Abstract
Myotonic dystrophy (DM) is a genetic disorder with multisystemic symptoms that is caused by expression (as RNA) of expanded repeats of CTG or CCTG in the genome. It is hypothesized that the RNA splicing factor muscleblind-like (MBNL) is sequestered to the expanded CUG or CCUG RNAs. Mislocalization of MBNL results in missplicing of a subset of pre-mRNAs that are linked to the symptoms found in DM patients. We demonstrate that MBNL can bind short structured CUG and CCUG repeats with high affinity and specificity. Only 6 base pairs are necessary for MBNL binding: two pyrimidine mismatches and four guanosine–cytosine base pairs in a stem. MBNL also has a preference for pyrimidine mismatches, but many other mismatches are tolerated with decreased affinity. We also demonstrate that MBNL binds the helical region of a stem–loop in the endogenous pre-mRNA target, the cardiac troponin T (cTNT) pre-mRNA. The stem–loop contains two mismatches and resembles both CUG and CCUG repeats. In vivo splicing results indicate that MBNL-regulated splicing is dependent upon the formation of stem–loops recognized by MBNL. These results suggest that MBNL may bind all of its RNA substrates, both normal and pathogenic, as structured stem–loops containing pyrimidine mismatches.
Keywords: zinc fingers, mismatched base pairs, triplet repeat expansions, cardiac troponin T, protein RNA interactions
INTRODUCTION
Myotonic dystrophy (DM) is a genetic disorder with multisystemic symptoms that include myotonia, cardiac arrhythmia, insulin resistance, and muscular weakness. There are two subtypes of myotonic dystrophy: DM1 and DM2. DM1 has been linked to a (CTG)n repeat expansion in the 3′ untranslated region (3′ UTR) of the DMPK gene. DM2 has been linked to (CCTG)n repeats expansion in intron 1 of the ZNF9 gene (for review, see Nykamp and Swanson 2004; Ranum and Cooper 2006). The genetic mutations in each subtype are in two unrelated genes on different chromosomes. The symptoms observed in the two subtypes are remarkably similar, with the only molecular commonality being the repeat expansions. The similarity in symptoms and sequence motifs as well as the fact that both repeats are noncoding indicate a common mechanism for both subtypes.
CUG expanded repeats fold into extended stem–loop structures, with guanosines and cytosines forming base pairs, while the uridines form mismatches (Michalowski et al. 1999; Tian et al. 2000; Mooers et al. 2005). Biochemical and structural studies have shown that extended helical regions of the stem-loops are primarily A-form in structure and are thermodynamically stable (Napierala and Krzyzosiak 1997; Mooers et al. 2005). The CCUG repeats of DM2 also fold into an extended stem–loop structure. It is currently thought this stem–loop consists of two adjacent guanosine–cytosine base pairs and two adjacent cytosine–uracil mismatches (Sobczak et al. 2003; Dere et al. 2004). However, it is possible for the RNA to anneal in another structure in which single guanosine–cytosine base pairs are interspersed with uracil–uracil and cytosine–cytosine mismatches (see Fig. 3A, below, for schematic) and it is unclear if one structure predominates. Thermodynamically, the first structure is the only one that is predicted (Zuker 2003).
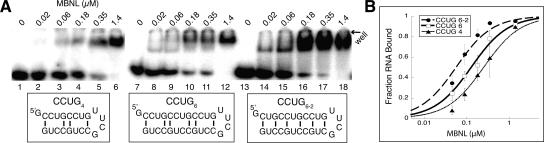
FIGURE 3.
MBNL binds CCUG expansions with high affinity, in two possible structures. (A) MBNL binding to CCUG4 (lanes 1–6), CCUG6 (lanes 7–12), and CCUG repeats in an alternate register (labeled CCUG6–2) in lanes 13–18. Concentration of MBNL is in micromolar labeled above each lane. (B) Binding curve for CCUG constructs.
One proposed mechanism for the disease is that, upon transcription, the CUG and CCUG repeats sequester RNA binding proteins from their normal cellular functions. It is hypothesized that the specific sequestration of the RNA binding protein muscleblind-like (MBNL) primarily leads to DM symptoms (for review, see Osborne and Thornton 2006; Ranum and Cooper 2006). Supporting this hypothesis is a mouse model in which expression of 250 noncoding CUG repeats causes symptoms similar to patients with DM (Mankodi et al. 2000). The link between MBNL1 and DM was strengthened with a mouse knockout model, where the MBNL1 gene was inactivated through deletion and the mice developed many key symptoms of DM (Kanadia et al. 2003a).
The muscleblind family of proteins was originally identified in Drosophila melanogaster as a gene required for muscle development and eye differentiation (Begemann et al. 1997). For a review of muscleblind see Pascual et al. (2006). There are three muscleblind paralogs in human, named MBNL1–3. Of the three human MBNL proteins, MBNL1 and MBNL2 are more abundant and have been shown to colocalize with CUG and CCUG repeats in the nucleus, forming nuclear foci in both DM1 and DM2 (Miller et al. 2000; Mankodi et al. 2001, 2003; Fardaei et al. 2002; Kanadia et al. 2003b; Jiang et al. 2004; Ho et al. 2005b; Lin et al. 2006). All three muscleblind proteins are similar in sequence and appear to have similar functions, as they can regulate alternative splicing in tissue culture (Ho et al. 2004; Dansithong et al. 2005; Paul et al. 2006). However, MBNL2 has also been shown to function in RNA localization in the cytoplasm (Adereth et al. 2005). Less is known about MBNL3, but it is possible that MBNL1 and MBNL3 may act antagonistically to each other when regulating gene expression that is involved in muscle differentiation (Squillace et al. 2002).
The RNA binding protein CUG-BP also has an important role in DM pathogenesis. CUG-BP and hnRNP H are overexpressed in the presence of expanded CUG repeats and are involved in controlling the alternative spicing of many of the same genes regulated by MBNL (Philips et al. 1998; Savkur et al. 2001; Charlet et al. 2002; Paul et al. 2006). An antagonistic relationship exists between MBNL and CUG-BP for several regulated exons, where one protein acts as a positive regulator while the other protein acts as a negative regulator. However, the actual role of each protein appears to depend on the specific pre-mRNA and splice junction in question. For example, for the fifth exon of the cardiac troponin T (cTNT) pre-mRNA, CUG-BP is a positive regulator of exon five inclusion while MBNL is a negative regulator of exon five inclusion (Philips et al. 1998; Ho et al. 2004, 2005a). Conversely, for exon 11 of the insulin receptor pre-mRNA, the roles of CUG-BP and MBNL have been reversed and CUG-BP is a negative regulator while MBNL is a positive regulator (Savkur et al. 2001, 2004; Dansithong et al. 2005; Paul et al. 2006).
It is currently thought that the relative levels of these two splicing factors and other splicing factors leads to specific pre-mRNA splice patterns (Ladd et al. 2005). As these two factors compete in their regulation of splicing, the missplicing seen in DM has been hypothesized to be due to both decreased levels of MBNL and/or increased levels of CUB-BP. Supporting both models, mice in which MBNL is knocked out show DM symptoms, as do mice in which CUG-BP is overexpressed (Timchenko et al. 2001; Kanadia et al. 2003a; Ho et al. 2005a). However, it has recently been shown in a tissue culture model that loss of MBNL has a more drastic effect on missplicing, while the increased levels of CUG-BP had only a secondary effect (Dansithong et al. 2005). This suggests that sequestration of MBNL to the CUG and CCUG repeats is the primary cause for the missplicing observed in DM.
To understand the function of MBNL1 in the disease state and during pre-mRNA splicing, we characterized the RNA binding activity of purified recombinant MBNL1. We tested binding to CUG and CCUG repeats, to a series of RNAs where portions of the CUG repeat are mutated, and to a fragment from the cTNT pre-mRNA. We found that MBNL1 preferentially binds short helical A-form RNA regions in both the CUG and CCUG repeats and the cTNT pre-mRNA structure. In all cases the helical structures contain similar pyrimidine–pyrimidine nucleotide mismatches. This suggests that MBNL1 may recognize and bind similar structures in both the pathogenic repeats and in its pre-mRNA targets.
