h) or 7%–50% (17,000 rpm, 18 h) sucrose gradients containing 100 mM KCl, 5 mM MgCl2, 20 mM HEPES•KOH, pH 7.4, and 2 mM dithiothreitol using a Beckman SW32.1 rotor. Gradients were collected using the ISCO Programmable Density Gradient System with continuous monitoring at 254 nm using an ISCO UA-6 absorbance detector.
Individual 40S and 60S ribosomal subunits were purified as follows: Yeast cells were grown overnight in 1–1.5 L of YEPD medium to an OD600=1.5–2, pelleted at 6,000g for 10 min at 4°C and resuspended on ice in 10 mL cold breaking buffer (20 mM Tris/HCl pH 7.6, 50 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 1 mg/mL Heparin) and containing one complete EDTA-free protease inhibitor cocktail tablets (Roche)/50 mL buffer. Cells were disrupted using glass beads in a BeadBeater (BioSpec) for 3 × 1 min with intermittent cooling on ice. Cell debris was removed by centrifugation at 14,000g for 20 min at 4°C. The supernatant was layered on top of a 2 M sucrose solution in breaking buffer and the ribosomes were pelleted at 50,000 rpm for 18 h at 4°C using a Beckman 70Ti rotor. The ribosome pellet was resuspended in buffer A (20 mM Tris/HCl pH 7.6, 50 mM KCl, 4 mM MgCl2, 2 mM EDTA) containing 0.25 M sucrose to an OD260 ∼100–150 a.u., and the KCl concentration was gradually increased to a final concentration of 0.5 M by adding 4 M KCl under conditions of continuous mixing for 30 min at 4°C using a magnet stirrer. The ribosomes were pelleted again at 60,000 rpm for 4.5 h at 4°C using a Beckman TLA 110 rotor. The pellet was resuspended in buffer A to an OD ∼50–100 a.u., incubated with 1 mM puromycin for 10 min at 4°C, and subsequently for 10 min at 37°C. 40S and 60S ribosomal subunits were resolved by centrifugation in 5%–25% sucrose gradients containing 500 mM KCl, 4 mM MgCl2, 2 mM EDTA, 20 mM Tris/HCl pH 7.6 (20,000 rpm, 18 h, Beckman SW32.1 rotor). Fractions containing 40S and 60S ribosomes were collected, concentrated using Centricon 50 microconcentrators (Millipore) and resuspended in buffer (20 mM Tris/HCl pH 7.6, 50 mM KCl, 2.5 mM MgCl2, 2 mM EDTA) containing 0.25 M sucrose. For Western blotting, proteins collected from sucrose gradient fractions were precipitated with 10% trichloroacetic acid (TCA) and resolved by 10% Laemmli SDS-polyacrylamide gel electrophoresis and then transferred onto Immobilon (Millipore) membranes.
Disruption of the YEF3 gene encoding eEF3
For the YEF3 gene disruption, the HIS gene was used. The cassette was amplified from the pRS423 plasmid using 5′-CTTTCCTTAATTGTTTTCTAAAGAACCGTGTATTTTTCTAGTTCGGGAGACGGTCACAGCTTGTCT-3′ and 5′-ATTACAAAAACATAGAAATTAAAATATACATAAATTATTAGATCACGCCTCGTTCAGAATGACACGT-3′ primers. The PCR fragment was transformed into BY47hS5 strain and disruptants were selected on C-His medium. The disruption was verified by PCR analysis of chromosomal DNA using primers 5′-GACTCCGTTTAATCACTTTCAACCGC-3′ and 5′-GGGTATGAGGCAATGCTCAATTTG-3′. PCR amplification using these primers yielded a 3698 base-pair (bp) fragment if the WT gene was present and a 1765 bp fragment when disruption was successful and the YEF3 gene had been replaced with the HIS3 cassette (not shown).
