Abstract
Splicing of introns from mRNA precursors is a two-step reaction performed by the spliceosome, an immense cellular machine consisting of over 200 different proteins and five small RNAs (snRNAs). We previously demonstrated that fragments of two of these RNAs, U6 and U2, can catalyze by themselves a splicing-related reaction, involving one of the two substrates of the first step of splicing, the branch site substrate. Here we show that these same RNAs can catalyze a reaction between RNA sequences that resemble the 5′ splice site and the branch site, the two reactants of the first step of splicing. The reaction is dependent on the sequence of the 5′ splice site consensus sequence and the catalytically essential domains of U6, and thus it resembles the authentic splicing reaction. Our results demonstrate the ability of protein-free snRNAs to recognize the sequences involved in the first splicing step and to perform splicing-related catalysis between these two pre-mRNA-like substrates.
Keywords: snRNAs, U6, ribozyme, spliceosome, splicing
INTRODUCTION
Splicing of mRNA precursors is not only an essential and nearly ubiquitous step in eukaryotic gene expression, but it is also an important mechanism for generation of proteomic diversity and regulation of cellular function. There are numerous examples where splicing and its regulation play key roles in cell growth control, differentiation, and disease. Given the complexity and crucial role of splicing, it is not surprising that the spliceosome, the multimegadalton ribonucleoprotein complex that performs splicing, is the largest and most complicated cellular machine known, comprising over 200 proteins and five RNA molecules, the snRNAs (Rappsilber et al. 2002; Zhou et al. 2002; Will and Luhrmann 2006).
Considerable data points to the existence of an extensive network of RNA–RNA and RNA–protein interactions in the spliceosome (for review, see Moore et al. 1993; Nilsen 1998; Brow 2002). Through these interactions, the boundaries between the intervening sequences (introns) and the coding regions (exons), termed the 5′ and 3′ splice sites, and a conserved region of the intron close to the 3′ splice site, called the branch site, are recognized in a number of steps. Through a number of conformational rearrangements, the active site of the spliceosome is formed and the 5′ and 3′ splice sites and the branch site are positioned for catalysis of the first step of splicing. Of the five spliceosomal snRNAs, U1 and U4 leave the spliceosome during the assembly process, and U5 snRNA is largely dispensable for at least the first step of splicing in vitro (O'Keefe et al. 1996; Segault et al. 1999). Thus, U2 and U6 snRNAs are the only spliceosomal RNAs that are absolutely required for both steps of splicing. In the catalytically active spliceosome, U2 snRNA is base-paired to the branch site, and U6 snRNA coordinates the 5′ splice site in part by base pairing to this region. At the same time, U2 and U6 are extensively base-paired to each other through three helices (Fig. 1). The base-paired U6/U2 complex not only helps to juxtapose the 5′ splice site and the branch site, it also likely provides a scaffold for the assembly of the other active site factors.
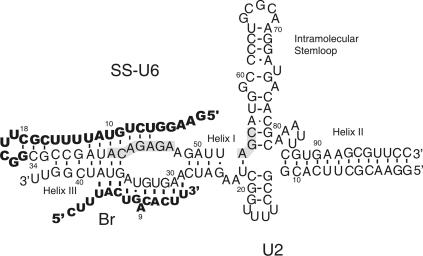
FIGURE 1.
U6/U2 base-paired complex and splicing substrates. The splicing substrates are shown in bold letters. Base-pairing interactions between Br and U2 and the bulged A are shown. SS is covalently attached to the 5′ end of U6 via a linker sequence and a hairpin (nt 11–25 of the bold region of SS-U6). The positions of the U6 intramolecular stem–loop and U6/U2 helices I, II, and III are shown. The highlighted areas mark the invariant domains of U6. The numbering of the U6 part of SS-U6 and U2 refers to human numbering. Base-pairing interactions are shown by black bars. The mutually exclusive base-pairing interactions shown between U6, U2, and SS in helix III region do not form concurrently.
Splicing proceeds through two transesterification reactions. For the first step of splicing, the 2′OH group of a conserved adenosine in the branch site makes a nucleophilic attack on the phosphodiester bond at the 5′ splice site, resulting in the formation of a branched (lariat) intron and release of the 5′ exon. During the second step, the 3′ OH group of the released first exon is activated to attack the 3′ splice site, resulting in ligation of exons and release of the lariat intron. Extensive mechanistic and structural similarities between self-splicing group II introns and the spliceosomal snRNAs have suggested that the spliceosome, similarly, might be an RNA enzyme, and domain-swapping experiments have indicated that a number of structural elements in snRNAs and group II introns are functionally interchangeable (Hetzer et al. 1997; Shukla and Padgett 2002). Since U2 and U6 snRNAs are the only spliceosomal RNAs required for both steps of splicing, if the splicing reactions are indeed RNA-catalyzed, then these two snRNAs should form the catalytic active site of the spliceosome (Collins and Guthrie 2000; Valadkhan and Manley 2002). Consistent with having a crucial role in splicing catalysis, both U6 and U2 snRNAs are highly conserved, and mutagenesis and cross-linking studies have not only proved the physical proximity between two invariant domains in U6, the ACAGAGA box and the AGC triad (Nilsen 1998), and the 5′ and 3′ splice sites but have also indicated a crucial role for these domains in splicing catalysis (Nilsen 1998; Rhode et al. 2006). However, direct proof for RNA catalysis in the spliceosome has been lacking.
We previously showed that a protein-free, in vitro assembled complex containing the central domains of human U2 and U6 snRNAs can recognize and bind a short RNA oligonucleotide that resembled the intron branch site. Under appropriate conditions, the U6/U2 complex catalyzed a reaction between the branch site adenosine and the backbone of U6, resulting in formation of an X-shaped, branched RNA species (Valadkhan and Manley 2000, 2001, 2003). Further characterization of this reaction, including ionic requirements and an important role for conserved sequence elements, revealed intriguing similarities to the first step of splicing. However, the chemistry of the reaction was different, and the 5′ splice site substrate was absent in this reaction. Thus, although the spliceosomal snRNAs showed splicing-related catalytic activity, their ability to perform the authentic splicing reaction remained uncertain. Specifically, it was not clear if the complex formed by U6 and U2 could recognize and position the other substrate of the first step of splicing, the 5′ splice site, in the active site in such a way that would allow a transesterification reaction between the branch site substrate and the 5′ splice site to occur.
Here we describe a novel U6/U2-catalyzed reaction between an RNA sequence containing the 5′ splice site consensus (5′SS) and the branch site substrate oligonucleotide. For this reaction, an RNA containing the 5′SS linked to the 5′ end of U6 snRNA via a short RNA hairpin was designed to help recruit and orient the 5′SS toward the catalytically important ACAGAGA domain of U6. This reaction, similar to the previously described U6/U2-catalyzed reaction, is dependent on the presence of Mg2+ in the buffer and on the ACAGAGA and AGC domains in U6 and, importantly, shows a strict dependence on the sequence of the 5′SS substrate. Similar to the removal of the 5′ exon during the first step of splicing, the 5′ end of the 5′SS is removed during this reaction, further strengthening the similarities between this reaction and the authentic first step of splicing.