RESULTS
The elongated zinc finger domains of MBNL are sufficient for RNA binding
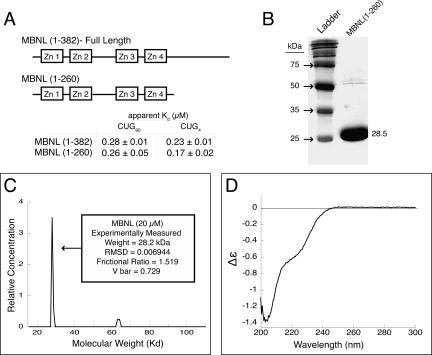
To characterize the RNA binding of MBNL1, we used a truncated version of MBNL1 (containing amino acids 1–260). Recombinant protein was expressed in Escherichia coli BL21* cells and purified using a GST affinity tag and ion exchange chromatography. When compared to full-length MBNL1(1–382), the truncated version of MBNL1(1–260) bound three different RNAs with similar affinity: an RNA with 90 CUG repeats, a shortened CUG4 construct, and to a region of the cardiac troponin T pre-mRNA (Fig. 1; Supplemental Fig. 1; for supplemental figure, e-mail: aberglund@molbio.uoregon.edu). Kino and colleagues (2004) also previously found that two truncated versions of MBNL1 (amino acids 1–248 and 1–269) bound RNA substrates in a three-hybrid assay better than a full-length version of MBNL1. MBNL1(1–260) contains all four zinc fingers of the full-length protein (Fig. 1A), and was used for all of the studies presented here and will be referred to as MBNL throughout the remainder of the text.
FIGURE 1.
Structural characterization of MBNL. (A) A schematic of MBNL showing the truncated form MBNL(1–260) used in these studies. (B) Recombinant expressed MBNL has a molecular weight of 28.5 kDa on a 10% SDS-PAGE gel. Note there is a small amount of co-purifying contaminant with a molecular weight of ∼55–60 kDa. (C) Analytical ultracentrifugation of MBNL shows that it sediments with the weight of a monomer. MBNL has a slightly elongated shape, as its frictional ratio is 1.519. (D) Circular dichroism spectra of MBNL, showing a major peak at 203 and a minor peak at 220 nm, indicating that a portion of MBNL is disordered and MBNL lacks significant α-helical content in its structure (units Δε are molar circular-dichroic absorption).
Biophysical methods were used to characterize MBNL. Analytical ultracentrifugation (AUC) with purified recombinant MBNL (Fig. 1B) was used to measure a molecular weight of 28.2 kDa (predicted molecular weight is 28.5 kDa). This indicates that MBNL is a monomer in solution (Fig. 1C), as the zinc finger domains do not seem to mediate any oligomerization. It may also be possible that the C-terminal domain (amino acids 261–382) may mediate oligomerization. The frictional ratio of MBNL was 1.5189. Globular proteins have a ratio of 1.2 (Dam and Schuck 2004), and an increased ratio is consistent with a protein that is elongated in solution. A copurifying contaminant (seen in Fig. 1B), of ∼65 kDa was also seen to sediment. However, the main peak at 28.2 kDa accounted for >95% of the sedimented protein signal (Fig. 1C).
Circular dichroism (CD) was used to probe the secondary structure of MBNL (Fig. 1D). The weak α-helical signal at 220 nm and the strong signal at 203 nm (indicative of disorder in protein structures) suggests that MBNL does not have significant α-helical structure, although it is possible that MBNL contains some or significant β-sheet structure, as the signal for this structure was undetectable due to interference with the buffer. The addition of a known RNA substrate did not alter the signal at 220 nm, suggesting no new α-helices were forming upon RNA binding (data not shown). Mutations of individual cysteines within the zinc finger domains reduce the stability of MBNL and reduce its RNA binding affinity (data not shown), indicating that zinc fingers competent to bind zinc are required for RNA binding. The addition of 5 μM zinc chloride did not alter the 203–220 nm ratio, indicating that the added Zn++ caused no structural changes in the protein and therefore that the four zinc finger domains were already saturated with zinc ions (data not shown).
MBNL binds moderate and short CUG expansions with similar affinity
To identify a minimal CUG repeat construct that could be used for characterizing the binding specificity of MBNL, we started with an RNA substrate containing 90 CUG repeats (CUG90); this RNA was then truncated until a minimal RNA substrate capable of binding MBNL was identified. In all assays, the protein was always in at least a 10-fold excess of the total number of CUG triplets, to ensure that multiple binding sites on the longer CUG expansions would not artificially enhance the apparent affinity of MBNL for the longer repeats. To stabilize the shorter CUG repeats that contained eight repeats or fewer, an ultrastable UUCG tetraloop was used to cap the short stem–loops (Molinaro and Tinoco 1995). Stabilizing the short CUG repeats was likely necessary because Miller and colleagues (2000) did not observe binding with short CUG repeats, possibly because in their experiments the shorter repeats were not forming stem–loops.
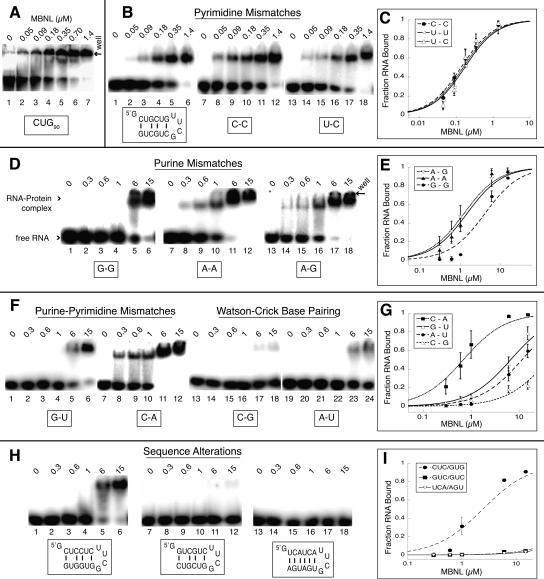
A stem–loop containing two pairs of CUG repeats separated by the tetraloop (CUG4) bound MBNL with similar affinity compared to CUG90 (Fig. 2, cf. A and B). The slightly reduced affinity of MBNL for CUG90 compared to CUG4 may be because multiple MBNL proteins need to bind to CUG90 to cause a shift, suggesting the apparent Kd for this RNA is likely to be lower than 260 nM. Our truncation studies of the CUG repeats indicate that the minimal binding site for MBNL is ≤6 base pairs (bp).
FIGURE 2.
MBNL binds short CUG repeats with affinity similar to longer repeats, and the presence of mismatches is necessary for binding. The concentration of MBNL is shown above each lane in micromolar in the gel shift assay. (A) MBNL binding CUG90 repeats (lanes 1–6) with an apparent Kd of 230 nm. (B) MBNL binding to pyrimidine–pyrimidine mismatches. The sequence of the (CUG)4 RNA is shown below lanes 1–6. The boxed C-C (lanes 7–12) and U-C (lanes 13–18) represent the replacement of the U-U mismatches with these mismatches. (D) MBNL binding to purine–purine mismatches in place of the U-U mismatch; boxed sequence indicates mismatch replacement base pairs. (F) MBNL binding to G-U (lanes 1–6) and C-A (lanes 7–12) mismatches and Watson–Crick base pairs in place of the U-U mismatches. (H) Binding of MBNL to RNAs containing sequence alterations to the cytosine and guanine positions in CUG4 and a control RNA with no sequence similarity to CUG repeats. (C,E,G,I) Binding curves of the different mismatch and sequence alteration RNAs.
Thermal melts were performed to verify that the small RNA constructs formed the predicted stem–loop structures. Discrete thermal transitions were seen for all RNAs constructs, indicating that a stable stem–loop structure was indeed formed (see Table 1 for a listing of estimated Tm values). In addition, the Tm of CUG4 was tested and found to be concentration independent up to 2 μM, indicating that only an intramolecular structure formed at the RNA concentrations used during our experiments (as opposed to intermolecular structures that may start to form at higher concentrations and would likely have a different melting curve).
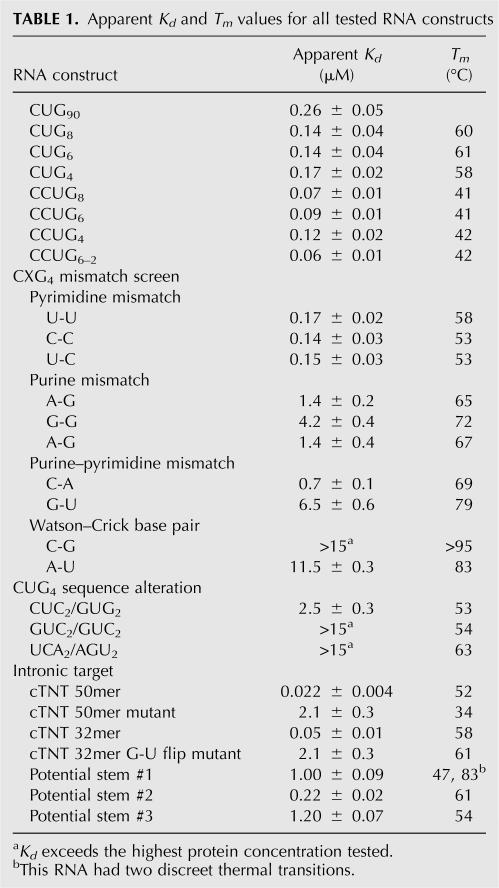
TABLE 1.