Toeprinting
For toeprinting analysis, ribosomal complexes were assembled essentially as described (Pestova and Hellen 2003); 2.5 pmol of CrPV IRES-containing mRNA were incubated with 3.5 pmol WT or mutant yeast 40S subunits in the presence or in the absence of 3.5 pmol yeast 60S subunits in 40 μL buffer containing 20 mM Tris pH 7.5, 100 mM potassium acetate, 2.5 mM magnesium acetate, 2 mM DTT, and 0.25 mM spermidine for 10 min at 37°C. Primer extension was done using AMV reverse transcriptase (Promega) and 32P-phosphorylated primer complementary to nt 6341–6359 of CrPV RNA. cDNA products were analyzed in a 6% polyacrylamide sequencing gel.
Western blotting
Western blotting was performed following standard procedures (Towbin et al. 1979). Western blots were decorated with rabbit polyclonal anti-eEF3, anti-eEF1A, anti-rpS5, or anti-rpS2 antibodies followed by incubation with goat anti-rabbit HRP-conjugated antibodies. The anti-human S5 antibodies were raised in rabbits by United States Biological using a 16-mer peptide (MTEWETAAPAVAETPD) as an antigen. The antibodies to yeast rpS5 directed against the conserved C-terminal peptide (AIKKKDELERVAKSNRC) were kindly provided by Dr. Shuetsu Fukushi (BioMedical Laboratories, Saitama, Japan). The anti-S2 antibodies were kindly provided by Dr. Jonathan Warner (Albert Einstein College of Medicine, New York). The blots were then detected with an enhanced chemiluminescence detection kit (ECL™, GE Healthcare).
Miscellaneous
Molecular cloning was performed following the general procedures described in Sambrook et al. (1989). DNA sequencing was accomplished by the Molecular Biology Core Laboratory at Cleveland State University. Sequencing was performed with custom synthesized oligonucleotides using the fluorescently labeled dideoxy terminator methodology. SDS-PAGE was performed according to Laemmli (1970). Yeast genomic DNA was isolated using the DNA-Pure™ Yeast Genomic Kit (PureBiotech) and following the manufacturer's protocol. β-galactosidase activity was measured following the protocol described in the Clontech Yeast Protocols Handbook with o-nitrophenyl β-D-galactopyranoside as a substrate. Cell extracts were prepared by subsequent cycles of cell freezing in liquid nitrogen and thawing at 37°C. Luciferase activities were measured using a dual-luciferase assay kit (Promega) as described by Dinman and colleagues (Harger and Dinman 2003, 2004; Jacobs and Dinman 2004).
ACKNOWLEDGMENTS
We thank Drs. Jonathan Warner, Thomas Dever, Jonathan Dinman, and Shuetsu Fukushi for their generous gifts of antibodies and plasmids used in this study as well as for their helpful advice. Dr. Alan Tartakoff is gratefully acknowledged for his helpful advice regarding tetrad dissection analysis. We are also grateful to Dr. Martin Snider for his helpful comments and the critical reading of the manuscript. This work was supported by Cleveland State University start-up and research challenge funds and National American Heart Association grant (0730120N) to A.A.K., an NIH grant (AI-51340) to C.U.T.H., an NIH grant (DK60596) to M.H., an NIH grant (HL79164) to B.M., an NIH grant (GM68079) to W.C.M., and an NIH grant (GM57483) to T.G.K.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.688207.