RESULTS
To examine further the potential of U6 and U2 snRNAs for performing catalysis in the spliceosome, we first set out to determine if these two snRNAs are capable of simultaneously binding the two regions in pre-mRNA that are involved in the first step of splicing: the branch site and the 5′ splice site consensus sequences. To this end, we used in vitro transcribed RNAs consisting of the central domains of human U6 (nucleotides 37–99) and U2 (nucleotides 1–44). Two short RNA oligonucleotides, one containing the consensus branch site (Br) and the other the 5′SS, were used as substrates. Incubation of these two RNAs with the preformed U2/U6 complex followed by analysis on nondenaturing gels indicated that, although Br efficiently bound the U6/U2 complex, both in the presence and absence of 5′SS, the 5′SS RNA did not stably interact with the U6/U2 complex under all conditions tested (data not shown). Analysis of the RNAs on denaturing gels indicated that no 5′SS-dependent products were formed, although the previously described Br-dependent product, RNA X (Valadkhan and Manley 2001), was present (data not shown).
While the branch site consensus sequence in both yeast and mammalian systems is capable of forming 4–7 base pairs (bp) with U2, the sequence complementarity between the 5′SS and the domain in U6 with which it interacts, the ACAGAGA box, is very limited. It is likely that in the authentic spliceosome, tertiary interactions with other RNA/protein factors play a major role in the binding and positioning of the 5′ splice site sequence. In an effort to compensate for the lack of such stabilizing interactions in our system, we covalently attached the 5′SS to the 5′ end of U6 by a linker sequence containing a hyperstable hairpin loop (Varani 1995) in such a way that the 5′ splice site would be positioned in the vicinity of the catalytically important ACAGAGA domain of U6 (Fig. 1). A number of such RNAs that differed in the length of the linker region and the exact sequence surrounding the 5′ splice site were synthesized.
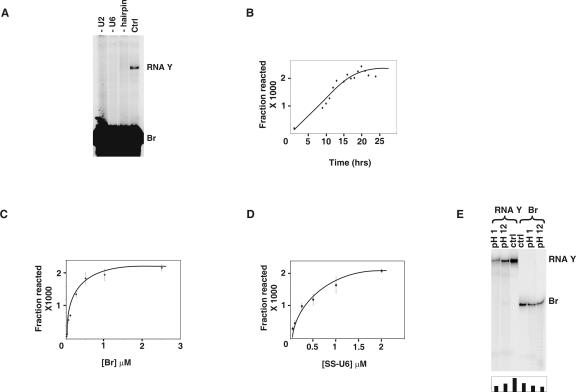
We next tested the above U2/U6 complexes formed with these chimeric constructs (SS-U6) for catalytic activity after the addition of 32P-labeled Br. Interestingly, after 24 h of incubation at room temperature in the presence of MgCl2, a novel RNA species was formed in reactions containing one of the SS-U6 constructs (Fig. 2A). The new RNA, which we named RNA Y, was resistant to denaturation at 85°C in 10 M urea and 20 mM EDTA and was dependent on the presence of both U2 and SS-U6 for its formation (Fig. 2A). RNA Y accumulated to ∼0.2% of the input after 15 h, with an apparent reaction rate of 0.3/h (Fig. 2B). Increasing the concentration of Br and SS-U6 from 0.001 to 5 μM and from 0.05 to 5 μM, respectively, showed an initial linear concentration dependence followed by a plateau (Fig. 2C,D). Incubation of RNA Y formation reactions in the dark or replacing Tris with HEPES in reaction buffer did not alter the efficiency of the reaction, ruling out the possibility that RNA Y is a photo-cross-linked product or a Tris adduct (data not shown). To ensure that the low yield of reaction reflects the intrinsic folding and catalytic properties of the RNAs, and not depletion of a rare subset of SS-U6/U2 or Br that contains a certain unknown chemical or sequence modification, we purified unreacted Br and SS-U6/U2s from RNA Y formation reactions and reused them in new reactions. The yield of RNA Y in these reactions was identical to reactions with previously unreacted substrates (data not shown).
FIGURE 2.
Characterization of RNA Y formation. (A) Requirements for RNA Y formation. Position of RNA Y and Br is shown to the right. The reactions in lanes marked “−U2” and “−U6” lack U2 and U6, respectively. Lane marked “−hairpin” contains a SS-U6 in which the hyperstable hairpin structure linking SS to U6 is replaced by a random sequence. Lane marked “Ctrl” contains a typical RNA Y formation reaction. (B) Time course of RNA Y formation. (C) Dependence of RNA Y formation on Br concentration. (D) Dependence of RNA Y formation on the concentration of SS-U6. (E) Effect of alkaline and acidic pH on formed RNA Y. Locations of RNA Y and Br are shown to the right. The pH of the buffer used is shown on top. The amount of RNA Y in each lane is shown in the graph at the bottom of the gel.
To investigate the chemical nature of the linkage in RNA Y, and to define its relationship to the previously characterized RNA X, we determined the effect of acid or alkali pH on purified RNA Y. Incubation of RNA Y at pH 1.0 or pH 12 at 37°C did not result in significant separation of Br and SS-U6, although this treatment resulted in a reduction in the amount of RNA Y due to phosphodiester bond cleavage (Fig. 2E). This indicates that, unlike the case in RNA X, where the linkage was ∼100 times more sensitive than a phosphodiester bond when treated with alkali under similar conditions (Valadkhan and Manley 2003), the linkage between SS-U6 and Br in RNA Y does not exhibit a higher acid or alkali sensitivity than a normal phosphodiester bond. The results also show that the chemistry of RNA Y formation is distinct from that of RNA X and also rule out a number of possible chemical linkages between the two, including for example several involving nucleobases (Shabarova and Bogdanov 1994).
Requirements of RNA Y formation
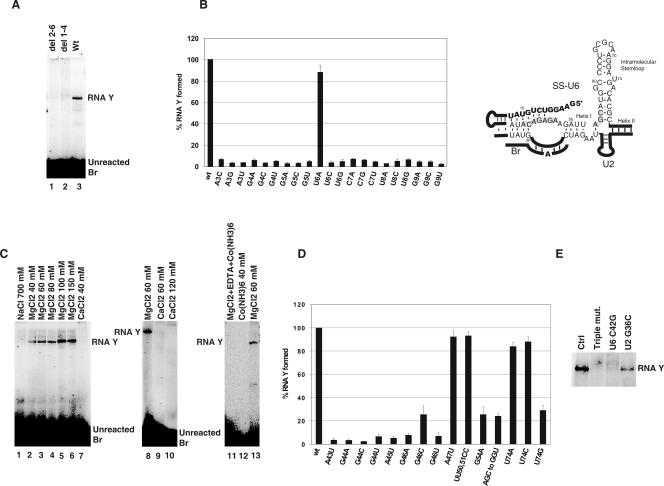
We next wished to determine if RNA Y formation is dependent on the 5′ splice site sequence in SS-U6. We first tested the effect of truncation mutations that removed nucleotides 1–4 or 2–6 of SS-U6 (del 1–4 and del 2–6 mutants; Fig. 3A, lanes 1 and 2). Interestingly, neither of the mutants formed RNA Y, even after extended incubation (Fig. 3A). To examine further the effect of specific mutations in the 5′ splice site domain of SS-U6 on RNA Y formation, we tested a number of point mutations in this region (Fig. 3B). Strikingly, almost all these mutations were incompatible with formation of RNA Y, underlining the crucial role of this domain in the reaction. Replacing the hyperstable stem–loop connecting the 5′SS to the 5′ end of U6 with a random sequence resulted in complete inhibition of RNA Y formation (Fig. 2A, −hairpin). In addition, changing the length of the linker sequence by deletion of three nucleotides (nucleotides 12–14 of the SS sequence) resulted in a block to RNA Y formation (data not shown). In contrast, mutations in the helix I region of U6 that changed the identity of the bases while keeping base pairing intact did not affect the efficiency of the reaction (Fig. 3D). Thus, not only the sequence of the 5′ splice site domain but also the correct positioning of this sequence in relationship to U6 is required for RNA Y formation.
