Abstract
Metal ions play a key role in RNA folding and activity. Elucidating the rules that govern the binding of metal ions is therefore an essential step for better understanding the RNA functions. High-resolution data are a prerequisite for a detailed structural analysis of ion binding on RNA and, in particular, the observation of monovalent cations. Here, the high-resolution crystal structures of the tridecamer duplex r(GCGUUUGAAACGC) crystallized under different conditions provides new structural insights on ion binding on GAAA/UUU sequences that exhibit both unusual structural and functional properties in RNA. The present study extends the repertory of RNA ion binding sites in showing that the two first bases of UUU triplets constitute a specific site for sodium ions. A striking asymmetric pattern of metal ion binding in the two equivalent halves of the palindromic sequence demonstrates that sequence and its environment act together to bind metal ions. A highly ionophilic half that binds six metal ions allows, for the first time, the observation of a disodium cluster in RNA. The comparison of the equivalent halves of the duplex provides experimental evidences that ion binding correlates with structural alterations and groove contraction.
Keywords: X-ray crystallography, asymmetry, electrostatic, monovalent cations
INTRODUCTION
Metal ions play an essential role in RNA folding and activity (Pyle 2002). Two modes of metal ion binding contribute to stabilize RNA secondary and tertiary structures (Draper et al. 2005). In diffuse binding, the ions interact with RNA through long-range electrostatic interactions. Site bound metal ions are, on the other hand, bound directly via RNA. The two modes of ion binding both contribute to stabilize the structure of RNA. In particular, divalent cations are essential components of RNA tertiary structure and are required for both stability and catalytic activity. RNA folds can form a specific pocket for their tight binding. Crystallographic studies have shown that magnesium ions have a marked preference for binding Hoogsteen edges of guanine (Jack et al. 1977; Scott et al. 1995; Ennifar et al. 2003). In addition, some specific sequences such as the tandem wobble G.U base pairs in RNA also constitute magnesium binding sites (Cate and Doudna 1996). On the other hand, monovalent metal ions stabilize specific RNA motifs (Draper and Misra 1998) and can support folding of catalytically active tertiary structure at high concentrations (Murray et al. 1998). Certain RNA motifs and pockets were found to specifically chelate potassium ions (Basu et al. 1998; Batey et al. 2000; Conn et al. 2002). Crystallographic studies of DNA oligonucleotides have also demonstrated that monovalent cations play an essential role in DNA structure and compaction (McFail-Isom et al. 1999). However, locating monovalent cation in nucleic acid structures requires accurate crystallographic data and great care (Tereshko et al. 2001). Due to the lack of high-resolution data, the role of RNA sequence on the binding of monovalent ions and in particular sodium ions that are more difficult to identify crystallographically is sparsely documented. In addition, how sequence and other factors act together to influence the specific binding of metal ions is still not well understood. Indeed, a comparative study of ion binding in two similar RNA sequences has noted that “subtle changes that are not immediately rationalized” affect ion binding in RNA (Ennifar et al. 2003).
On the basis of the high-resolution crystallographic structures of the native and brominated palindromic RNA tridecamer r(GCGUUUGAAACGC) bearing the GAAA tetranucleotide, we attempt to answer the following questions: (1) Are there specific sites along the sequence for the binding of monovalent cations in RNA? (2) How do sequence and electrostatic environment act together to bind ions specifically? GAAA sequences in RNA are of particular interest since they exhibit unusual functional and structural features. First, they constitute a minimal Mn2+ ribozyme when paired to the UUU triplet (Dange et al. 1990; Kazakov and Altman 1992; Kuo and Herrin 2000). In catalyzing the specific cleavage between the GpA step, they represent the smallest ribozyme observed to date and may constitute an interesting model for active prebiotic molecules. However, structural information about ion binding in this sequence and the catalytic mechanism still remains to be elucidated. GAAA sequences are also part of the polypurine tracts (PPT) of retroviruses that are resistant to degradation by retroviral RNase H (Telesnitsky and Goff 1997). Crystallographic studies have shown that in their RNA/DNA hybrid form, they adopt an unusual conformation that prevents them from being hydrolyzed by enzymes (Safiranos et al. 2001; Kopka et al. 2003). Interestingly, GAAA tetranucleotide is very rarely observed within a regular double in RNA structures available in the structural data banks (Berman et al. 2000). However, they are frequently found in GNRA tetraloops, and they also constitute a conserved part of the hammerhead ribozyme where they adopt an irregular structure containing noncanonical base pairs (Scott et al. 1995; Martick and Scott 2006).