Apparent Kd and Tm values for all tested RNA constructs
MBNL recognizes both the mismatch and Watson–Crick base pairs within the CUG repeat stem–loop
To determine the importance of the U-U mismatch in the CUG stem–loop RNA, we replaced the U-U mismatch with other mismatches or placed Watson–Crick base pairs in this position (see Fig. 2). MBNL binds to sequences containing any pryrimidine–pyrimidine mismatch in any combination with similar affinity (apparent Kd values range from 140 to 170 nM) (Fig. 2B,C; Table 1). The substitution of purine–purine mismatches (A-A, G-G, and A-G) for U-U mismatches all moderately inhibit the binding of MBNL by 10–20-fold (Fig. 2D,E; Table 1). Replacement of the U-U mismatch with C-A mismatches only reduces binding approximately fourfold while a G-U wobble base pair reduces binding more than 20-fold (Fig. 2F,G; Table 1). Replacing the U-U mismatches with Watson–Crick base pairs almost completely abolishes MBNL binding (Fig. 2F), indicating the mismatch plays an essential role in MBNL's binding.
The identity and location of the C-G and G-C base pairs in the CUG repeats are also important for MBNL binding. When the G-C base pairs in CUG4 were changed such that cytosines were all one strand and the guanosines were all one strand, MBNL binding was reduced 15-fold (Fig. 2H, lanes 1–6). Changing the polarity of the RNA from CUG to GUC completely eliminated binding (Fig. 2H), demonstrating that the polarity and order of the base pairs in the sequence is important. Finally, an RNA with UCA repeats that form all Watson–Crick base pairs capped with a UUCG loop does not interact with MBNL (Fig. 2H).
Thermal melts were again performed on all RNA constructs, to verify secondary structures formed (Table 1). All RNA structures were relatively stable, with Tm > 50°C. Analysis of the range of the transitions indicates that all RNA structures should be fully intact at 25°C, the temperature at which all binding studies were performed.
MBNL binds short CCUG stem–loops
A series of short CCUG RNA constructs with four, six, and eight repeats were designed and named CCUG4, CCUG6, and CCUG8, respectively (all of which also contained a tetraloop to act as a cap). MBNL bound all three RNA constructs with similar affinity and on average with twofold stronger affinity compared to CUG repeats (Fig. 3; Table 1). CCUG expansions can anneal in two possible structures (Fig. 3A). Presently, CCUG expansions are thought to be in the first structure (Sobczak et al. 2003; Dere et al. 2004), where two C-G base pairs are interspaced by two U-C mismatches. CCUG repeats are thermodynamically predicted by Mfold to be in this structure (Zuker 2003).
However, another structure is possible in which G-C base pairs are flanked by alternating U-U and C-C mismatches. A CCUG6 construct was designed that forced the stem–loop to anneal in this alternate structure (CCUG6–2, Fig. 3A). MBNL bound this construct with slightly enhanced affinity compared to CCUG6, indicating that MBNL binds CCUG expansions in either configuration but may have a slight preference for the conformation containing the alternating U-U and C-C mismatches. Melting studies performed on the CCUG stem–loops showed lower Tm than those for the CUG stem–loops (Table 1), which reflect the decreased stability of these structures due to the increased proportion of mismatches in the structure.
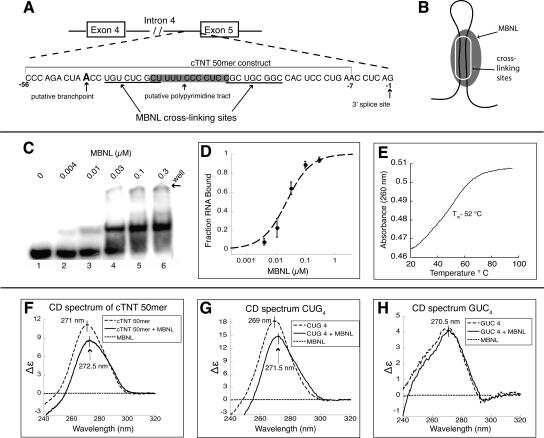
MBNL binds a helical A-form structure within the cTNT pre-mRNA
It has been previously shown that MBNL can be cross-linked to the 3′ end of the fourth intron of the cardiac troponin T (cTNT) pre-mRNA in vitro (Ho et al. 2004). However, quantitative binding to this substrate has not been performed. We found that MBNL binds a 50 nucleotide (nt) fragment from the 3′ end of the intron (cTNT 50mer) with the highest affinity of any of the RNAs we tested, with an apparent Kd of 22 nM (Fig. 4C,D). This 50mer spans nucleotide residues 8 through 58 directly upstream of exon 5 (Fig. 4A). The full-length MBNL1 (1–382) bound the cTNT 50mer with similar affinity to MBNL(1–260) (Supplemental Fig. 1).
FIGURE 4.
MBNL binds a structured region in the 3′ end of intron 4 in the human cardiac troponin T (cTNT) pre-mRNA. (A) A schematic of intron 4 of the cTNT pre-mRNA. The cTNT 50mer used for binding studies is indicated. (B) A schematic model of MBNL binding this RNA as stem–loop. (C) Binding of MBNL to cTNT 50mer with the concentration of protein labeled above each lane. (D) Binding curve of cTNT 50mer. (E) Thermal melt of the cTNT 50mer, with a Tm of 52°C. (F) Circular dichroism (CD) spectra of the cTNT 50mer with and without MBNL present (units Δε are molar circular-dicroic absorption). (G) CD spectra of CUG4 with and without MBNL present. (H) CD spectra of GUC4 with and without MBNL present.
Because MBNL was found to bind short stem–loop RNAs, we hypothesized that the cTNT 50mer might also contain a short stem–loop recognized by MBNL (Fig. 4B). UV melting shows that the cTNT 50mer unfolded with a single transition and a Tm of 52°C (Fig. 4E), indicating at least one structural element within the RNA. As CUG repeats are known to be A-form (Mooers et al. 2005), we analyzed the CD spectrum of the cTNT 50mer and the CUG4 stem–loop RNA to determine if this structure was also A-form in nature. A-form duplex RNA has a characteristic peak between 260 and 270 nm in a CD spectrum, while single-stranded RNA has a peak at ∼275 nm (Hung et al. 1994; Ivanov et al. 2003). The cTNT 50mer (1 μM) had a peak at 271 nm and CUG4 (1 μM) had a strong peak at 269 nm (Fig. 4F). These values are in the higher range of the duplex A-form range, but currently only the CD spectrum of purely duplex or single-stranded RNA has been characterized, and the mismatches in the CUG4 stem and potential mismatches in the cTNT stem structures may shift the peak. Because the cTNT 50mer has a peak at a similar wavelength, it indicates that its stem structure may be similar to that of the CUG4 stem.
Addition of MBNL (1 μM) to the cTNT 50mer decreases the signal by 23%, with the peak shifting to 272.5 nm. Both the reduction in signal and shift suggest that MBNL binding alters the RNA structure in some way that may reduce the base stacking of the RNA. MBNL has no signal at 270 nm (Fig. 4F), meaning the change in signal is due solely to structural changes in the RNA. Approximately 85% of the RNA is predicted to be bound at these RNA and protein concentrations, indicating that the peak shift and the 23% decrease in signal is not stoichiometric, but only a partial decrease in signal by the majority of the RNA population.
The addition of 1 μM MBNL to CUG4 similarly decreased the CD signal by 20% and caused a peak shift of 2.5 nm, from 269 to 271.5 nm (Fig. 4G). The GUC4 (1 μM) construct was tested and found to have a signal at 270 nm, which was unchanged in the presence of 1 μM MBNL, as this RNA is not bound by MBNL (Fig. 4H). The relative amplitude differences between cTNT 50mer, CUG4, and GUC4 are likely due to differences in base stacking due to sequence differences.