FIGURE 3.
Sequence and ionic requirements for RNA Y formation. (A) Requirement of SS for RNA Y formation. The position of RNA Y and unreacted Br is shown to the right (Wt, wild type). (B) Effect of point mutations in SS on RNA Y formation (wt, wild type). Values represent the average of at least three independent experiments. The diagram illustrates the potential base-pairing interactions between SS and ACAGAGA in SS-U6. (C) Cationic requirement for RNA Y formation. Position of RNA Y and unreacted Br is shown to the side of gel panels. Identity and concentration of the sole cation used in the reaction are shown above each lane. Lane labeled “MgCl2+EDTA+Co(NH3)6” was initially folded in 60 mM MgCl2 and later received 0.5 M NaCl, 5 mM Co(NH3)6, and 40 mM EDTA. (D) Effect of mutations in U6 on RNA Y formation (wt, wild type). Values represent the average of three independent experiments. (E) Mutational study of the role of ACAGAGA in RNA Y formation. Position of RNA Y is shown to the right. RNA species used in each reaction is shown above each lane (Ctrl, control).
We next tested the effects of various cations on RNA Y formation. Increasing the concentration of MgCl2 from 20 to 100 mM resulted in a fourfold increase in RNA Y formation (Fig. 3C, lanes 2–6). RNA Y did not form in the presence of 40 mM cobalt hexamine, a Mg2+ mimic that can perform most of the structural but not the catalytic roles of Mg2+ (Cowan 1993), as the sole cation, or in buffers containing various ratios of cobalt hexammine and NaCl (Fig. 3C, lane 12; data not shown). Monovalent cations NaCl, KCl, and LiCl at molar concentrations were not active in promoting RNA Y formation (Fig. 3C, lane 1 and data not shown). Replacing Mg2+ with up to 120 mM Ca2+ also did not result in RNA Y formation, although addition of Mg2+ to reactions initially set-up with Ca2+ as the sole cation did result in formation of RNA Y (Fig. 3C, lanes 7, 9, and 10 and data not shown). Thus, RNA Y formation, similar to authentic splicing, requires Mg2+ ions. In an attempt to replace Mg2+ with cobalt hexammine in prefolded SS-U6/U2 complexes, we performed the initial assembly and folding of the RNAs in a Mg2+-containing buffer, followed by addition of 0.5 M NaCl, 5 mM cobalt hexammine, and 40 mM EDTA (final concentrations). We reasoned that this treatment should result in replacement of Mg2+ with Na+ and cobalt hexammine in the catalytically competent SS-U6/U2 complexes without significant structural perturbation. However, RNA Y formation was undetectable in these reactions, indicating that inner-sphere coordination of Mg2+, likely in a catalytic role, was required for the reaction (Fig. 3C, lane 11).
We also investigated the sequence requirements in U6 RNA for RNA Y formation. Point mutations in the CAGAG nucleotides of the essential ACAGAG box (Fabrizio and Abelson 1990; Madhani et al. 1990; Datta and Weiner 1993; Wolff et al. 1994; Sun and Manley 1995; McPheeters 1996) were incompatible with RNA Y formation (Fig. 3D,E). In contrast, mutations outside of this domain, such as A47U or UU50–51CC, did not significantly affect the reaction. Thus, similar to authentic splicing, RNA Y formation is dependent on the conserved ACAGAG domain of U6. Mutations in the AGC triad similarly significantly reduced RNA Y formation (Fig. 3D; data not shown), further strengthening the similarity between authentic splicing and RNA Y formation. To determine if the requirement for the ACAGAGA sequence stems from its potential for base pairing with SS, we tested a double mutant of SS-U6 containing a G to C mutation in SS at position 9 and a C to G mutation in position 42 of U6 (ACAGAGA), which restores the base-pairing potential between SS and U6 at this position. Either of the point mutations by itself was incompatible with RNA Y formation (Fig. 3D,E). The double mutant was also inactive, and a third mutation at position 36 of U2 (G36 to C), which restores the base-pairing potential between U6 nucleotide 42 and U2 nucleotide 36, did not rescue the activity (Fig. 3E and data not shown). Thus, the nucleotides at the critical ACAGAGA region of U6 are required for RNA Y formation for reasons beyond their potential ability to base-pair with SS.
A U to G mutation at position U74, which has been shown to provide the binding site for a functionally important metal ion in the authentic spliceosome (Yean et al. 2000), also resulted in significant reduction in RNA Y production, while the other point mutations at this position had no effect (Fig. 3D). This could result from the ability of a G at position 74 to form a base pair with C61, resulting in A73 being bulged out of the duplex, which in turn would disturb the local geometry of the U6 intramolecular stem–loop. Interestingly, mutation of U74 to G in the authentic spliceosome results in a block to splicing in vitro and lethality in vivo, while, at least in yeast, other point mutations at this position have no effect (Madhani et al. 1990; McPheeters 1996; Sun and Manley 1997).
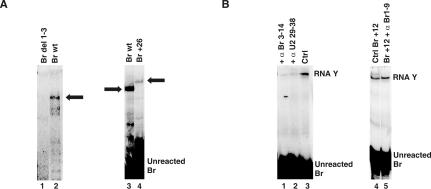
We also determined the effect of alterations of Br on RNA Y formation. Removal of the first three nucleotides of Br (Br del 1–3) blocked RNA Y formation (Fig. 4A, lane 1). Addition of extra nucleotides at the 5′ or 3′ end, though, did not prevent RNA Y formation, although significantly, products made with these larger Br species migrated as larger RNAs (e.g., Fig. 4A, lane 4). Importantly, a DNA version of Br was inactive in RNA Y formation (data not shown), indicating the requirement for one or more 2′OH.
FIGURE 4.
Br requirements in RNA Y formation. (A) Effect of changing the length of Br on RNA Y formation. The positions of RNA Y made with wild type (wt) and a Br species that has 26 extra nucleotides added to the 5′ end (Br+26) is marked by arrows. Location of unreacted Br is indicated. (B) Effect of blocking the base-pairing interaction between Br and U2 on RNA Y formation. Positions of RNA Y and unreacted Br are indicated to the right (Ctrl, control).
We next asked if binding of Br to the branch binding domain of U2 is required for RNA Y formation. To test this, we used short DNA oligonucleotides that were complementary to the branch-binding domain of U2 (nucleotides 29–38) to block the ability of Br to bind U2. In the presence of the blocking oligo, RNA Y formation was significantly reduced (Fig. 4B, lane 2). Addition of a DNA oligonucleotide complementary to nucleotides 3–14 of Br resulted in a similar decrease in RNA Y formation (Fig. 4B, lane 1). To ensure that inhibition was not due to a nonspecific effect caused by the addition of the complementary DNA oligos, we used a Br species containing a 12-nucleotide (nt) addition at its 5′ end (Br+12). DNA oligonucleotides that were complementary to the first 9 nt of Br+12 (α Br 1–9), which are upstream of the U2-binding region, did not affect RNA Y formation with Br+12 (Fig. 4B, cf. lanes 4 and 5).