The present study provides detailed structural information on the hydration of GAAA/UUU sequences and the localization of divalent and monovalent cations in four isomorphous structures obtained in different crystallization conditions. The high-resolution crystallographic data have allowed the unambiguous identification of sodium ions specifically bound through their inner sphere to groups lining the major groove of RNA. A marked asymmetry in ion binding is observed in the two equivalent halves of the duplex with respect to the palindromic axis. The highly ionophilic half consisting of six base pairs that binds five metal ions is reminiscent of the loop E of the 5S ribosomic RNA (Correl et al. 1997). The equivalent half only chelates one sodium ion. Comparison of their hydration in the context of the crystal packing environment shows how the sequence and the electrostatic environment influence the specific binding of the metal ions. This study also shows that GAAA tetranucleotides inserted in a regular RNA duplex present an unusual structural plasticity that correlates with metal ion binding. These results support previous structural studies that observed unusual conformations in GAAA sequences of RNA/DNA hybrids (Safiranos et al. 2001; Kopka et al. 2003) and show that the structural alterations correlate with cation binding.
RESULTS AND DISCUSSION
Overall structure
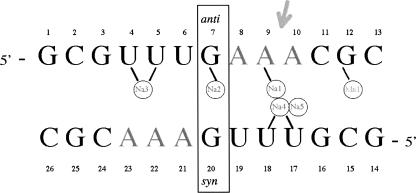
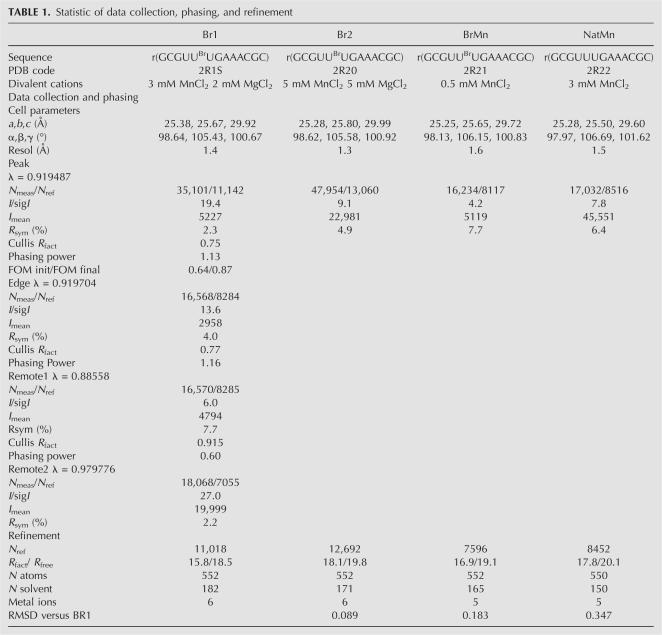
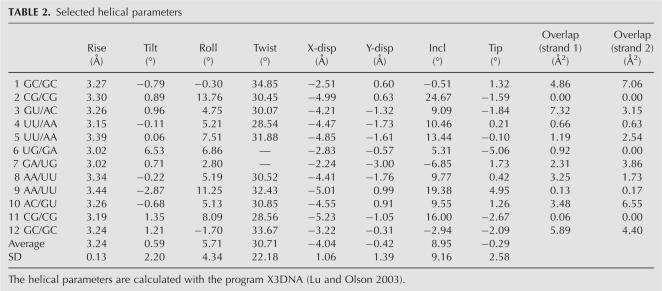
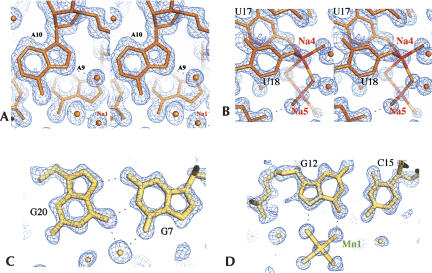
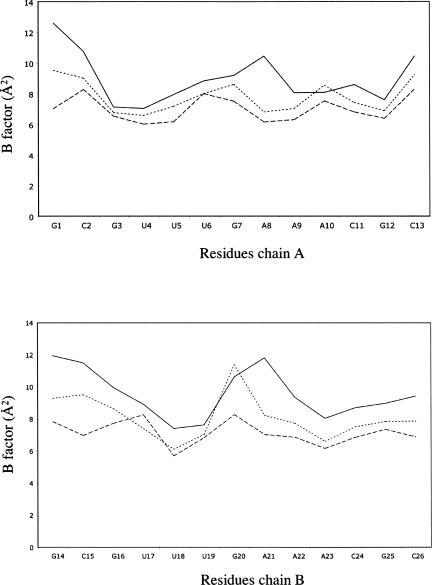
In order to analyze the influence of metal ions on RNA structure, four isomorphous structures of the tridecamer r(GCGUUUGAAACGC) (Fig. 1) were crystallized with different relative amounts of divalent cations. The Br1 and Br2 brominated duplexes grew in crystallization drops containing both magnesium and manganese ions (Table 1). In contrast, the brominated BrMn and the native NatMn structures were crystallized with manganese ion alone as a divalent cation (0.5 mM and 3 mM, respectively). In addition, a concentration of 100 mM of sodium ions (NaCl) was used for the crystallization of the four duplexes. The four duplex structures do not display any catalytic activity. The RNA tridecamers Br1, Br2, BrMn, and NatMn adopt a similar geometry. The two palindromic halves that contain the GAAA tetranucleotide surround a central well-ordered Ganti:Gsyn mispair. With an RMSD value of 0.089 Å, the two brominated structures Br1 and Br2 obtained in similar crystallization conditions are identical. The brominated BrMn and native NatMn