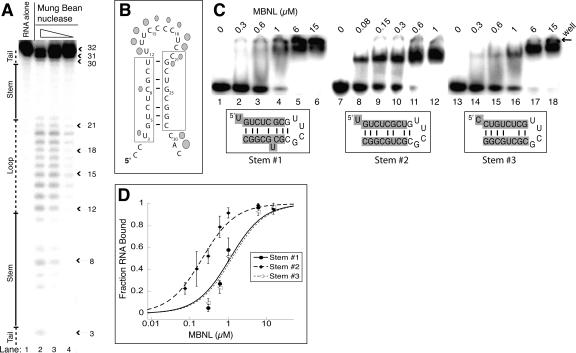
MBNL binds a stem–loop containing mismatches at the 3′ end of the fourth cTNT intron
To determine the structure of the putative stem–loop in intron 4 of the cTNT pre-mRNA, mung bean nuclease was used to probe the secondary structure. A shortened version of the cTNT RNA (cTNT 32mer) was used in the structure probing assay because the cTNT 50mer was less stable in these assays and degraded more easily. MBNL binds the cTNT 32mer with high affinity, with an apparent Kd of 50 nM (Table 1), indicating most of the binding determinants for MBNL are present in this shortened RNA.
Mung bean nuclease is a nonspecific single-stranded cutter that cleaved residues 12–22 and 30–31, while most of the other residues were not. This suggests that residues 12–22 are likely single-stranded and located within a loop, with the final three residues being in a 3′ tail (Fig. 5A). Residues 4–11 and 22–29 are protected, except that position 8 is cleaved at a low level (lane 3), indicating these two regions form the stem (Fig. 5B).
FIGURE 5.
MBNL binds the cTNT 32mer as a stem–loop. (A) Mung bean nuclease cleavage pattern of cTNT 32mer. The concentration of mung bean nuclease is 100, 10, and 1 units per microliter, in lanes 2–4, respectively. (B) Schematic of the likely stem–loop within cTNT intron 4, showing cleavage locations of mung bean nuclease. Larger shapes indicate strong cleavage events while smaller shapes represent weak cleavage events. The cross-linking sites determined previously are boxed (Ho et al. 2004). (C) Binding curve of the three potential stem structures of the cTNT intron 4, showing MBNL prefers structure #2. (D) Gel shifts showing MBNL binding to the three potential stems of cTNT intron 4. Concentration of MBNL is labeled above each lane; note the lower concentrations in lanes 8–12 compared to lanes 2–6 and lanes 14–18. Sequences highlighted in gray are sequences from the cTNT intron in the three different potential base pair and mismatch configurations. The nongray sequence is the tetraloop cap and an additional base pair to stabilize the structure if necessary.
This cleavage pattern suggests the two MBNL sites identified through cross-linking (Ho et al. 2004) come together to form the stem. Therefore, it appears that these two separate sites, underlined in Figure 5B, are actually one individual site (Fig. 5B). Three different base-pairing configurations are possible for this stem containing slightly different base-pairing and mismatch possibilities (Fig. 5C,D). To determine which (if any) of the three stems is favored by MBNL, we created three stems with the different base-pairing configurations capped with the UUCG tetraloop (Fig. 5D). MBNL bound all three RNA structures, but it clearly prefers stem #2. MBNL bound this RNA with five- to sixfold greater affinity compared to stems #1 and #3, with an apparent Kd of 0.22 μM for stem #2. This Kd is in the same range as the Kd for the CUG4 RNA, although it is fourfold weaker than the endogenous cTNT 32mer. This suggests that Stem #2 is the preferred structure that MBNL binds in the endogenous pre-mRNA target but that loop or tail regions may make additional contacts with MBNL. Alternatively, the UUCG cap may perturb the structure of RNA in a way that negatively effects MBNL binding.
Thermal melts were performed on these stem–loops, and stem #2 was found to have the highest Tm, suggesting it is the most stable stem and likely the biologically relevant structure. However, the stabilities of these stems were likely altered by the presence of the UUCG cap. For instance, stem #1 had a second discreet transition at 83°C (Table 1). It is likely that stem #1 has two separable structural elements, separated by the C bulge and U-C mismatch (Fig. 5C). The UUCG cap is known to form a stable fold with just two adjacent G-C base pairs (Molinaro and Tinoco 1995), making it likely that the second structural element of stem #1 is strongly stabilized by this cap, which is not part of the wild-type sequence.
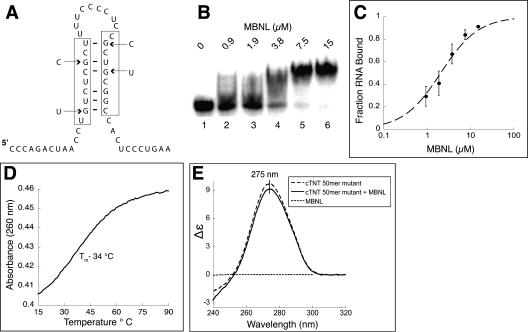
Mutations that destabilize the stem in the cTNT fourth intron also significantly reduce binding of MBNL
We hypothesized that the stem–loop structure in the cTNT RNA was critical for MBNL binding and that mutations that destabilize the stem–loop would abolish MBNL binding. Previously, Ho and colleagues (2004) found that four simultaneous guanosine point mutations (which will be referred to as the 4G construct) in this region of the cTNT fourth intron reduce MBNL's ability to cross-link to this RNA, and the ability of MBNL to negatively regulate the inclusion of the downstream exon is eliminated. These four point mutations change four of the six guanosines that participate in base pairing or wobble base pairing in the stem–loop we identified (Fig. 6A).
FIGURE 6.
Four point mutations destabilize the stem in the cTNT 50mer and reduce MBNL binding. (A) Schematic of the stem–loop with the four point mutations indicated by arrows. (B) Gel shift assay shows MBNL binding to the mutated 50mer. (C) Binding curve of MBNL to the mutant cTNT 50mer. (D) UV melt of the cTNT 50mer mutant, with a Tm of 34°C. (E) Circular dichroism spectra of the cTNT 50mer mutant alone (1 μM) and with MBNL (1 μM).
The four mutations in the cTNT 50mer reduced binding of MBNL to the RNA ∼100-fold, the Kd changed from 23 nM for the wild-type cTNT RNA to 2.1 μM for the mutant RNA (Fig. 6B,C; Table 1). This result is consistent with the nearly complete reduction of MBNL cross-linking to this mutant RNA (Ho et al. 2004). A UV melt showed that these mutations significantly reduced the stability of the RNA structure within the cTNT RNA, as expected. The Tm was shifted from 52°C to 34°C for the mutant RNA (Fig. 6D). The CD spectra of the cTNT 50mer mutant was also different from the wild-type cTNT RNA, with the mutant having a reduced signal and the peak shifted to 275 nm, into the known single-stranded RNA wavelength region (Fig. 6E). Addition of MBNL (1 μM) reduced the peak only 4% (compared to a 23% reduction of the wild-type sequence) and did not shift the peak wavelength. These results suggest that the stem–loop structure within the cTNT pre-mRNA is the recognition site for MBNL.
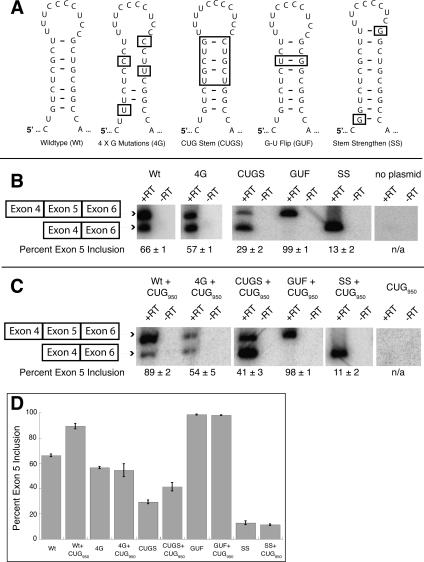
Regulated splicing by MBNL requires a stem–loop containing a MBNL binding site
To test the role of the stem–loop in vivo, a cTNT minigene that includes exons 4–6 (Ho et al. 2004) was used to make mutations in the MBNL binding site and monitored for changes in the splicing of the cTNT minigene in HeLa cells containing MBNL (Miller et al. 2000). To determine if splicing of the mutated MBNL binding sites changed upon MBNL sequestration, a plasmid expressing 950 CUG repeats, called DMPK-CUG950 (Ho et al. 2005b) was co-transfected into the HeLa cells. We reproduced the results of Ho and colleagues showing that MBNL regulates the splicing of the wild-type cTNT minigene (Fig. 7B,C), finding that exon 5 inclusion shifts from 66% to 89% when MBNL is sequestered by the CUG repeats. When the four guanosines are mutated to cytosines (4G mutations) MBNL no longer regulates the splicing of this cTNT minigene.