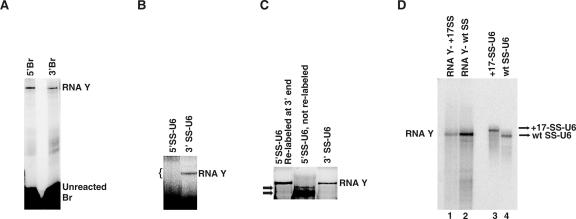
Characterization of RNA Y
We next wished to determine the RNA composition of RNA Y. Br, SS-U6, and U2 were labeled at either the 3′ or 5′ end and used individually in RNA Y formation reactions. Use of 3′- or 5′-labeled U2 did not result in the formation of a labeled RNA Y (data not shown), indicating that, although U2 is required for RNA Y formation, it is not present in RNA Y. Labels at either end of Br appeared in RNA Y (Fig. 5A), proving the presence of full-length Br. Similarly, reactions performed with 3′-labeled SS-U6 resulted in labeling of RNA Y (Fig. 5B). However, 5′-end labeled SS-U6 did not form a labeled product (Fig. 5B). This could result from one of two possibilities: either 5′ labeled SS-U6 was unable to form RNA Y, or part of the 5′ end of SS-U6, which carries the 5′ splice site domain, was removed during the formation of RNA Y. To distinguish between these two possibilities, we excised the region of the gel that corresponded to the expected location of RNA Y (region marked by bracket in Fig. 5B) and, after elution, relabeled any RNA in the eluate at the 3′ end and electrophoresed it on another gel along with an RNA Y control. Significantly, a band corresponding to RNA Y was visualized (Fig. 5C), proving that RNA Y was indeed formed with 5′-labeled SS-U6, but the 5′ end of SS-U6 was removed during the formation of RNA Y. This is analogous to the cleavage of the 5′ exon during authentic splicing, and indicates a parallel between splicing and RNA Y formation. In addition, the presence or absence of a mono- or triphosphate group at the 5′ end of SS-U6 did not alter the efficiency of RNA Y formation (data not shown), indicating that 5′-labeled SS-U6 was able to form RNA Y. This result also proved that the 5′ phosphate of SS-U6 cannot be the site of linkage in RNA Y.
FIGURE 5.
Determining the RNA sequences present in RNA Y. (A) 5′ or 3′ end-labeled Br in RNA Y formation. Positions of RNA Y and unreacted Br are shown to the right. (B) 5′ or 3′ end-labeled SS-U6 in RNA Y formation; bracket indicates the position of the gel piece that was cut to analyze for the presence of RNA Y. Position of RNA Y is shown to the right. (C) Relabeling of the RNAs eluted from the region marked by bracket in (B). Position of RNA Y is shown to the right. The SS-U6 used is shown above each lane. Arrows point to unreacted 5′-labeled SS-U6 that was co-purified with the unlabeled sample. (D) RNA Y formation with SS-U6 containing a 17-nt extension. Position of wild type (wt) SS-U6 and the SS-U6 containing a 17-nt extension at the 5′ end are shown to the right. Position of RNA Y formed is shown to the left.
To confirm and extend the above data, we added 17 extra nucleotides to the 5′ end of SS and tested the ability of this SS-U6 construct in RNA Y formation. The added nucleotides did not block the formation of RNA Y, although they reduced the efficiency of the reaction (Fig. 5D, cf. lanes 1 and 2). Importantly, although this SS-U6 construct migrated slower than the original SS-U6 on PAGE (Fig. 5D, lanes 3 and 4), the RNA Y formed had identical mobility to RNA Y formed with wt SS-U6, consistent with the removal of the 5′ end nucleotides of SS during RNA Y formation.
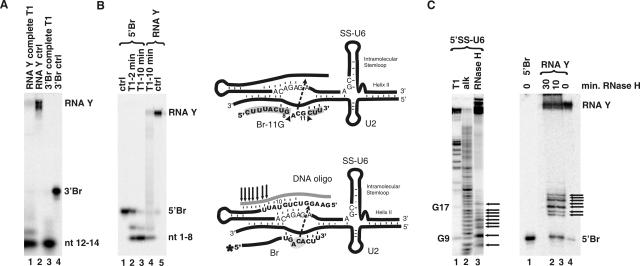
We next investigated the site of linkage between Br and SS-U6 in RNA Y on Br. Since the formation of the covalent bond between Br and SS-U6 is coupled to the removal of the 5′ end of SS-U6, it is possible that the 5′ end of SS-U6 was removed through a transesterification reaction that joins either the 3′ or 5′ end of Br, or a functional group on one of the internal nucleotides of Br, to the rest of the SS-U6. This would result in the formation of a linear species (if the 5′ or 3′ end of Br is linked to the rest of SS-U6) or a Y-shaped RNA (if an internal functional group on Br is linked to SS-U6). To differentiate between these two possibilities, we purified RNA Y made with a 5′ or 3′-labeled Br mutant that had a G substitution at position 11 (Br-11G). Br-11G showed the same reaction profile as the wild-type Br in terms of its ionic and U6 sequence requirements (data not shown). Next we subjected the purified RNA Y to complete RNase T1 digestion. The complete T1 digestion of RNA Y made with 3′-labeled Br-11G resulted in the release of a labeled fragment corresponding to nucleotides 12–14 of Br, which showed identical mobility to the fragment released from the unreacted 3′Br-11G (Fig. 6A, cf. lanes 1 and 3; see also Fig. 6, top diagram). This result indicated that the linkage between Br and SS-U6 did not involve the last three nucleotides of Br.
FIGURE 6.
Defining the sites of linkage in Br and SS in RNA Y. (A) Complete RNase T1 digestion of RNA Y made with 3′-labeled Br. Locations of 3′Br, RNA Y, and the nucleotides 12–14 fragment resulting from complete T1 digestion are shown to the right. (B) Complete RNase T1 digestion of RNA Y made with 5′-labeled Br. Positions of RNA Y, 5′Br, and the nucleotides 1–8 fragment resulting from T1 digestion are shown to the right (min, minutes of RNase T1 digestion). The identity of the RNA species is shown on top. Top diagram depicts the result of the complete RNA digestions. Arrowheads point to the sites of T1 digestion. Fragments released after complete T1 digestion are highlighted in gray. (C) RNase H digestion of 5′-labeled SS-U6 (left panel) and RNA Y (right panel) with a DNA oligo complementary to the first 18 nt of SS (T1, partial RNase T1 digestion; alk, partial alkali hydrolysis ladder). Positions of fragments released by RNase T1 are shown to the left. Positions of RNA Y and 5′Br are shown to the right. Fragments released by RNase H treatment are marked by arrows (min, minutes of RNase H digestion). The bottom diagram illustrates the outcome of RNase H digestion of RNA Y. The DNA oligonucleotide is shown in gray. The arrows indicate the 7 most downstream sites of RNase H digestion (see text). The location of the DNA oligo, SS-U6, U2 and Br is shown. The asterisk marks the location of the radioactive label. The dotted arrow shows the likely site of the linkage between the two molecules.