structures crystallized at lower concentrations of divalent ions differ slightly from Br1 (Table 1, S1). However, since, the four structures exhibit almost identical structural features, Br1 has been used as a representative model for the following structural analysis. Most of the helical values are typical for an A-form helix (Table 2). However, a marked unstacking of the adenines at step A9/A10 is observed (Fig. 2A; Table 2). The two complementary uracils U17 and U18 are also unstacked (Fig. 2B). On the contrary, the equivalent base pairs at step A22/A23 are well stacked. Another particularity of this structure is illustrated by the high values of the B-factor of the phosphate groups of the two central GpA steps (Fig. 3). It is rare to observe higher values of B-factors in the backbone of a residue located in the middle of a double helix duplex. This unusual feature may therefore reflect an intrinsic flexibility of the GpA step. The GpA step corresponds to the cleavable step in the Mn2+ ribozymes (Dange et al. 1990; Kazakov and Altman 1992; Kuo and Herrin 2000). A higher B-factor in the center of the duplex may be also related to the presence of the G7:G20 Hoogsteen base pair (Fig. 2C). This study reveals that the GAAA sequence presents an important structural plasticity when inserted within a regular RNA duplex. Previous studies have shown that a similar sequence within the polypurine tract of retrovirus exhibit important structural alterations in RNA/DNA hybrid duplexes (Safiranos et al. 2001; Kopka et al. 2003).
FIGURE 1.
Sequence of the tridecamer. In the brominated Br1, Br2, and BrMn structures, the U6 and U19 uracils are replaced with 5-bromo-uracils. The ions are schematically represented within circles. The arrow indicates the destacking between A9 and A10.
TABLE 1.
Statistic of data collection, phasing, and refinement
TABLE 2.
Selected helical parameters
FIGURE 2.
(A) Stereo view of the unstacking at the A9/A10 step. Electron density map 2fo-fc at 1.4 Å of resolution (Br1) and contoured at 1.4σ. (B) Stereo view of the disodium bound at the unstacked U17/U18 step. Electron density map 2fo-fc at 1.4 Å of resolution (Br1) and contoured at 1.4σ. (C) The Hoogsteen pair G7–G20. Electron density map 2fo-fc at 1.3 Å (Br2) of resolution and contoured at 1.5σ. (D) Binding of the divalent cation at G12. Electron density map 2fo-fc at 1.3 Å (Br2) of resolution and contoured at 1.5σ.
FIGURE 3.
Plot of the B-factors of strand 1 (A) and strand 2 (B) of the Br1 structure. Average B-factors of phosphate groups (uninterrupted line), sugar (dotted line), and bases (dashed line) are represented.