FIGURE 7.
The sequence and structure of the cTNT stem–loop upstream of exon 5 is important for regulated splicing by MBNL. (A) Schematic of mutations made to the stem–loop in intron 4 of the cTNT minigene. (B) RT-PCR results of the wild-type and mutant cTNT minigenes. (C) Cotransfection with a second minigene that contains 950 CUG repeats to test the role of MBNL sequestration on the splicing of the cTNT minigenes. (D) Graphical representation of exon 5 inclusion for the different constructs with and without the over expression of the 950 CUG repeats.
The 4G mutant had slightly reduced exon 5 inclusion, 57% compared to 66% for the wild-type sequence (Fig. 7B). This reduction in exon 5 inclusion is an unexpected result, as the point mutations both disrupt the stem–loop and MBNL binding, which should cause an increase in exon 5 inclusion. However, these point mutations create a large new single-stranded element in the pre-mRNA, and this structural change might have unanticipated effects, such as the recruitment of other protein factors, or the creation of an unknown silencing element. Upon cotransfection with the DMPK-CUG950 minigene, no change was seen in exon 5 inclusion with this construct (Fig. 7C,D), indicating that once MBNL's binding site is abolished, the splicing of this construct is independent of MBNL. Ho and colleagues (2004) also observed this surprising result with the 4G mutant minigene.
To determine if a stem–loop that contains CUG repeats similar to the CUG4 construct would function to recruit MBNL, the upper portion of the stem was replaced with two CUG repeats on each side (CUG stem [CUGS], Fig. 7A). This CUGS minigene was regulated by MBNL. In the presence of MBNL only 29% inclusion of exon 5 was observed (Fig. 7B) while sequestration of MBNL by the CUG950 RNA leads to 41% of the exon 5 inclusion. The lower percentage of exon 5 inclusion compared to wild type and the weaker effect of MBNL regulated is likely due to the stability of the stem–loop being different between these two RNAs.
Further strengthening the stem–loop with two additional base pairs, one at each end of the helix (Fig. 7A, stem strengthen [SS]), almost completely inhibited the use of exon 5 both in the presence of MBNL and when it was sequestered (Fig. 7B–D). This result indicates that a strong stem–loop inhibits the use of the 3′ splice site, and the binding of MBNL probably does not enhance the effect because the stem is sufficient on its own, while the weaker stems (wild type and CUGS) can be melted by the splicing factors binding at the 3′ end of the intron.
The in vitro binding experiments demonstrated that MBNL prefers to have purines on both sides of the helix, and therefore we predicted that altering the cTNT stem–loop in a manner that shifted all the purines to one side and the pyrimidines on the other side would result in an MBNL unregulated exon. This was done by flipping a G-U base pair (G-U flip [GUF], Fig. 7A). This change results in complete inclusion of exon 5 (99%) in the presence or sequestration of MBNL (Fig. 7B–D). As expected this RNA is bound weakly by MBNL (Table 1, a 42-fold decrease in binding compared to cTNT 32mer). The complete inclusion of exon 5 is likely the result of strengthening the polyprimidine tract, which is recognized by the constitutive splicing factor U2AF65.
DISCUSSION
MBNL's structure and its lack of cooperative RNA binding
The AUC and CD experiments with MBNL suggest the protein adopts a slightly elongated structure. When monitored by CD, the α-helical structure of MBNL does not appear to significantly alter upon the addition of CUG4, although other structural components have yet to be measured upon RNA binding. We propose MBNL, like other zinc finger proteins, is organized into domains around each zinc ion, and that each domain contacts the RNA in a relatively independent manner (Hudson et al. 2004; Brown 2005; Hall 2005; Auweter et al. 2006).
The lack of significantly higher affinity binding to longer CUG repeats suggests that MBNL does not bind in a highly cooperative manner to CUG repeats. An analysis of our binding data supports this conclusion. Scatchard and Hill plots (data not shown) reveal MBNL binds CUG90 with only minimal cooperativity (Hill constant of 1.4) under the binding conditions used in these studies. Analysis of the other repeat RNA substrates indicates only very weak cooperativity, and for cTNT 50mer there is no evidence for cooperative binding at all. These results suggest this version of MBNL recognizes the many binding sites on long CUG repeats as independent binding sites.
It should be noted that the apparent Kd reported here are dependent on our binding conditions: 175 mM NaCl, 5 mM MgCl2, 20 mM Tris (pH 7.5), 1.25 mM BME, 12.5% glycerol, 2 mg/mL BSA, and 0.1 mg/mL heparin. These conditions are quite stringent and the apparent Kd are lowered if NaCl, heparin concentrations, or temperature are decreased in the binding conditions. If we perform binding studies under these less stringent conditions, our apparent Kd for CUG repeats are similar to those measured by Y. Yuan and M.S. Swanson (pers. comm.).
The RNA binding specificity of MBNL for CUG and CCUG repeats
Analysis of RNA substrates in which the CUG repeat tract sequences are modified in various ways (Fig. 2) clearly shows the preference of MBNL for pyrimidine–pyrimidine mismatches as well as the Watson–Crick base pairs in their particular positions. The requirement of mismatches for MBNL binding could be due to two different reasons; either the mismatches are directly recognized by MBNL or the mismatches allow the helix to be easily distorted so MBNL can gain access to the C-G and G-G base pairs.
Both the specific recognition and structural distortion are likely playing a role in MBNL binding the CUG repeats. The distortion is supported by the decrease in CD signal and the peak shift upon MBNL binding to the CUG repeats and the cTNT 50mer (Fig. 4). Furthermore, there also appears to be a general inverse trend between MBNL binding affinity and the stability of the RNA structure. All the RNA substrates to which MBNL binds with strongest affinity generally have lower Tm values (Table 1). The CCUG substrates, as a group, have the strongest affinity for MBNL and also have the lowest Tm values. The pyrimidine–pyrimidine mismatches also, as a group (Table 1), have the lowest Tm values for RNAs containing a mismatch, as well as the strongest binding affinity for MBNL. This trend further suggests that MBNL alters the structures of the RNAs when it binds. If more energy is required to distort or disrupt the base pairing interactions of the RNA, it might result in weaker binding by MBNL.
MBNL binds CCUG repeats with approximately twofold stronger affinity in both structural conformations (Fig. 3) compared to the CUG repeats. Previously, Kino and colleagues also qualitatively found that MBNL preferred CCUG repeats over CUG repeats. These results are surprising because patients with DM2 tend to have more CCUG repeats compared to patients with CUG repeats (Liquori et al. 2001). Additionally, the ZNF9 pre-mRNA containing the CCUG repeats appears to be expressed at similar or even higher levels than the DMPK pre-mRNA (Kanadia et al. 2003b; Shimizu et al. 2003). Yet, the symptoms of DM2 patients are less serious than those of DM1 patients (Meola and Moxley 2004). This supports the model that the expanded CUG repeats, unlike the CCUG repeats, are affecting transcription or other processes in the cell and create another layer of misregulation in DM1 compared to DM2 (Cho and Tapscott 2007).
Recognition of a helical element within the cTNT pre-mRNA by MBNL
Our observation that MBNL binds short stem–loops prompted us to consider that MBNL might recognize a short stem–loop within the cTNT pre-mRNAs as well. We chose to study the cTNT intron 4 because it was the only available pre-mRNA substrate with a well-identified binding site (Ho et al. 2004). The combination of UV melting of the cTNT 50mer RNA (Fig. 4E), CD of this RNA in the absence and presence of MBNL (Fig. 4F), and structure probing of the cTNT 32mer (Fig. 5A,B) show that this RNA folds into a stem–loop structure that appears to be partially A-form, which was also found for the CUG repeats. This proposed helix has some similarities to the CUG and CCUG helices in that one of the pyrimidine–pyrimidine mismatches is bracketed by G-C base pairs (though the other mismatch is flanked by G-U wobble base pairs). The similarities between the CUG, CCUG, and cTNT stems suggest MBNL recognizes both its pathogenic and natural RNA targets through an analogous mode of recognition.