REFERENCES
- Altmann, M., Muller, P.P., Wittmer, B., Ruchti, F., Lanker, S., Trachsel, H. A Saccharomyces cerevisiae homolog of mammalian translation initiation factor 4B contributes to RNA helicase activity. EMBO J. 1993;12:3997–4003. doi: 10.1002/j.1460-2075.1993.tb06077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand, M., Balar, B., Ulloque, R., Gross, S.R., Kinzy, T.G. Domain and nucleotide dependence of the interaction between Saccharomyces cerevisiae translation elongation factors 3 and 1A. J. Biol. Chem. 2006;281:32318–32326. doi: 10.1074/jbc.M601899200. [DOI] [PubMed] [Google Scholar]
- Andersen, C.B., Becker, T., Blau, M., Anand, M., Halic, M., Balar, B., Mielke, T., Boesen, T., Pedersen, J.S., Spahn, C.M., et al. Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature. 2006;443:663–668. doi: 10.1038/nature05126. [DOI] [PubMed] [Google Scholar]
- Baxter-Roshek, J.L., Petrov, A.N., Dinman, J.D. Optimization of ribosome structure and function by rRNA base modification. PLoS ONE. 2007;2:e174. doi: 10.1371/journal.pone.0000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringer, M., Rodnina, M.V. The ribosomal peptidyl transferase. Mol. Cell. 2007;26:311–321. doi: 10.1016/j.molcel.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Bertram, G., Bell, H.A., Ritchie, D.W., Fullerton, G., Stansfield, I. Terminating eukaryote translation: Domain 1 of release factor eRF1 functions in stop codon recognition. RNA. 2000;6:1236–1247. doi: 10.1017/s1355838200000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen, D.E., Nissen, P. The social life of ribosomal proteins. FEBS J. 2005;272:2098–2108. doi: 10.1111/j.1742-4658.2005.04651.x. [DOI] [PubMed] [Google Scholar]
- Chakraburtty, K. Translational regulation by ABC systems. Res. Microbiol. 2001;152:391–399. doi: 10.1016/s0923-2508(01)01210-4. [DOI] [PubMed] [Google Scholar]
- Dinman, J.D. Ribosomal frameshifting in yeast viruses. Yeast. 1995;11:1115–1127. doi: 10.1002/yea.320111202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doring, T., Mitchell, P., Osswald, M., Bochkariov, D., Brimacombe, R. The decoding region of 16S RNA: A cross-linking study of the ribosomal A, P, and E sites using tRNA derivatized at position 32 in the anticodon loop. EMBO J. 1994;13:2677–2685. doi: 10.1002/j.1460-2075.1994.tb06558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresios, J., Panopoulos, P., Synetos, D. Eukaryotic ribosomal proteins lacking a eubacterial counterpart: Important players in ribosomal function. Mol. Microbiol. 2006;59:1651–1663. doi: 10.1111/j.1365-2958.2006.05054.x. [DOI] [PubMed] [Google Scholar]
- elBaradi, T.T., van der Sande, C.A., Mager, W.H., Raue, H.A., Planta, R.J. The cellular level of yeast ribosomal protein L25 is controlled principally by rapid degradation of excess protein. Curr. Genet. 1986;10:733–739. doi: 10.1007/BF00405095. [DOI] [PubMed] [Google Scholar]
- Fernandez, J., Yaman, I., Mishra, R., Merrick, W.C., Snider, M.D., Lamers, W.H., Hatzoglou, M. Internal ribosome entry site-mediated translation of a mammalian mRNA is regulated by amino acid availability. J. Biol. Chem. 2001;276:12285–12291. doi: 10.1074/jbc.M009714200. [DOI] [PubMed] [Google Scholar]
- Fleming, G., Belhumeur, P., Skup, D., Fried, H.M. Functional substitution of mouse ribosomal protein L27′ for yeast ribosomal protein L29 in yeast ribosomes. Proc. Natl. Acad. Sci. 1989;86:217–221. doi: 10.1073/pnas.86.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrick, K., Dunny, G.M., Noller, H.F. Tagging ribosomal protein S7 allows rapid identification of mutants defective in assembly and function of 30 S subunits. J. Mol. Biol. 2000;298:379–394. doi: 10.1006/jmbi.2000.3563. [DOI] [PubMed] [Google Scholar]
- Fukushi, S., Okada, M., Stahl, J., Kageyama, T., Hoshino, F.B., Katayama, K. Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J. Biol. Chem. 2001;276:20824–20826. doi: 10.1074/jbc.C100206200. [DOI] [PubMed] [Google Scholar]