The G7anti:G20syn mispair
In the center of the duplex, the G7 and G20 form a Hoogsteen-type asymmetric base pair stabilized by G7(N2)–G20(N7) and G7(N1)–G20(O6) hydrogen bonds (Fig. 2C). A water molecule bridges the two O6 carbonyl groups and stabilizes the interaction. The G20 adopts a syn conformation with a glycosidic torsion angle χ of 5.7°. A hydrogen bond between the G20 phosphate oxygen and the G20(N2) amino nitrogen also stabilizes the nucleotide conformation. The Gsyn:Ganti mispair is rare in RNA. It has been observed in the HIV Rev response element (RRE) (Hung et al. 2000; Ippolito and Steitz 2000) in which it is required for specific recognition by the protein Rev. The C1′–C1′ distance is 11.4 Å, a value close to that observed in the two RRE structures. However, in the present structure, the sugar pucker remains in C3′-endo while it adopts the C2′-endo, C4′-exo, or C1′-exo forms in the RRE structures.
Ion binding to RNA duplex
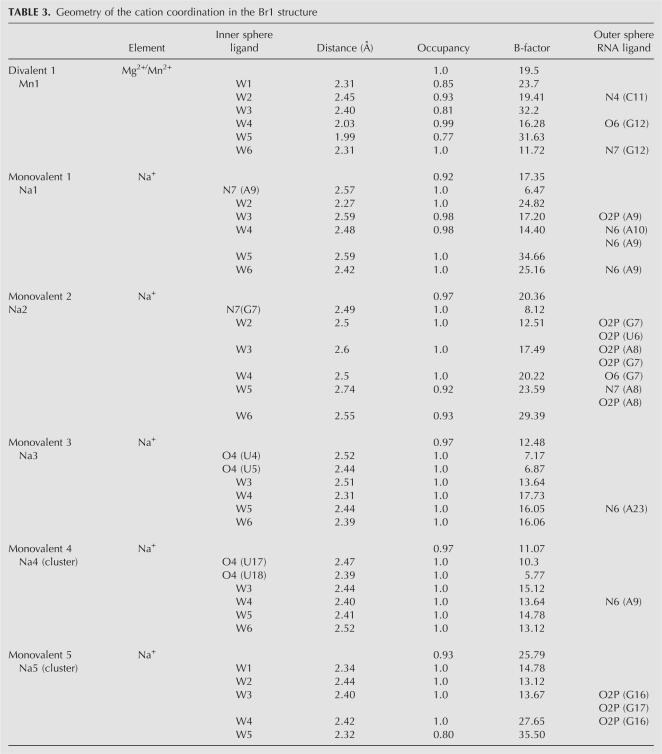
Monovalent and divalent cations have been unambiguously identified in the high-quality electron density maps (Fig. 2) of the four structures according to geometric and crystallographic criteria (Table 3).
TABLE 3.
Geometry of the cation coordination in the Br1 structure
Divalent cations
A divalent cation has been found at G12 in all four structures. The average distance of 2.2 Å between the metal ion and the water molecules of its coordination sphere indicates that it could be either a magnesium or a manganese ion. As described in Table 3 and Figure 2D, this metal ion interacts with the O6 and N7 atoms of the guanine G12 through two water molecules of its coordination sphere. Such a binding mode is common in A-form DNA or RNA duplexes (Mayer-Jung et al. 1998). In the BrMn and the NatMn structures whose crystal grew only in the presence of Mn2+, this site must correspond to a hexahydrated manganese ion.
Monovalent cations
A total of five monovalent cations assigned to sodium ions have been located in the electron density maps. These ions have been independently found in the four refined structures except Na5, which is not observed in either BrMn or NatMn. The metal ions common to the four structures (Na1–Na4) chelate the N7 atoms of purines or O4 carbonyl groups of uracils in the major groove. The fifth one (Na5), which is involved in a bimetallic cluster, binds anionic oxygen atoms through its inner coordination sphere (Fig. 2B). Two modes of direct coordination of sodium ions are observed. In the first one, sodium ions directly coordinate the N7 of purines. Na1, which is located at the unstacked step A9/A10, binds directly to N7 of A9 and indirectly to N6 of A10. Its hydration sphere also interacts with the phosphate group of A9. In a similar manner, Na2 binds directly to N7 of G7. The second binding mode involves the coordination of the O4 groups of the consecutive uracils. Figure 4, A and B, shows that sodium ions Na3 and Na4 chelate the carbonyl groups of the two uracils U4/U5 and their equivalent step U17/U18, with respect to the palindromic axis. It has been shown previously that monovalent metal ions coordinate RNA atoms and stabilize RNA motifs such as the AA platform motif of the RNA tetraloop receptor (Basu et al. 1998), noncanonical base pairs of the RNA in the signal recognition particle (Batey et al. 2000), and an RNA pocket in a ribosomal RNA fragment (Conn et al. 2002). Monovalent ions also stabilize RNA tetrad in chelating carbonyl groups of guanines and uracils (Pan et al. 2003). Analysis of ions around 23S RNA has shown that monovalent ions modeled as Na+ bind to the electronegative edge of guanosines in the context of G-U wobble base pairs (Klein et al. 2004). Interestingly, the monovalent ions