To determine if nucleotides at the 5′ end of Br were involved in the linkage, we subjected RNA Y made with 5′-labeled Br-11G to complete RNase T1 digestion. The results showed that the first 8 nt at the 5′ end of Br were released from RNA Y and had identical mobility to the 8-nt fragment released from unreacted Br (Fig. 6B, lanes 3 and 4; see also Fig. 6, top diagram). Since a linkage between SS-U6 and the first 8 nt of Br would have resulted in either a retardation in gel mobility or, in the case of a fragment of SS-U6 ending in a G being attached to the 5′ end of Br, the removal of the radioactive label at the 5′ end of Br, the above results indicate that the first 8 nt of Br were free and not involved in the linkage to SS-U6. Confirming the above results, we were able to radiolabel RNA Y after purification at either 5′ or 3′ ends of Br, indicating that the hydroxyl groups at the 5′ and 3′ ends of Br were not involved in the formation of the linkage (data not shown). In addition, the presence or absence of a mono- or triphosphate group at the 5′ end of Br, or a 2′–3′ cyclic end, did not interfere with RNA Y formation (data not shown), suggesting that the identities of the functional groups at the ends of Br are not important in the reaction. Taken together, the above results prove that an internal functional group located between nucleotides 9 and 11 of Br, perhaps on the putative bulged A at position 9, is the site of linkage with SS-U6. RNA Y is thus indeed a branched, Y-shaped molecule.
To determine the location of the linkage in SS-U6, we subjected purified RNA Y made with 5′-labeled Br to RNase H digestion using a DNA oligonucleotide complementary to nucleotides 1–18 of SS. If the site of linkage to Br was located in the region targeted by this oligonucleotide, incomplete base pairing of the oligonucleotide to SS and thus incomplete RNase H digestion would likely occur (see Fig. 6, bottom diagram). The result of RNase H reactions with this oligonucleotide is shown in Figure 6C. Release of a number of radiolabeled fragments moving slightly above Br (Fig. 6C, right panel, lanes 2 and 3) was detected, indicating that the site of linkage to Br is located 5′ to the area digested during the RNase H reaction. The presence of at least seven fragments resulting from incomplete RNase H digestion indicates that the site of linkage is at least seven nucleotides upstream of the 3′ boundary of RNase H digestion, that is, 7 nt upstream of nucleotide 18 in SS (see Fig. 6, bottom diagram). Not only do these results confirm that the 3′ end of SS-U6 is free and not involved in the formation of the linkage to Br, they also map the site of linkage between the two RNAs to the first 12 nt of the SS region. We attempted to further pinpoint the location of the junction between Br and SS-U6 by RT-PCR (Vogel et al. 1997). Although low yield and other technical difficulties prevented us from determining the precise location of the junction using this approach, these experiments nonetheless confirmed the existence in RNA Y of a covalent linkage between the Br and SS-U6 (data not shown). Taken together, the above data indicate that RNA Y is a Y-shaped molecule with an internal functional group in Br forming a covalent linkage to one of the first 12 nt of SS-U6 (Fig. 7). This results in loss of several nucleotides from the 5′ end of SS and formation of a branched RNA species. RNA Y thus resembles the lariat RNA that results from the first step of pre-mRNA splicing.
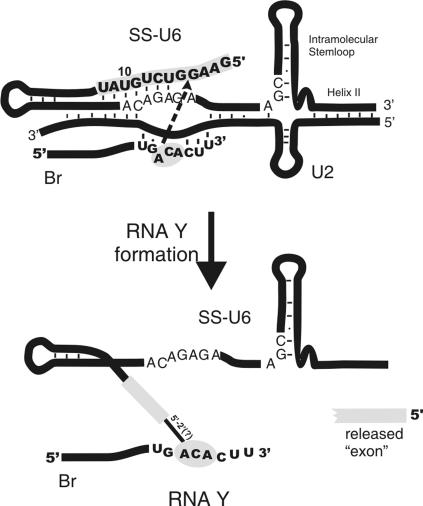
FIGURE 7.
RNA Y. Locations of Br, SS-U6, U2, U6 intramolecular stem–loop, and U6/U2 helix II are shown. Gray areas mark the regions that contain the site of linkage between Br and SS. Arrow indicates the direction of the nucleophilic reaction between putative reacting nucleotides involved in RNA Y formation.
DISCUSSION
We have investigated further the catalytic activity of spliceosomal snRNAs and have shown that they indeed have the capability to perform a reaction between two RNAs that resemble the 5′ splice site and branch site of introns. Our data indicate that, similar to authentic splicing, the reaction performed by the in vitro-assembled U6/U2 complex is dependent on the invariant domains of U6 and the presence of Mg2+ in the buffer. Importantly, the 5′ end of the SS substrate is removed, similar to the release of the 5′ exon during the first step of splicing. The removal of this exon-like fragment occurs concomitantly with the joining of Br and SS together, forming a Y-shaped RNA that resembles the lariat introns forming after the first step of splicing (Fig. 7). These results provide new evidence for the catalytic potential of spliceosomal RNAs to perform the first step of splicing and strengthen the possibility of RNA catalysis in the spliceosome.
Protein-free catalysis by spliceosomal snRNAs
The similarities between the first step of splicing and RNA Y formation are remarkable. However, an important question is whether RNA Y formation is identical to the first step of splicing in terms of chemistry; that is, whether it is a transesterification between the 2′OH group of an internal adenosine in Br and an internal phosphate in SS, leading to the formation of a 2′–5′ phosphodiester linkage. Such a scenario is consistent with all our data, and also explains the release of the exon-like RNA fragment during the reaction. While it is conceivable that reactions involving nucleobases, or a nucleobase and a 2′OH, might result in the release of an exon-like fragment and formation of a linkage between Br and SS in RNA Y, such reactions are unusual compared to the more common RNA-catalyzed reactions between 2′OH and phosphates (Shabarova and Bogdanov 1994). In addition, if the linkage between Br and SS was mediated via a nucleobase, then incubation of RNA Y in acidic pHs should have led to separation of Br and SS due to hydrolysis of the acid-labile glycosidic bond of the nucleobase(s) involved in the linkage, which would likely have been enhanced by the presence of the added covalent bond on the nucleobase, and this was not observed.
Performing a thorough analysis of the chemistry of the reaction, similar to what was originally done for the authentic splicing reaction (Padgett et al. 1984; Konarska et al. 1985), was hindered by the low efficiency of the reaction, which prevented us from obtaining sufficient amounts of pure RNA Y. Similarly, our attempt at pinpointing the site of reaction in Br by using a Br species containing a deoxy substitution at the putative branch site, A9, was not successful. This deoxy substitution did not completely abolish RNA Y formation (data not shown), which can be due to the activation of one of the neighboring residues as the reactive nucleotide when the original reactive group, 2′OH of A9, is absent. Thus, this result does not rule out the participation of 2′OH of A9 in formation of the chemical linkage in RNA Y. Further analysis of the small amount of RNA Y formed with the deoxy-containing Br was not possible due to the low yield of the reaction. Although these technical issues prevented us from determining the chemistry of RNA Y unambiguously, the reaction nonetheless bears significant similarity with the first step of splicing.