The stem–loop structure in the fourth intron of the cTNT pre-mRNA is not predicted by Mfold, perhaps due to its multiple mismatches and the minimal number of consecutive base pairs. It was therefore previously predicted to be single stranded. As stated above, our data strongly support the formation of this stem–loop and that MBNL recognizes this RNA structure. The lack of binding to the cTNT 50mer containing the four mutations due to the disruption of the stem–loop and potential removal of key nucleotides for MBNL recognition (Fig. 6) further support the model that MBNL binds this stem–loop. The lack of cross-linking by MBNL to a cTNT RNA containing these four mutations as well as the elimination of regulated splicing of the cTNT exon 5 when these mutations are introduced into a cTNT minigene (Ho et al. 2004) suggest that the presence of this stem–loop within intron 4 of the cTNT pre-mRNA is important for the regulated splicing of exon 5 by MBNL.
Clearly MBNL can bind a range of RNA stems, but the common theme of pyrimidine–pyrimidine mismatches and presence of G-C and C-G base pairs indicates these are requirements for binding by MBNL. Further studies are necessary to fully define the specificity of MBNL before predictions can be made to identify binding sites in other MBNL regulated pre-mRNAs. Another challenge is that these potential regulatory stem–loops will not necessarily be predicted by folding programs due to the lack of consecutive base pairs and presence of mismatches as observed for the cTNT intron 4 stem–loop.
Significance of MBNL's binding to a helical element in its role as a splicing regulator
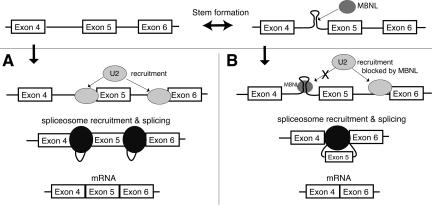
A possible model for the mechanism through which MBNL regulates the exclusion of exon 5 in the cTNT mRNA is that MBNL competes for binding of the intron with other splicing factors. When MBNL binds the stem–loop at the 3′ end of the intron, the presence of MBNL or the stabilization of the stem–loop may inhibit recognition of this 3′ splice site by the splicing machinery, causing exon skipping to occur. If the stem–loop does not form and MBNL is not present, the splicing machinery recognizes this site and exon 5 is included (Fig. 8).
FIGURE 8.
Model of MBNL's regulation of the cTNT pre-mRNA splicing. (A) If the stem–loop does not form, the splicing machinery represented by U2 (U2 snRNP) recognizes this 3′ splice site and exon 5 is included. (B) When the stem–loop forms and MBNL binds, this 3′ splice site is not recognized by the splicing machinery and exon 5 is skipped.
The mutations made to the cTNT splicing minigene support the model that recruitment of MBNL to a stem–loop causes repression of exon 5. The replacement of the endogenous binding site with another sequence that MBNL binds causes MBNL dependent repression of exon 5, while minor mutations that significantly reduce MBNL's binding abolish MBNL ability to repress inclusion of exon 5. These results indicate that direct binding of MBNL to this stem–loop is required for the repression of exon 5. Mutations that add additional base pairs that stabilize the stem–loop also strongly cause repression of exon 5, showing that increased stability of the stem correlates with repression of exon 5. It appears that if the stem is strengthened enough, MBNL is no longer needed for repression, as sequestering MBNL function does not affect splicing of this construct.
This mechanism may have two levels of regulation. First, the stem–loop alone might inhibit the use of this 3′ splice site; second, the addition of MBNL might further stabilize this stem–loop and increase the block at this 3′ splice site. Previously, stem–loop formation has been shown to reduce the use of 5′ and 3′ splice sites (Watakabe et al. 1989; Libri et al. 1991), but this is the first example to our knowledge in which the stem–loop is specifically recognized by a factor that regulates splicing. Therefore MBNL may be a member of a new second class of splicing regulators utilizing secondary structure, while other splicing regulators have previously been shown to recognize single-stranded motifs, such as the SR splicing factors (for review, see Blencowe et al. 1999; Singh and Valcarcel 2005).
Like other factors regulating splicing, MBNL functions in one context to exclude an exon as described for the cTNT exon 5, while for other exons the presence of MBNL enhances the inclusion of a particular exon. One possible mechanism through which MBNL could enhance exon inclusion would be to sequester exonic or intronic splicing silencers in a stem–loop. Alternatively the location of MBNL binding (upstream or downstream of the exon or within the exon) may determine if it acts as splicing enhancer or repressor as has been found for other splicing factors, such as NOVA (Ule et al. 2006).
MATERIALS AND METHODS
Cloning and protein purification
MBNL1 was PCR amplified and was cloned into GST fusion vector pGEX-6P-1 (Amersham), using DNA (MBNL isoform with amino acids 1–382) provided by Maury Swanson (Ho et al. 2004). Both the full-length MBNL1 and MBNL1(1–260) constructs were cloned using BamHI and NotI restriction sites.
Using BL21-Star expression cells (Invitrogen), protein expression was induced with 0.25 mM IPTG at an OD600 ∼0.5–1, for 3–4 h at 37°C. Cells were lysed in 30 mL of buffer (500 mM NaCl, 25 mM Tris at pH 7.5, 10 mM β-mercaptoethanol [BME], and 5% glycerol) using 1 mg/mL of lysozyme followed by sonication (3×30 sec). Cell extract was centrifuged for 15 min at 17,000 rpm, and lysate that contained GST-MBNL was collected. GST-MBNL was bound to GST affinity beads for 30 min at 4°C. Beads were washed five times with buffer (1 M NaCl, 25 mM Tris at pH 7.5, and 5 mM BME); MBNL was cleaved from the affinity tag with Precision Protease (Amersham) and collected from the beads. The protein was then run over an anion exchange (Q) column. Copurifying contaminants bind the column, but MBNL does not. MBNL was collected in the column flowthrough, concentrated, and dialyzed into storage buffer (50% glycerol, 500 mM NaCl, 20 mM Tris at pH 7.5, 5 mM BME) and stored at −20°C.
RNA synthesis, labeling, and purification
The RNA substrate CUG90 and CUG54 was transcribed with T7 polymerase off the pCTG54 and pCTG90 plasmids, respectively, provided by Maury Swanson (University of Florida College of Medicine, Gainesville, FL) (Miller et al. 2000). During transcription, RNAs were radiolabeled using [α-32P]CTP. All other RNA substrates were ordered from IDT DNA, and 5′-end labeled using [γ-32P]ATP. All RNAs were purified on 8% polyacrylamide denaturing gels.
Gel shift assay
Solutions for the protein–RNA binding experiments contained 175 mM NaCl, 5 mM MgCl2, 20 mM Tris (pH 7.5), 1.25 mM BME, 12.5% glycerol, 2 mg/mL BSA, and 0.1 mg/mL heparin. Prior to incubation, RNA substrates were annealed by incubation at 95°C for 2 min and then placed directly on ice for 20 min in 66 mM NaCl, 6.7 mM MgCl2, and 27 mM Tris (pH 7.5). Protein was then added to the RNA. The binding reaction was 10 μL volume and was incubated for 20 min at room temperature before 3–5 μL were loaded on a prechilled 4°C gel. RNA and RNA–protein complexes were separated on 3% acrylamide (37.5:1, or 80:1), 0.3% agarose, 0.5× Tris-Borate (TB) gels, run for ∼30 min at 4°C, 175 V. Gels were dried and autoradiographed.
For binding curves, gels were quantified using ImageQuant (Molecular Dynamics). The percent RNA bound was determined by taking the ratio of RNA:protein complex to total RNA, per lane. Binding curves were graphed, and apparent Kd values were determined with KaleidaGraph (Synergy) software using the following equation: y= [(m2+m1+m0)−(−m2−m1−m0)2−(4×m1×m0)]0.5/(2×m1), where y=% bound, m2= Kd, m1=total RNA concentration, and m0=protein concentration. This equation assumes a 1:1 interaction between the RNA and protein, which allows only an apparent Kd to be determined for CUG90 and RNAs containing more than one binding site. To determine the standard error of the apparent dissociation constants, three to five binding titrations were performed with each substrate and the apparent Kd values determined for each titration separately, prior to averaging. The error bars on the binding curve were obtained by averaging the individual titration points and calculating the standard deviation. Data points greater than two standard deviations from the average were discarded, with at least three data points remaining for standard deviation analysis.