described in these studies as well as the sodium ions reported in the MeRNA database (Stefan et al. 2006) display larger distances between the Na+ ions and their ligands (between 2.8 and 3.2 Å). These distances are significantly larger than the ones observed in our structure and in other high-resolution structures of nucleotides (Kennard et al. 1971). It is therefore possible that the longer distances that are observed at a lower resolution reflect the presence of hybrid binding sites with partial occupancies that combine water and sodium ions (McFail-Isom et al. 1999). By showing that the two first bases of the UUU triplet constitute a specific site for a sodium ion, our study adds a new sequence to the repertory of RNA ion binding sites. One should note that in both equivalent sites, the metal ions bind the first two bases of the triplet. It is likely that the electronegative potential generated contiguous carbonyl groups in the GUUU tetranucleotide favors ion binding. Similarly, it has been observed that tandem G-U wobble base pairs in group I intron structure constitute specific divalent binding sites due to their high electronegative potential (Cate and Doudna 1996; Keel et al. 2007).
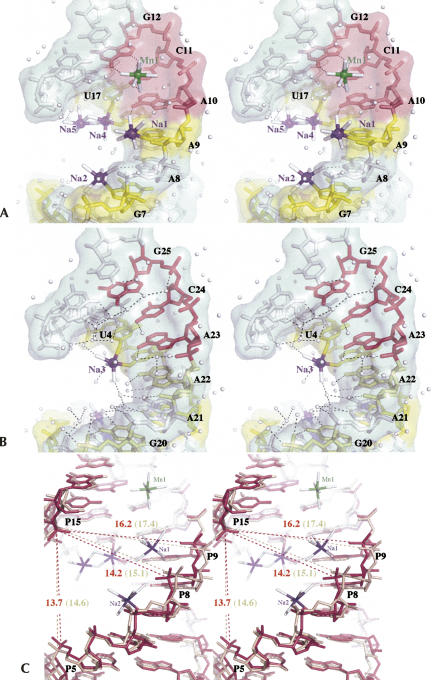
FIGURE 4.
Ion binding sites in the Br1 tridecamer. (A) Ionophilic half (G7–C13). (B) Hydrophilic half (G20–C26) (C) Superimposition of the ionophilic half (dark pink) and the hydrophilic half (gold). The distances between the phosphorus atoms are reported in ångstroms. Distances between the corresponding phosphorus atoms of the hydrophilic half are depicted in parentheses.
Disodium cluster
Our structure also provides the first example of a disodium bound to an RNA molecule. The two sodium ions Na4 and Na5 share two water molecules of their hydration sphere (Fig. 2B). Na4 coordinates the O4 carbonyl groups of U17 and U18 and Na5 interacts through its outer sphere with the anionic oxygen atoms of G16 and G17. Since the occupancies of both sodium cations are close to unity, it is likely that they simultaneously occupy this binding site. The geometry of the disodium coordination is close to that observed in the structure of ATP (Kennard et al. 1971). However, the binding environment is different. In the ATP structure, each sodium ion of the cluster is coordinated in a symmetric manner. The coordination sphere of a Na+ ion is composed of five anionic oxygen atoms and a N7 atom. The two sodium ions share two anionic oxygens and are surrounded by a more electronegative potential than in the present structure. Disodium that shares two water molecules is observed in the structure of the salt Na2H2P2O6•6H2O (Collin and Willis 1971). Disodium that shares three ligands is observed in the structure of monosodium inosine 5′-phosphate (Rao and Sundaralingam 1969) and of sodium guanylyl-3′, 5′-cytidine nonahydrate (Rosenberg et al. 1976). In contrast to the present structure, the binuclear clusters are coordinated by a symmetric environment. Thus, the present structure shows that disodium can also exist in an asymmetric environment. Interestingly, the formation and the binding of a disodium ion has been observed in a recent molecular dynamics study of a DNA dodecamer (Várnai and Zakrzewska 2004). During the simulation, two sodium ions that share two water molecules of their inner coordination spheres bind to the anionic oxygen of a phosphate group (P. Várnai, pers. comm.). Other bimetallic clusters have been observed in nucleic acids. In the Z-DNA duplex d(m5CGTAm5CG), a mixture of magnesium and sodium ions was identified in a polynuclear cluster (Wang et al. 1984). In the structure of the loop E of 5S RNA, a magnesium binuclear cluster has been observed (Correl et al. 1997).