The role of invariant domains of U6 in RNA Y formation
A number of hairpin ribozymes contain an identical ACAGAGA box. It has been shown that, while the first 4 nt form canonical base pairs with the substrate close to the cleavage site, the last 2 nt are highly conserved and play an important role not only in sculpting the active site of this ribozyme but also in catalysis (Anderson et al. 1994; Burke et al. 1996; Rupert and Ferre-D'Amare 2001). Additionally, in the crystal structure of the hairpin ribozyme, the last G of this sequence is involved in hydrogen bonding to both the pro-Rp oxygen of the scissile phosphate and the 2′OH nucleophile (Rupert and Ferre-D'Amare 2001). Since the last G of ACAGAGA in spliceosomal U6 is located across the duplex from the 5′ splice site, it is tantalizing to speculate that this G (which is extremely sensitive to mutation in RNA Y formation) plays a similarly critical role in the activation of the nucleophile or stabilization of the developing negative charge on the scissile phosphate. Interestingly, the last nucleotides of this motif are also conserved in U6atac, the U6 counterpart in the minor, atac spliceosome, which has a GGAGAGA motif instead of the ACAGAGA (Patel and Steitz 2003). Similar to the case in the major spliceosome, the 5′ half of this motif is involved in base pairing to the substrate (Incorvaia and Padgett 1998; Patel and Steitz 2003), and it is conceivable that the 3′ half of this conserved domain might be playing a role in catalysis.
Positioning of the 5′ splice site substrate by U6/U2
Multiple factors contribute to proper positioning of the 5′SS in the spliceosome. Canonical and noncanonical base-pairing interactions between the ACAGAGA domain of U6 and the conserved G at position +1 and the GU dinucleotide at positions +5 and +6 of introns (Kandels-Lewis and Seraphin 1993; Lesser and Guthrie 1993; Hwang and Cohen 1996; Luukkonen and Seraphin 1998) play significant roles. Indeed it is likely that the whole intronic section of the 5′ splice site region is in close contact with the ACAGAGA domain. These interactions are further bolstered by interaction between U5 and the nucleotides at the end of the 5′ exon, and the RNA–protein interactions that help position the 5′ splice site at the catalytic active site of the spliceosome (Nilsen 1998).
The stabilizing function of U5 and spliceosomal proteins, which are of course absent in our minimal U6/U2 system, was partially compensated for by the covalent attachment of the 5′ splice site sequence to the 5′ end of U6 via a hairpin loop. This was designed to orient the 5′SS toward the ACAGAGA to facilitate the interaction between the ACAGAGA and 5′SS. To ensure further that the intronic part of the 5′SS was positioned close to the ACAGAGA, we increased the base-pairing potential between the two by making mutations in the less-conserved positions +3 and +4 of the 5′SS from AA to CU, thus creating a 5-nt base-pairing potential between ACAGAGA and 5′ SS. Interestingly, in the atac spliceosome, U6 atac forms a 5-nt base-pairing interaction with the 5′ splice site (Incorvaia and Padgett 1998; Patel and Steitz 2003). Thus, more extensive base-pairing interactions do not interfere with the positioning of the 5′ splice site for catalysis in the spliceosome.
Magnesium-dependent catalysis by U6/U2
Divalent cations play diverse catalytic and structural roles in the active site of a number of larger ribozymes that catalyze splicing reactions (Lehmann and Schmidt 2003; Stahley and Strobel 2006), and it is likely that inner-sphere Mg2+ coordination at the 5′ and 3′ splice sites plays a similar role in the catalytic strategy of the spliceosome by stabilizing the developing negative charge on the leaving oxyanion (Sontheimer et al. 1997; Gordon et al. 2000). RNA Y formation is also strictly Mg2+-dependent, and substitution studies with cobalt hexamine suggest that at least one inner-sphere–coordinated Mg2+ is required for the reaction. It is likely that Mg2+ performs the same function in RNA Y formation as in the authentic spliceosome; however, a structural requirement for an inner-sphere Mg requirement cannot be ruled out. Recent data suggest that a functionally important Mg2+ coordination by the intramolecular stem–loop of U6 might have a structural rather than a catalytic role (Yean et al. 2000; Huppler et al. 2002; Rhode et al. 2006).
RNA-based catalysis in the spliceosome
We have provided evidence that a reaction very similar if not identical to the first splicing step can be performed by protein-free spliceosomal RNAs, providing further evidence for the hypothesis that pre-mRNA splicing occurs by RNA-based catalysis. The fragments of U6 and U2 used in RNA Y formation are both structurally and functionally closely analogous to the catalytically essential domain V and domain VI of group II introns (Valadkhan and Manley 2002; Lehmann and Schmidt 2003). However, group II introns contain additional domains which help the assembly of the active site and the positioning of the substrates. The RNA and protein factors that are thought to play the same role in the spliceosome are absent in our minimal system, which provides at least a partial explanation for the low yield of the reaction. The absence of these supporting elements can result in a high degree of structural flexibility, misfolding, or incorrect substrate positioning in the in vitro-assembled U6/U2 complex and the minimal splicing substrates used in RNA Y formation, as has been observed for other ribozymes (e.g., Fedorova et al. 2003; Penedo et al. 2004). Comparison of the known structural features of the spliceosome and the minimal U6/U2 system (Madhani and Guthrie 1994; Valadkhan and Manley 2000; data not shown) indicates that misfolding of a functionally crucial part of the molecule, rather than global misfolding, is likely to be the underlying basis for the low yield of the reaction.
An important remaining question is whether these same RNAs can catalyze a reaction resembling the second step of splicing, or if there is direct participation of protein factors in second step catalysis. In vivo, almost all RNAs, including natural ribozymes, exist as ribonucleoprotein complexes. However, in the case of previously studied larger natural ribozymes, the association with proteins does not seem to change the catalytic strategy used by these RNAs; the reactions remain RNA-catalyzed even in the context of a ribonucleoprotein complex, with proteins playing supportive, and not catalytic, roles (Hsieh et al. 2004). Whether the spliceosomal RNAs similarly remain the sole catalyst in the activated spliceosome remains to be established.
MATERIALS AND METHODS
Preparation of RNA constructs
U2 and U6 snRNA fragments, the SS-U6 construct, and all mutant constructs were made by in vitro transcription of PCR-generated templates, as described (Valadkhan and Manley 2000). The U2 construct contained nucleotides 1–44 of human U2 snRNA with the following modifications: three point mutations were induced to improve base pairing to U6 (U2A, U9A, and U41G), and two G residues were added at the 5′ end to improve base pairing to U6 and transcription efficiency. A short RNA sequence containing the human 5′ splice site consensus (the less conserved nucleotides +3 and +4 were changed to CU to improve the base-pairing potential to U6) was covalently linked to the 5′ end of U6 via a UUCG hairpin (Fig. 1). SS and Br oligos were made both by chemical synthesis (Dharmacon Biotech and Invitrogen) and by in vitro transcription as described (Valadkhan and Manley 2001). Br substrates used in RNA Y formation reactions had the following sequences: CUUUACUGACACUU (wt), CUUUACUGACGCUU (Br, A11G), CUUUACUGACACUUC (Br+C15), GGCCUGGGUUUUCUUUACUGACACUU (Br+12), UACUGACACUU (Br del 1–3), and GGCCGUUCGCGGCCGGCCUGGGUUUUCUUUACUGACACUUA (Br+26). 3′ and 5′ end-labeling reactions were performed as previously described (Valadkhan and Manley 2000).