Structure probing assay
End-labeled RNA was annealed and placed in the same binding conditions used for the gel shift assay (except BSA was excluded and 0.1 mg/mL tRNA was used instead of heparin, and the volume of the binding reaction was 9 μL). RNases (1 μL) were added, and the RNA digested for 2 min at room temperature. The reaction was quenched with phenol; RNA was collected through ethanol precipitation, resuspended in denaturing dye, and heated at 95°C for 10 min. The sample was then run on a 15% polyacrylamide (19:1), 8 M Urea, 1× TBE gel.
Analytical ultracentrifugation
AUC runs with MBNL (20 μM) were made in solutions containing 170 mM NaCl, 20 mM Tris (pH 7.5), BME 0.2 mM, and 1% glycerol (v/v). Data were collected on an Optima XL-I (Beckman), for ∼10 h at 60,000 rpm, 4°C. Data were analyzed using SedFit 97 (NIH).
Circular dichroism and thermal melts
The CD spectra of MBNL (2 μM) were measured in 50 mM NaCl and 10 mM sodium phosphate buffer (pH 7.5). Data were collected on a J-720 spectropolarimeter (Jasco) for 0.5–1 h at room temperature at a rate of 5 nm/min. For the CD spectra of RNA, the samples were annealed by snap-cooling at 95°C for 2 min, then directly on ice for 15 min in 50 mM NaCl, 5 mM MgCl2, and 10 mM phosphate (pH 7.5); lower salt and phosphate were used to reduce interference at lower wavelengths. CD measurements were made at room temperature over a period of 2–3 h, at spectral scanning rates of 5 or 10 nm/min. For measurements on protein–RNA complexes the components were mixed and incubated for 20 min prior to data collection.
For UV thermal melts, RNAs (1 μM) were snap-annealed and melted at a rate of 2°C per minute, monitored at 260 nm. Tm values were calculated by determining the inflection point for each thermal transition, or the midpoint of the transition if the inflection point was not apparent.
In vivo splicing
Wild-type cTNT, the 4G cTNT mutant, and the DMPK-CUG950 minigenes were obtained from the laboratory of Thomas Cooper (Ho et al. 2004). All additional mutations were made using PCR from the WT sequence. Sal1 and Spe1 restriction sites were used to subclone all mutants after PCR.
HeLa cells were grown in monolayers in DMEM with GLUTAMAX (Gibco) and supplemented with 10% fetal bovine serum (Gibco). Approximately 2.0 (± 0.2)×105 cells were plated in six well plates and transfected 18–20 h later at 70%–90% confluency. One microgram of plasmid was transfected into each well of cells, using 5 μL of Lipofectin2000 (Invtitrogen) according to the manufacturer's protocols. For cotransfection, 1 μg of total plasmid was transfected, at 500 ng of each construct. Cells were harvested 20–24 h after transfection using triplE reagent (Gibco). Immediately following harvesting, RNA was isolated from the cell pellets using an RNeasy kit (Qiagen). Five hundred nanograms of isolated RNA were DNased with RQI DNase (Promega) according to the manufacturer's protocol. One hundred nanograms of DNAsed RNA were reverse transcribed with Superscript II and a cTNT-specific reverse primer according to the manufacturer's protocols. Thirty nanograms of the RT reaction were subjected to 22–25 rounds of PCR amplification using cTNT-specific primers spiked with a kinased cTNT forward primer. The linear range for PCR was determined for the wild-type and 4G construct and found to be between 20 and 26 cycles. The resulting PCR products were run on a 6% (19:1) polyacrylamide denaturing gel at 6 W for 2 h. The gel was subsequently autoradiographed, and quantitation of the radioactive bands was performed using ImageQuant software.
ACKNOWLEDGMENTS
We thank Alice Barkan and members of the Berglund and von Hippel laboratories for experimental advice and comments on the manuscript, especially Sandra Grieves, Emily Goers, Amy Mahady, Kristy Henscheid, and Danielle Cass. We thank Charles Thornton and Tom Cooper for plasmids and Maury Swanson for the MBNL plasmid, helpful suggestions on the manuscript, and communicating results prior to publication. This work was supported by grants from the Muscular Dystrophy Association and NIH (AR053903) to J.A.B., and NIH (GM-15792) to Peter H. von Hippel, who serves as coadvisor (with J.A.B.) to M.B.W. M.B.W. was supported by NIH training grant GM-07759 to the Institute of Molecular Biology.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.610607.
REFERENCES
- Adereth, Y., Dammai, V., Kose, N., Li, R., Hsu, T. RNA-dependent integrin α3 protein localization regulated by the muscleblind-like protein MLP1. Nat. Cell Biol. 2005;7:1240–1247. doi: 10.1038/ncb1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auweter, S.D., Oberstrass, F.C., Allain, F.H. Sequence-specific binding of single-stranded RNA: Is there a code for recognition? Nucleic Acids Res. 2006;34:4943–4959. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begemann, G., Paricio, N., Artero, R., Kiss, I., Perez-Alonso, M., Mlodzik, M. Muscleblind, a gene required for photoreceptor differentiation in Drosophila, encodes novel nuclear Cys3His-type zinc-finger-containing proteins. Development. 1997;124:4321–4331. doi: 10.1242/dev.124.21.4321. [DOI] [PubMed] [Google Scholar]
- Blencowe, B.J., Bowman, J.A., McCracken, S., Rosonina, E. SR-related proteins and the processing of messenger RNA precursors. Biochem. Cell Biol. 1999;77:277–291. [PubMed] [Google Scholar]
- Brown, R.S. Zinc finger proteins: Getting a grip on RNA. Curr. Opin. Struct. Biol. 2005;15:94–98. doi: 10.1016/j.sbi.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Charlet, B.N., Savkur, R.S., Singh, G., Philips, A.V., Grice, E.A., Cooper, T.A. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell. 2002;10:45–53. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- Cho, D.H., Tapscott, S.J. Myotonic dystrophy: Emerging mechanisms for DM1 and DM2. Biochim. Biophys. Acta. 2007;1772:195–204. doi: 10.1016/j.bbadis.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Dam, J., Schuck, P. Calculating sedimentation coefficient distributions by direct modeling of sedimentation velocity concentration profiles. Methods Enzymol. 2004;384:185–212. doi: 10.1016/S0076-6879(04)84012-6. [DOI] [PubMed] [Google Scholar]
- Dansithong, W., Paul, S., Comai, L., Reddy, S. MBNL1 is the primary determinant of focus formation and aberrant insulin receptor splicing in DM1. J. Biol. Chem. 2005;280:5773–5780. doi: 10.1074/jbc.M410781200. [DOI] [PubMed] [Google Scholar]
- Dere, R., Napierala, M., Ranum, L.P., Wells, R.D. Hairpin structure-forming propensity of the (CCTG•CAGG) tetranucleotide repeats contributes to the genetic instability associated with myotonic dystrophy type 2. J. Biol. Chem. 2004;279:41715–41726. doi: 10.1074/jbc.M406415200. [DOI] [PubMed] [Google Scholar]
- Fardaei, M., Rogers, M.T., Thorpe, H.M., Larkin, K., Hamshere, M.G., Harper, P.S., Brook, J.D. Three proteins, MBNL, MBLL, and MBXL, colocalize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum. Mol. Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- Hall, T.M. Multiple modes of RNA recognition by zinc finger proteins. Curr. Opin. Struct. Biol. 2005;15:367–373. doi: 10.1016/j.sbi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Ho, T.H., Charlet, B.N., Poulos, M.G., Singh, G., Swanson, M.S., Cooper, T.A. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, T.H., Bundman, D., Armstrong, D.L., Cooper, T.A. Transgenic mice expressing CUG-BP1 reproduce splicing misregulation observed in myotonic dystrophy. Hum. Mol. Genet. 2005a;14:1539–1547. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- Ho, T.H., Savkur, R.S., Poulos, M.G., Mancini, M.A., Swanson, M.S., Cooper, T.A. Colocalization of muscleblind with RNA foci is separable from misregulation of alternative splicing in myotonic dystrophy. J. Cell Sci. 2005b;118:2923–2933. doi: 10.1242/jcs.02404. [DOI] [PubMed] [Google Scholar]