It is probable that the electronegative potential of O6 and O4 carbonyl groups of the GUUU sequence along an RNA strand has contributed to the binding of a bimetallic cluster. The disodium that bridges the consecutive uracil bases to their phosphate groups may also be a factor to rigidify the RNA structure. This would explain why the two structures, BrMn and NatMn, that lack Na5 display higher thermal B-factors than Br1 and Br2.
Asymmetry of ion binding
Despite their equivalent sequences, the two halves of the duplex display an asymmetric pattern of metal ion binding. In the ionophilic half, G7–C13, that binds five cations, a metal ion is found every two base pairs (Fig. 4A). The base pair A9:U18 exhibits the unusual property of being able to bind three metal ions. Interestingly, this correlates with the unstacking of the adenines at the A9/A10 step (Fig. 2A). Although this region adopts the geometry of a standard A-form RNA, its ionophilic character is reminiscent of the deep groove binding sites seen in the 5S rRNA loop E motif (Correl et al. 1997; Auffinger et al. 2004). However, despite a similar sequence, the equivalent half (G20–G26) only chelates one sodium ion at the step U4/U5. This spectacular asymmetry provides the experimental evidence that factors other than the base sequence influence ion binding in RNA. This contrasts with the observation of symmetric ion binding sites within the 5S rRNA loop E motif (Auffinger et al. 2004). Two structural features seem to correlate with the ionophilic character of the half G7–C13/G14–G20 part of the tridecamer. First, the geometry of the ionophilic half is slightly more compact than the hydrophilic half. Figure 4C shows that the groove is narrower in the ionophilic half than in its symmetric counterpart. The contraction of the ionophilic groove is irregular and is indicated by the shorter distances between the phosphate groups of U5, A8, and A9 and that of C15, on the opposite strand. It is likely that the higher concentration of the negative charges around the metal ion binding sites in the ionophilic half has increased the residence time and favored the inner sphere binding of the metal ions on the RNA. However, it is difficult to determine if the helix contraction is the cause or the consequence of its ionophilic character. The second structural feature associated with the ion binding is the marked unstacking of adenines A9 and A10.
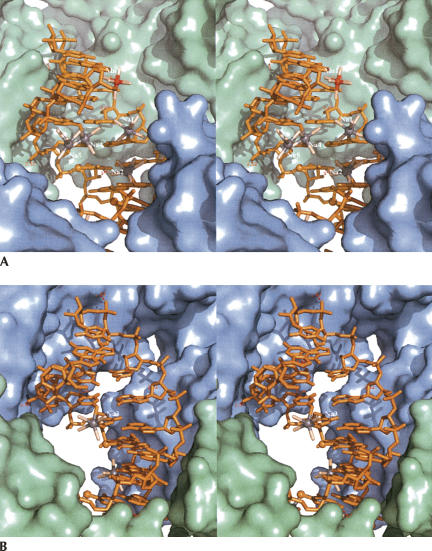
In space group P1 the two halves of the duplex are different by definition. Although the two halves are both surrounded by three equivalent RNA duplexes (S5, S6), they have a different electrostatic environment. The ionophilic one is locked between three molecules arranged in a more compact manner (Fig. 5). This analysis demonstrates that the electrostatic environment plays an important role for the binding of metal ions to RNA bases. This provides experimental support for the current view that the occupancies of metal binding sites in RNA are strongly influenced by negative electrostatic potential that is created by the folded RNA structures (Hermann and Westhof 1998; Banato et al. 2003). This study also reveals that the UUU triplet constitutes a strong binding site for monovalent cation that is less influenced by the environment.
FIGURE 5.
Crystal packing environment around the ionophilic half (green) (A) and around the hydrophilic half (blue) (B).