RNA Y formation reactions
The assembly and folding of the U6/U2 complex were done as described for RNA X formation reactions (Valadkhan and Manley 2003), using SS-U6 instead of U6, followed by the addition of 5′- or 3′-labeled Br (50–100 nM final concentration). Reaction mixtures were incubated at room temperature for 24 h followed by loading onto 12%–20% denaturing PAGE. Purified RNA Y was tested for resistance to denaturation as described (Valadkhan and Manley 2001). Reactions with repurified, unreacted SS-U6, U2, and Br were set up as described (Valadkhan and Manley 2003). To investigate the binding of Br to the branch-binding domain of U2, an oligo that was complementary to either Br nucleotides 3–14 or U2 nucleotides 29–38 was added to the reaction mixture at 0.5 μM final concentration. To determine the observed rate of the reaction, time points were taken every hour for 24 h, and the amount of RNA Y formed and Br precursor in each sample was quantitated using ImageQuant. The amount of RNA Y was normalized to the amount of Br in each sample, and the resulting values were fit to an exponential curve.
SS-U6 mutants were made by PCR using oligos that contained the desired mutation, followed by in vitro transcription. All RNAs were tested for quality by end labeling and RNase T1 digestion to ensure the presence of a homogeneous RNA population. Mutant RNAs were used in RNA Y formation reactions as detailed above. Each reaction was repeated at least three times, and the amount of RNA Y formed was normalized to the amount of input Br. The average of three independent experiments was graphed, with two standard deviations as error bars.
Reactions with end-labeled SS-U6
To separate RNA Y from the 500-fold excess of unreacted SS-U6 precursor, which migrates very close to the formed RNA Y on PAGE, the region of the gel that contained RNA Y was excised and the RNA contained within the gel piece was eluted and precipitated. A DNA oligo complementary to Br was annealed to the eluate, and the mixture was loaded onto PAGE containing 2 M urea. The binding of the complementary oligo to RNA Y resulted in a gel shift that separated RNA Y from the excess SS-U6 precursor. The shifted band was eluted from the gel, and the gel shift procedure was repeated to completely separate the unreacted precursor from RNA Y. Purity of the resulting RNA Y was tested by relabeling and limited RNase T1 digestion.
RNase digestion reactions
Limited RNase T1 digestion reactions on purified, end-labeled RNA Y were performed as previously described (Valadkhan and Manley 2000). For complete RNase T1 reactions, the conditions were similar to the limited reactions except that 10- to 50-fold more RNase was added and the reactions were incubated for 10 min at 50°C.
RNase H reactions
Purified RNA Y that contained a radioactive label at the 5′ end of Br was annealed to a DNA oligo that was complementary to nucleotides 1–18 of SS at 5 μM final oligonucleotide concentration. The annealing reaction was performed in 50 mM Tris, pH 7.2, and 40 mM MgCl2 by heating to 75°C and slow cooling to 30°C. RNase H reactions were performed in a buffer that contained 50 mM Tris, pH 7.5, 10 mM MgCl2, 100 mM KCl, 0.5 mM EDTA, 0.5 mM DTT, and 1 U of RNase H (USB). The reactions were incubated at 37°C for 10–30 min followed by analysis on a 16% denaturing PAGE.
ACKNOWLEDGMENTS
We thank Richard Padgett and Tim Nilsen for helpful discussions and comments on the manuscript and Beate Schwer and Jef Boeke for providing reagents. We also thank Nima Shah and Catherine Tran for technical assistance. This work was supported by an NIH grant to J.L.M., a Searle Scholar Award from the Kinship Foundation, an IRG award from the American Cancer Society, and NIH grant GM078572 to S.V.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.626207.
REFERENCES
- Anderson, P., Monforte, J., Tritz, R., Nesbitt, S., Hearst, J., Hampel, A. Mutagenesis of the hairpin ribozyme. Nucleic Acids Res. 1994;22:1096–1100. doi: 10.1093/nar/22.6.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow, D.A. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- Burke, J.M., Butcher, S.E., Sargueil, B. Structural analysis and modification of the hairpin ribozyme. In: Eckstein F., Lilley D.M.J., editors. Nucleic acids and molecular biology. Springer; Berlin, Germany: 1996. pp. 129–143. [Google Scholar]
- Collins, C.A., Guthrie, C. The question remains: Is the spliceosome a ribozyme? Nat. Struct. Biol. 2000;7:850–854. doi: 10.1038/79598. [DOI] [PubMed] [Google Scholar]
- Cowan, J.A. Metallobiochemistry of RNA. Co(NH3)6 3+ as a probe for Mg2+(aq) binding sites. J. Inorg. Biochem. 1993;49:171–175. doi: 10.1016/0162-0134(93)80002-q. [DOI] [PubMed] [Google Scholar]
- Datta, B., Weiner, A.M. The phylogenetically invariant ACAGAGA and AGC sequences of U6 small nuclear RNA are more tolerant of mutation in human cells than in Saccharomyces cerevisiae . Mol. Cell. Biol. 1993;13:5377–5382. doi: 10.1128/mcb.13.9.5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio, P., Abelson, J. Two domains of yeast U6 small nuclear RNA required for both steps of nuclear precursor messenger RNA splicing. Science. 1990;250:404–409. doi: 10.1126/science.2145630. [DOI] [PubMed] [Google Scholar]
- Fedorova, O., Mitros, T., Pyle, A.M. Domains 2 and 3 interact to form critical elements of the group II intron active site. J. Mol. Biol. 2003;330:197–209. doi: 10.1016/s0022-2836(03)00594-1. [DOI] [PubMed] [Google Scholar]
- Gordon, P.M., Sontheimer, E.J., Piccirilli, J.A. Metal ion catalysis during the exon-ligation step of nuclear pre-mRNA splicing: Extending the parallels between the spliceosome and group II introns. RNA. 2000;6:199–205. doi: 10.1017/s1355838200992069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer, M., Wurzer, G., Schweyen, R.J., Mueller, M.W. Trans-activation of group II intron splicing by nuclear U5 snRNA. Nature. 1997;386:417–420. doi: 10.1038/386417a0. [DOI] [PubMed] [Google Scholar]
- Hsieh, J., Andrews, A.J., Fierke, C.A. Roles of protein subunits in RNA-protein complexes: Lessons from ribonuclease P. Biopolymers. 2004;73:79–89. doi: 10.1002/bip.10521. [DOI] [PubMed] [Google Scholar]
- Huppler, A., Nikstad, L.J., Allmann, A.M., Brow, D.A., Butcher, S.E. Metal binding and base ionization in the U6 RNA intramolecular stem–loop structure. Nat. Struct. Biol. 2002;9:431–435. doi: 10.1038/nsb800. [DOI] [PubMed] [Google Scholar]
- Hwang, D.Y., Cohen, J.B. U1 snRNA promotes the selection of nearby 5′ splice sites by U6 snRNA in mammalian cells. Genes & Dev. 1996;10:338–350. doi: 10.1101/gad.10.3.338. [DOI] [PubMed] [Google Scholar]
- Incorvaia, R., Padgett, R.A. Base pairing with U6atac snRNA is required for 5′ splice site activation of U12-dependent introns in vivo. RNA. 1998;4:709–718. doi: 10.1017/s1355838298980207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandels-Lewis, S., Seraphin, B. Involvement of U6 snRNA in 5′ splice site selection. Science. 1993;262:2035–2039. doi: 10.1126/science.8266100. [DOI] [PubMed] [Google Scholar]