- Gaccioli, F., Huang, C.C., Wang, C., Bevilacqua, E., Franchi-Gazzola, R., Gazzola, G.C., Bussolati, O., Snider, M.D., Hatzoglou, M. Amino acid starvation induces the SNAT2 neutral amino acid transporter by a mechanism that involves eukaryotic initiation factor 2α phosphorylation and cap-independent translation. J. Biol. Chem. 2006;281:17929–17940. doi: 10.1074/jbc.M600341200. [DOI] [PubMed] [Google Scholar]
- Gazda, H.T., Sieff, C.A. Recent insights into the pathogenesis of Diamond-Blackfan anemia. Br. J. Haematol. 2006;135:149–157. doi: 10.1111/j.1365-2141.2006.06268.x. [DOI] [PubMed] [Google Scholar]
- Grondek, J.F., Culver, G.M. Assembly of the 30S ribosomal subunit: Positioning ribosomal protein S13 in the S7 assembly branch. RNA. 2004;10:1861–1866. doi: 10.1261/rna.7130504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harger, J.W., Dinman, J.D. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA. 2003;9:1019–1024. doi: 10.1261/rna.5930803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harger, J.W., Dinman, J.D. Evidence against a direct role for the Upf proteins in frameshifting or nonsense codon readthrough. RNA. 2004;10:1721–1729. doi: 10.1261/rna.7120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatovich, O., Cooper, M., Kulesza, H.M., Beggs, J.D. Cloning and characterisation of the gene encoding the ribosomal protein S5 (also known as rp14, S2, YS8) of Saccharomyces cerevisiae . Nucleic Acids Res. 1995;23:4616–4619. doi: 10.1093/nar/23.22.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels, J., Friedlander, G., Bergmann, S., Sarig, O., Ziv, Y., Barkai, N. Revealing modular organization in the yeast transcriptional network. Nat. Genet. 2002;31:370–377. doi: 10.1038/ng941. [DOI] [PubMed] [Google Scholar]
- Ito, H., Fukuda, Y., Murata, K., Kimura, A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;53:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, J.L., Dinman, J.D. Systematic analysis of bicistronic reporter assay data. Nucleic Acids Res. 2004;32:e160. doi: 10.1093/nar/gnh157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeeninga, R.E., Venema, J., Raue, H.A. Rat RL23a ribosomal protein efficiently competes with its Saccharomyces cerevisiae L25 homologue for assembly into 60 S subunits. J. Mol. Biol. 1996;263:648–656. doi: 10.1006/jmbi.1996.0605. [DOI] [PubMed] [Google Scholar]
- Kapasi, P., Chaudhuri, S., Vyas, K., Baus, D., Komar, A.A., Fox, P.L., Merrick, W.C., Mazumder, B. L13a blocks 48S assembly: Role of a general initiation factor in mRNA-specific translational control. Mol. Cell. 2007;25:113–126. doi: 10.1016/j.molcel.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar, A.A., Lesnik, T., Cullin, C., Merrick, W.C., Trachsel, H., Altmann, M. Internal initiation drives the synthesis of Ure2 protein lacking the prion domain and affects [URE3] propagation in yeast cells. EMBO J. 2003;22:1199–1209. doi: 10.1093/emboj/cdg103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar, A.A., Gross, S.R., Barth-Baus, D., Strachan, R., Hensold, J.O., Goss Kinzy, T., Merrick, W.C. Novel characteristics of the biological properties of the yeast Saccharomyces cerevisiae eukaryotic initiation factor 2A. J. Biol. Chem. 2005;280:15601–15611. doi: 10.1074/jbc.M413728200. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laletina, E., Graifer, D., Malygin, A., Ivanov, A., Shatsky, I., Karpova, G. Proteins surrounding hairpin IIIe of the hepatitis C virus internal ribosome entry site on the human 40S ribosomal subunit. Nucleic Acids Res. 2006;34:2027–2036. doi: 10.1093/nar/gkl155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maicas, E., Pluthero, F.G., Friesen, J.D. The accumulation of three yeast ribosomal proteins under conditions of excess mRNA is determined primarily by fast protein decay. Mol. Cell. Biol. 1988;8:169–175. doi: 10.1128/mcb.8.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder, B., Sampath, P., Fox, P.L. Translational control of ceruloplasmin gene expression: Beyond the IRE. Biol. Res. 2006;39:59–66. doi: 10.4067/s0716-97602006000100007. [DOI] [PubMed] [Google Scholar]
- Mitra, K., Frank, J. Ribosome dynamics: Insights from atomic structure modeling into cryo-electron microscopy maps. Annu. Rev. Biophys. Biomol. Struct. 2006;35:299–317. doi: 10.1146/annurev.biophys.35.040405.101950. [DOI] [PubMed] [Google Scholar]