MATERIALS AND METHODS
The brominated r(GCGUUBr−UGAAACGC) and native r(GCGUUUGAAACGC) tridecamer were synthesized and purified by Dharmacon. Crystals were grown by the vapor diffusion method at 21°C. Hanging drops were made by mixing 2 μL of RNA solution (0.5 mM) to an equivalent volume of well solution containing 50 mM Tris (pH 7), 100 mM NaCl, various concentrations of MnCl2 or MgCl2 (Table 1) and 30% of 2-methyl-2,4-pentane-diol (MPD). The data sets were collected at B30A beamline (ESRF). Crystals of the brominated molecule were used to collect data at three wavelengths for multiple-wavelength anomalous dispersion (MAD) experiments. Triclinic crystals diffracted up to 1.3 Å resolution. Data were processed with MOSFLM and scaled with SCALA of the CCP4 package (CCP4 1994). The heavy atom search, phasing, and solvent flattening were performed with CNS (Brunger et al. 1998). Two peaks were found by anomalous Patterson map analysis in all the crystals of the brominated tridecamer. However, the first phasing attempts using the data sets collected from a single crystal (Br2) diffracting up to 1.3 Å failed due to X-ray-induced debromination during data collection (Ennifar et al. 2002). Growing larger crystals and collecting data at the different wavelengths from distinct crystals helped us to overcome this obstacle (Br1). The first experimental electron density maps obtained after solvent flattening showed that the asymmetric unit is composed of an RNA duplex. Subsequent cycles of building and refinement were performed with the program O (Jones et al. 1991) and CNS. The isomorphous crystal structures of the brominated (BrMn) and the native (NatMn) tridecamer grown only in the presence of manganese ions were refined independently. The statistics of phasing and refinement are reported in Table 1. Figures were made with the program Pymol (http://www.pymol.org). The coordinates and the structure factors are deposited in the PDB (Table 1). The helical parameters were analyzed with the program X3DNA (Lu and Olson 2003).
Supporting information
Supporting information is available at Timsit@ibpc.fr.
ACKNOWLEDGMENTS
We thank Peter Várnai for fruitful discussions and the careful reading of the manuscript. We are grateful to Philippe Dumas for helpful advice concerning brome photolysis during data collection. We thank the people of the beamline BM30A team (European Synchrotron Radiation Facility, Grenoble, France).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.730207.
NOTE ADDED IN PROOF
The influence of sodium ions on RNA structure has been recently illustrated by the paper of Casiano-Negroni et al. (2007).
REFERENCES
- Auffinger, P., Bielecki, L., Westhof, E. Symmetric K+ and Mg2+ ion-binding sites in the 5S rRNA loop E inferred from molecular dynamics simulations. J. Mol. Biol. 2004;335:555–571. doi: 10.1016/j.jmb.2003.10.057. [DOI] [PubMed] [Google Scholar]
- Banatao, D.R., Altman, R.B., Klein, T.E. Microenvironment analysis and identification of magnesium binding sites in RNA. Nucleic Acids Res. 2003;31:4450–4460. doi: 10.1093/nar/gkg471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu, S., Rambo, R.P., Strauss-Soukup, J., Cate, J.H., Ferré-D'Amaré, A.R., Strobel, S.A., Doudna, J.A. A specific monovalent metal ion integral to the AA platform of the RNA tetraloop receptor. Nat. Struct. Biol. 1998;5:986–992. doi: 10.1038/2960. [DOI] [PubMed] [Google Scholar]
- Batey, R.T., Rambo, R.P., Lucast, L., Rha, B., Doudna, J.A. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science. 2000;287:1232–1239. doi: 10.1126/science.287.5456.1232. [DOI] [PubMed] [Google Scholar]
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Casiano-Negroni, A., Sun, X., Al-Hashimi, H.M. Probing Na+-induced changes in the HIV-1 TAR conformational dynamics using NMR residual dipolar coupling; new insights into the role of the counterions and electrostatic interactions in adaptative recognition. Biochemistry. 2007;46:6525–6535. doi: 10.1021/bi700335n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate, J.H., Doudna, J. Metal-binding sites in the major groove of a large ribozyme domain. Structure. 1996;4:1221–1229. doi: 10.1016/s0969-2126(96)00129-3. [DOI] [PubMed] [Google Scholar]