- Konarska, M.M., Grabowski, P.J., Padgett, R.A., Sharp, P.A. Characterization of the branch site in lariat RNAs produced by splicing of mRNA precursors. Nature. 1985;313:552–557. doi: 10.1038/313552a0. [DOI] [PubMed] [Google Scholar]
- Lehmann, K., Schmidt, U. Group II introns: Structure and catalytic versatility of large natural ribozymes. Crit. Rev. Biochem. Mol. Biol. 2003;38:249–303. doi: 10.1080/713609236. [DOI] [PubMed] [Google Scholar]
- Lesser, C.F., Guthrie, C. Mutations in U6 snRNA that alter splice site specificity: Implications for the active site. Science. 1993;262:1982–1988. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- Luukkonen, B.G., Seraphin, B. Genetic interaction between U6 snRNA and the first intron nucleotide in Saccharomyces cerevisiae . RNA. 1998;4:167–180. [PMC free article] [PubMed] [Google Scholar]
- Madhani, H.D., Guthrie, C. Randomization–selection analysis of snRNAs in vivo: evidence for a tertiary interaction in the spliceosome. Genes & Dev. 1994;8:1071–1086. doi: 10.1101/gad.8.9.1071. [DOI] [PubMed] [Google Scholar]
- Madhani, H.D., Bordonne, R., Guthrie, C. Multiple roles for U6 snRNA in the splicing pathway. Genes & Dev. 1990;4:2264–2277. doi: 10.1101/gad.4.12b.2264. [DOI] [PubMed] [Google Scholar]
- McPheeters, D.S. Interactions of the yeast U6 RNA with the pre-mRNA branch site. RNA. 1996;2:1110–1123. [PMC free article] [PubMed] [Google Scholar]
- Moore, M.J., Query, C.C., Sharp, P.A. Splicing of precursors to messenger RNAs by the spliceosome. In: Gesteland R.F., Atkins J.F., editors. The RNA world. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1993. pp. 303–357. [Google Scholar]
- Nilsen, T.W. RNA–RNA interactions in nuclear pre-mRNA splicing. In: Simons R., Grunberg-Manago M., editors. RNA structure and function. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1998. pp. 279–307. [Google Scholar]
- O'Keefe, R.T., Norman, C., Newman, A.J. The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell. 1996;86:679–689. doi: 10.1016/s0092-8674(00)80140-3. [DOI] [PubMed] [Google Scholar]
- Padgett, R.A., Konarska, M.M., Grabowski, P.J., Hardy, S.F., Sharp, P.A. Lariat RNA's as intermediates and products in the splicing of messenger RNA precursors. Science. 1984;225:898–903. doi: 10.1126/science.6206566. [DOI] [PubMed] [Google Scholar]
- Patel, A.A., Steitz, J.A. Splicing double: Insights from the second spliceosome. Nat. Rev. Mol. Cell Biol. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- Penedo, J.C., Wilson, T.J., Jayasena, S.D., Khvorova, A., Lilley, D.M. Folding of the natural hammerhead ribozyme is enhanced by interaction of auxiliary elements. RNA. 2004;10:880–888. doi: 10.1261/rna.5268404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber, J., Ryder, U., Lamond, A.I., Mann, M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode, B.M., Hartmuth, K., Westhof, E., Luhrmann, R. Proximity of conserved U6 and U2 snRNA elements to the 5′ splice site region in activated spliceosomes. EMBO J. 2006;25:2475–2486. doi: 10.1038/sj.emboj.7601134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupert, P.B., Ferre-D'Amare, A.R. Crystal structure of a hairpin ribozyme–inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- Segault, V., Will, C.L., Polycarpou-Schwarz, M., Mattaj, I.W., Branlant, C., Luhrmann, R. Conserved loop I of U5 small nuclear RNA is dispensable for both catalytic steps of pre-mRNA splicing in HeLa nuclear extracts. Mol. Cell. Biol. 1999;19:2782–2790. doi: 10.1128/mcb.19.4.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabarova, Z., Bogdanov, A. Advanced organic chemistry of nucleic acids. VCH; New York: 1994. [Google Scholar]
- Shukla, G.C., Padgett, R.A. A catalytically active group II intron domain 5 can function in the U12-dependent spliceosome. Mol. Cell. 2002;9:1145–1150. doi: 10.1016/s1097-2765(02)00505-1. [DOI] [PubMed] [Google Scholar]
- Sontheimer, E.J., Sun, S., Piccirilli, J.A. Metal ion catalysis during splicing of premessenger RNA. Nature. 1997;388:801–805. doi: 10.1038/42068. [DOI] [PubMed] [Google Scholar]
- Stahley, M.R., Strobel, S.A. RNA splicing: Group I intron crystal structures reveal the basis of splice site selection and metal ion catalysis. Curr. Opin. Struct. Biol. 2006;16:319–326. doi: 10.1016/j.sbi.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Sun, J.S., Manley, J.L. A novel U2–U6 snRNA structure is necessary for mammalian mRNA splicing. Genes & Dev. 1995;9:843–854. doi: 10.1101/gad.9.7.843. [DOI] [PubMed] [Google Scholar]
- Sun, J.S., Manley, J.L. The human U6 snRNA intramolecular helix: Structural constraints and lack of sequence specificity. RNA. 1997;3:514–526. [PMC free article] [PubMed] [Google Scholar]
- Valadkhan, S., Manley, J.L. A tertiary interaction detected in a human U2–U6 snRNA complex assembled in vitro resembles a genetically proven interaction in yeast. RNA. 2000;6:206–219. doi: 10.1017/s1355838200992197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadkhan, S., Manley, J.L. Splicing-related catalysis by protein-free snRNAs. Nature. 2001;413:701–707. doi: 10.1038/35099500. [DOI] [PubMed] [Google Scholar]
- Valadkhan, S., Manley, J.L. Intrinsic metal binding by a spliceosomal RNA. Nat. Struct. Biol. 2002;9:498–499. doi: 10.1038/nsb0702-498. [DOI] [PubMed] [Google Scholar]
- Valadkhan, S., Manley, J.L. Characterization of the catalytic activity of U2 and U6 snRNAs. RNA. 2003;9:892–904. doi: 10.1261/rna.5440303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani, G. Exceptionally stable nucleic acid hairpins. Annu. Rev. Biophys. Biomol. Struct. 1995;24:379–404. doi: 10.1146/annurev.bb.24.060195.002115. [DOI] [PubMed] [Google Scholar]
- Vogel, J., Hess, W.R., Börner, T. Precise branch point mapping and quantification of splicing intermediates. Nucleic Acids Res. 1997;25:2030–2031. doi: 10.1093/nar/25.10.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will, C.L., Luhrmann, R. Spliceosome structure and function. In: Gesteland R.F., et al., editors. The RNA world. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 369–400. [Google Scholar]
- Wolff, T., Menssen, R., Hammel, J., Bindereif, A. Splicing function of mammalian U6 small nuclear RNA: Conserved positions in central domain and helix I are essential during the first and second step of pre-mRNA splicing. Proc. Natl. Acad. Sci. 1994;91:903–907. doi: 10.1073/pnas.91.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yean, S.L., Wuenschell, G., Termini, J., Lin, R.J. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature. 2000;408:881–884. doi: 10.1038/35048617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., Licklider, L.J., Gygi, S.P., Reed, R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]