- Morimoto, K., Lin, S., Sakamoto, K. The functions of RPS19 and their relationship to Diamond-Blackfan anemia: A review. Mol. Genet. Metab. 2006;90:358–362. doi: 10.1016/j.ymgme.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Mueller, P.P., Harashima, S., Hinnebusch, A.G. A segment of GCN4 mRNA containing the upstream AUG codons confers translational control upon a heterologous yeast transcript. Proc. Natl. Acad. Sci. 1987;84:2863–2867. doi: 10.1073/pnas.84.9.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg, D., Muller, R., Funk, M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Nierhaus, K.H. Decoding errors and the involvement of the E-site. Biochimie. 2006;88:1013–1019. doi: 10.1016/j.biochi.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Nilsson, J., Nissen, P. Elongation factors on the ribosome. Curr. Opin. Struct. Biol. 2005;15:349–354. doi: 10.1016/j.sbi.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Nishiyama, T., Yamamoto, H., Uchiumi, T., Nakashima, N. Eukaryotic ribosomal protein RPS25 interacts with the conserved loop region in a dicistroviral intergenic internal ribosome entry site. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkl1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller, H.F. RNA structure: Reading the ribosome. Science. 2005;309:1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- Nowotny, V., Nierhaus, K.H. Assembly of the 30S subunit from Escherichia coli ribosomes occurs via two assembly domains which are initiated by S4 and S7. Biochemistry. 1988;27:7051–7055. doi: 10.1021/bi00418a057. [DOI] [PubMed] [Google Scholar]
- Odreman-Macchioli, F.E., Tisminetzky, S.G., Zotti, M., Baralle, F.E., Buratti, E. Influence of correct secondary and tertiary RNA folding on the binding of cellular factors to the HCV IRES. Nucleic Acids Res. 2000;28:875–885. doi: 10.1093/nar/28.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle, J.M., Ramakrishnan, V. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- Otto, G.A., Lukavsky, P.J., Lancaster, A.M., Sarnow, P., Puglisi, J.D. Ribosomal proteins mediate the hepatitis C virus IRES-HeLa 40S interaction. RNA. 2002;8:913–923. doi: 10.1017/s1355838202022057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore, L.A., Schmeing, T.M., Maag, D., Applefield, D.J., Acker, M.G., Algire, M.A., Lorsch, J.R., Ramakrishnan, V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Pestova, T.V., Hellen, C.U. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes & Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Lomakin, I.B., Hellen, C.U.T. Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep. 2004;5:906–913. doi: 10.1038/sj.embor.7400240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfingsten, J.S., Costantino, D.A., Kieft, J.S. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science. 2006;314:1450–1454. doi: 10.1126/science.1133281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez, M., Wek, R.C., Vazquez de Aldana, C.R., Jackson, B.M., Freeman, B., Hinnebusch, A.G. Mutations activating the yeast eIF-2 α kinase GCN2: Isolation of alleles altering the domain related to histidyl-tRNA synthetases. Mol. Cell. Biol. 1992;12:5801–5815. doi: 10.1128/mcb.12.12.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, F., Brakier-Gingras, L. A functional interaction between ribosomal proteins S7 and S11 within the bacterial ribosome. J. Biol. Chem. 2003;278:44913–44920. doi: 10.1074/jbc.M306534200. [DOI] [PubMed] [Google Scholar]
- Rose, M.D., Winston, F., Heiter, P. Methods in yeast genetics: A laboratory course manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1990. [Google Scholar]
- Ross, C.L., Patel, R.R., Mendelson, T.C., Ware, V.C. Functional conservation between structurally diverse ribosomal proteins from Drosophila melanogaster and Saccharomyces cerevisiae: Fly L23a can substitute for yeast L25 in ribosome assembly and function. Nucleic Acids Res. 2007;35:4503–4514. doi: 10.1093/nar/gkm428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, F.F., Maniatis, T. Molecular cloning: A laboratory manual. 2d ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Sarnow, P., Cevallos, R.C., Jan, E. Takeover of host ribosomes by divergent IRES elements. Biochem. Soc. Trans. 2005;33:1479–1482. doi: 10.1042/BST0331479. [DOI] [PubMed] [Google Scholar]