- Hudson, B.P., Martinez-Yamout, M.A., Dyson, H.J., Wright, P.E. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat. Struct. Mol. Biol. 2004;11:257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- Hung, S.H., Yu, Q., Gray, D.M., Ratliff, R.L. Evidence from CD spectra that d(purine)·r(pyrimidine) and r(purine)·d(pyrimidine) hybrids are in different structural classes. Nucleic Acids Res. 1994;22:4326–4334. doi: 10.1093/nar/22.20.4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, S., Alekseev, Y., Bertrand, J.R., Malvy, C., Gottikh, M.B. Formation of stable triplexes between purine RNA and pyrimidine oligodeoxyxylonucleotides. Nucleic Acids Res. 2003;31:4256–4263. doi: 10.1093/nar/gkg443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, H., Mankodi, A., Swanson, M.S., Moxley, R.T., Thornton, C.A. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum. Mol. Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- Kanadia, R.N., Johnstone, K.A., Mankodi, A., Lungu, C., Thornton, C.A., Esson, D., Timmers, A.M., Hauswirth, W.W., Swanson, M.S. A muscleblind knockout model for myotonic dystrophy. Science. 2003a;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- Kanadia, R.N., Urbinati, C.R., Crusselle, V.J., Luo, D., Lee, Y.J., Harrison, J.K., Oh, S.P., Swanson, M.S. Developmental expression of mouse muscleblind genes Mbnl1, Mbnl2 and Mbnl3. Brain Res. Gene Expr. Patterns. 2003b;3:459–462. doi: 10.1016/s1567-133x(03)00064-4. [DOI] [PubMed] [Google Scholar]
- Kino, Y., Mori, D., Oma, Y., Takeshita, Y., Sasagawa, N., Ishiura, S. Muscleblind protein, MBNL1/EXP, binds specifically to CHHG repeats. Hum. Mol. Genet. 2004;13:495–507. doi: 10.1093/hmg/ddh056. [DOI] [PubMed] [Google Scholar]
- Ladd, A.N., Stenberg, M.G., Swanson, M.S., Cooper, T.A. Dynamic balance between activation and repression regulates pre-mRNA alternative splicing during heart development. Dev. Dyn. 2005;233:783–793. doi: 10.1002/dvdy.20382. [DOI] [PubMed] [Google Scholar]
- Libri, D., Piseri, A., Fiszman, M.Y. Tissue-specific splicing in vivo of the β-tropomyosin gene: Dependence on an RNA secondary structure. Science. 1991;252:1842–1845. doi: 10.1126/science.2063196. [DOI] [PubMed] [Google Scholar]
- Lin, X., Miller, J.W., Mankodi, A., Kanadia, R.N., Yuan, Y., Moxley, R.T., Swanson, M.S., Thornton, C.A. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- Liquori, C.L., Ricker, K., Moseley, M.L., Jacobsen, J.F., Kress, W., Naylor, S.L., Day, J.W., Ranum, L.P. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- Mankodi, A., Logigian, E., Callahan, L., McClain, C., White, R., Henderson, D., Krym, M., Thornton, C.A. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- Mankodi, A., Urbinati, C.R., Yuan, Q.P., Moxley, R.T., Sansone, V., Krym, M., Henderson, D., Schalling, M., Swanson, M.S., Thornton, C.A. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum. Mol. Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- Mankodi, A., Teng-Umnuay, P., Krym, M., Henderson, D., Swanson, M., Thornton, C.A. Ribonuclear inclusions in skeletal muscle in myotonic dystrophy types 1 and 2. Ann. Neurol. 2003;54:760–768. doi: 10.1002/ana.10763. [DOI] [PubMed] [Google Scholar]
- Meola, G., Moxley R.T., 3rd Myotonic dystrophy type 2 and related myotonic disorders. J. Neurol. 2004;251:1173–1182. doi: 10.1007/s00415-004-0590-1. [DOI] [PubMed] [Google Scholar]
- Michalowski, S., Miller, J.W., Urbinati, C.R., Paliouras, M., Swanson, M.S., Griffith, J. Visualization of double-stranded RNAs from the myotonic dystrophy protein kinase gene and interactions with CUG-binding protein. Nucleic Acids Res. 1999;27:3534–3542. doi: 10.1093/nar/27.17.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J.W., Urbinati, C.R., Teng-Umnuay, P., Stenberg, M.G., Byrne, B.J., Thornton, C.A., Swanson, M.S. Recruitment of human muscleblind proteins to (CUG)n expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinaro, M., Tinoco I., Jr Use of ultrastable UNCG tetraloop hairpins to fold RNA structures: Thermodynamic and spectroscopic applications. Nucleic Acids Res. 1995;23:3056–3063. doi: 10.1093/nar/23.15.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooers, B.H., Logue, J.S., Berglund, J.A. The structural basis of myotonic dystrophy from the crystal structure of CUG repeats. Proc. Natl. Acad. Sci. 2005;102:16626–16631. doi: 10.1073/pnas.0505873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napierala, M., Krzyzosiak, W.J. CUG repeats present in myotonin kinase RNA form metastable “slippery” hairpins. J. Biol. Chem. 1997;272:31079–31085. doi: 10.1074/jbc.272.49.31079. [DOI] [PubMed] [Google Scholar]
- Nykamp, K.R., Swanson, M.S. Toxic RNA in the nucleus: Unstable microsatellite expression in neuromuscular disease. Prog. Mol. Subcell. Biol. 2004;35:57–77. doi: 10.1007/978-3-540-74266-1_3. [DOI] [PubMed] [Google Scholar]
- Osborne, R.J., Thornton, C.A. RNA-dominant diseases. Hum. Mol. Genet. 2006;15((Suppl.)):R162–R169. doi: 10.1093/hmg/ddl181. [DOI] [PubMed] [Google Scholar]
- Pascual, M., Vicente, M., Monferrer, L., Artero, R. The Muscleblind family of proteins: An emerging class of regulators of developmentally programmed alternative splicing. Differentiation. 2006;74:65–80. doi: 10.1111/j.1432-0436.2006.00060.x. [DOI] [PubMed] [Google Scholar]
- Paul, S., Dansithong, W., Kim, D., Rossi, J., Webster, N.J., Comai, L., Reddy, S. Interaction of muscleblind, CUG-BP1 and hnRNP H proteins in DM1-associated aberrant IR splicing. EMBO J. 2006;25:4271–4283. doi: 10.1038/sj.emboj.7601296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips, A.V., Timchenko, L.T., Cooper, T.A. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- Ranum, L.P., Cooper, T.A. RNA-mediated neuromuscular disorders. Annu. Rev. Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- Savkur, R.S., Philips, A.V., Cooper, T.A. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- Savkur, R.S., Philips, A.V., Cooper, T.A., Dalton, J.C., Moseley, M.L., Ranum, L.P., Day, J.W. Insulin receptor splicing alteration in myotonic dystrophy type 2. Am. J. Hum. Genet. 2004;74:1309–1313. doi: 10.1086/421528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, K., Chen, W., Ashique, A.M., Moroi, R., Li, Y.P. Molecular cloning, developmental expression, promoter analysis and functional characterization of the mouse CNBP gene. Gene. 2003;307:51–62. doi: 10.1016/s0378-1119(03)00406-2. [DOI] [PubMed] [Google Scholar]
- Singh, R., Valcarcel, J. Building specificity with nonspecific RNA-binding proteins. Nat. Struct. Mol. Biol. 2005;12:645–653. doi: 10.1038/nsmb961. [DOI] [PubMed] [Google Scholar]
- Sobczak, K., de Mezer, M., Michlewski, G., Krol, J., Krzyzosiak, W.J. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 2003;31:5469–5482. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squillace, R.M., Chenault, D.M., Wang, E.H. Inhibition of muscle differentiation by the novel muscleblind-related protein CHCR. Dev. Biol. 2002;250:218–230. doi: 10.1006/dbio.2002.0798. [DOI] [PubMed] [Google Scholar]
- Tian, B., White, R.J., Xia, T., Welle, S., Turner, D.H., Mathews, M.B., Thornton, C.A. Expanded CUG repeat RNAs form hairpins that activate the double-stranded RNA-dependent protein kinase PKR. RNA. 2000;6:79–87. doi: 10.1017/s1355838200991544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko, N.A., Iakova, P., Cai, Z.J., Smith, J.R., Timchenko, L.T. Molecular basis for impaired muscle differentiation in myotonic dystrophy. Mol. Cell. Biol. 2001;21:6927–6938. doi: 10.1128/MCB.21.20.6927-6938.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule, J., Stefani, G., Mele, A., Ruggiu, M., Wang, X., Taneri, B., Gaasterland, T., Blencowe, B.J., Darnell, R.B. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- Watakabe, A., Inoue, K., Sakamoto, H., Shimura, Y. A secondary structure at the 3′ splice site affects the in vitro splicing reaction of mouse immunoglobulin μ chain pre-mRNAs. Nucleic Acids Res. 1989;17:8159–8169. doi: 10.1093/nar/17.20.8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]