- Schuler, M., Connell, S.R., Lescoute, A., Giesebrecht, J., Dabrowski, M., Schroeer, B., Mielke, T., Penczek, P.A., Westhof, E., Spahn, C.M. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat. Struct. Mol. Biol. 2006;13:1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S., Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn, C.M., Beckmann, R., Eswar, N., Penczek, P.A., Sali, A., Blobel, G., Frank, J. Structure of the 80S ribosome from Saccharomyces cerevisiae–tRNA-ribosome and subunit–subunit interactions. Cell. 2001a;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- Spahn, C.M., Kieft, J.S., Grassucci, R.A., Penczek, P.A., Zhou, K., Doudna, J.A., Frank, J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001b;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- Spahn, C.M., Gomez-Lorenzo, M.G., Grassucci, R.A., Jorgensen, R., Andersen, G.R., Beckmann, R., Penczek, P.A., Ballesta, J.P., Frank, J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvester, J.E., Fischel-Ghodsian, N., Mougey, E.B., O'Brien, T.W. Mitochondrial ribosomal proteins: Candidate genes for mitochondrial disease. Genet. Med. 2004;6:73–80. doi: 10.1097/01.gim.0000117333.21213.17. [DOI] [PubMed] [Google Scholar]
- Thompson, S.R., Gulyas, K.D., Sarnow, P. Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element. Proc. Natl. Acad. Sci. 2001;98:12972–12977. doi: 10.1073/pnas.241286698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin, H., Staehelin, T., Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay, Y.F., Thompson, J.R., Rotenberg, M.O., Larkin, J.C., Woolford J.L., Jr Ribosomal protein synthesis is not regulated at the translational level in Saccharomyces cerevisiae: Balanced accumulation of ribosomal proteins L16 and rp59 is mediated by turnover of excess protein. Genes & Dev. 1988;6:664–676. doi: 10.1101/gad.2.6.664. [DOI] [PubMed] [Google Scholar]
- Uritani, M., Miyazaki, M. Role of yeast peptide elongation factor 3 (EF-3) at the AA-tRNA binding step. J. Biochem. 1988;104:118–126. doi: 10.1093/oxfordjournals.jbchem.a122405. [DOI] [PubMed] [Google Scholar]
- Wilson, D.N., Nierhaus, K.H. Ribosomal proteins in the spotlight. Crit. Rev. Biochem. Mol. Biol. 2005;40:243–267. doi: 10.1080/10409230500256523. [DOI] [PubMed] [Google Scholar]
- Wilson, D.N., Nierhaus, K.H. The E-site story: The importance of maintaining two tRNAs on the ribosome during protein synthesis. Cell. Mol. Life Sci. 2006;63:2725–2737. doi: 10.1007/s00018-006-6125-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J.E., Powell, M.J., Hoover, S.E., Sarnow, P. Naturally occurring dicistronic Cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell. Biol. 2000;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wower, J., Scheffer, P., Sylvers, L.A., Wintermeyer, W., Zimmermann, R.A. Topography of the E-site on the Escherichia coli ribosome. EMBO J. 1993;12:617–623. doi: 10.1002/j.1460-2075.1993.tb05694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaman, I., Fernandez, J., Liu, H., Caprara, M., Komar, A.A., Koromilas, A.E., Zhou, L., Snider, M.D., Scheuner, D., Kaufman, R.J., et al. The zipper model of translational control: A small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell. 2003;113:519–531. doi: 10.1016/s0092-8674(03)00345-3. [DOI] [PubMed] [Google Scholar]
- Yonath, A. Ribosomal crystallography: Peptide bond formation, chaperone assistance and antibiotics activity. Mol. Cells. 2005;20:1–16. [PubMed] [Google Scholar]
- Yonath, A., Bashan, A. Ribosomal crystallography: Initiation, peptide bond formation, and amino acid polymerization are hampered by antibiotics. Annu. Rev. Microbiol. 2004;58:233–251. doi: 10.1146/annurev.micro.58.030603.123822. [DOI] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H., Noller, H.F. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]