- Collaborative Computing Project 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Collin, R.L., Willis, M. The crystal structure of disodium dihydrogen hypophosphate hexahydrate (Na2H2P2O6•6H2O) and disodium dihydrogen pyrophosphate hexahydrate (Na2H2P2O7•6H2O) Acta Crystallogr. B. 1971;27:291–302. [Google Scholar]
- Conn, G.L., Gittis, A.G., Lattman, E.E., Misra, V.K., Draper, D.E. A compact RNA tertiary structure contains a buried backbone-K+ complex. J. Mol. Biol. 2002;318:963–973. doi: 10.1016/S0022-2836(02)00147-X. [DOI] [PubMed] [Google Scholar]
- Correl, C.C., Freeborn, B., Moore, P.B., Steitz, T.A. Metals, motifs and recognition in the crystal structure of a 5S rRNA domain. Cell. 1997;91:705–712. doi: 10.1016/s0092-8674(00)80457-2. [DOI] [PubMed] [Google Scholar]
- Dange, V., Van Atta, R.B., Hecht, S.M. A Mn2+-dependent ribozyme. Science. 1990;248:585–588. doi: 10.1126/science.2185542. [DOI] [PubMed] [Google Scholar]
- Draper, D.E., Misra, V.K. RNA shows its metal. Nat. Struct. Biol. 1998;5:927–930. doi: 10.1038/2901. [DOI] [PubMed] [Google Scholar]
- Draper, D.E., Grilley, D., Soto, A.M. Ions and RNA folding. Annu. Rev. Biophys. Biomol. Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- Ennifar, E., Carpentier, P., Ferrer, J.-L., Walter, P., Dumas, P. X-ray induced debromination of nucleic acids at the Br K absorption edge and implication for MAD phasing. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1262–1268. doi: 10.1107/s0907444902009526. [DOI] [PubMed] [Google Scholar]
- Ennifar, E., Walter, P., Dumas, P. A crystallographic study of the binding of 13 metal ions to two related RNA duplexes. Nucleic Acids Res. 2003;10:2671–2682. doi: 10.1093/nar/gkg350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann, T., Westhof, E. Exploration of metal ion binding sites in RNA folds by Brownian-dynamics simulations. Structure. 1998;6:1303–1314. doi: 10.1016/s0969-2126(98)00130-0. [DOI] [PubMed] [Google Scholar]
- Hung, L.-W., Holbrook, E.L., Holbrook, S.R. The crystal structure of the Rev binding element of HIV-1 reveals novel base pairing and conformational variability. Proc. Natl. Acad. Sci. 2000;97:5107–5112. doi: 10.1073/pnas.090588197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippolito, J.A., Steitz, T. The structure of the HIV-1 RRE high affinity Rev binding site at 1.6 Å resolution. J. Mol. Biol. 2000;295:711–717. doi: 10.1006/jmbi.1999.3405. [DOI] [PubMed] [Google Scholar]
- Jack, A., Ladner, J.E., Rhodes, D., Brown, R.S., Klug, A. A crystallographic study of metal-binding to yeast phenylalanine transfer RNA. J. Mol. Biol. 1977;111:315–328. doi: 10.1016/s0022-2836(77)80054-5. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., Kjeldgaard, M. Improved methods for building models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kazakov, S., Altman, S. A trinucleotide can promote metal-ion-dependent specific cleavage of RNA. Proc. Natl. Acad. Sci. 1992;89:7939–7943. doi: 10.1073/pnas.89.17.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keel, A., Rambo, R., Batey, R., Kieft, J. A general strategy to solve the phase problem in RNA crystallography. Structure. 2007;15:761–772. doi: 10.1016/j.str.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennard, O., Isaacs, N.W., Motherwell, W.D.S., Coppola, J.C., Wampler, D.L., Larson, A.C., Watson, D.G. The crystal and molecular structure of adenosine triphosphate. Proc. R. Soc. Lond. A. 1971;325:401–436. [Google Scholar]
- Klein, D., Moore, P.B., Steitz, T.A. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA. 2004;10:1366–1379. doi: 10.1261/rna.7390804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka, M.L., Lavelle, L., Han, G.W., Ng, H.-L., Dickerson, R.E. An unusual sugar conformation in the structure of an RNA/DNA decamer of the polypurine tract may affect recognition by RNase H. J. Mol. Biol. 2003;334:653–665. doi: 10.1016/j.jmb.2003.09.057. [DOI] [PubMed] [Google Scholar]
- Kuo, T.C., Herrin, D.L. Quantitative studies of Mn2+-promoted specific and nonspecific cleavages of a large RNA: Mn2+-GAAA ribozymes and the evolution of small ribozymes. Nucleic Acids Res. 2000;28:4197–4206. doi: 10.1093/nar/28.21.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, X.-J., Olson, W.K. 3DNA: A software for the analysis, the rebuilding and the vizualisation of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martick, M., Scott, W.G. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer-Jung, C., Moras, D., Timsit, Y. Hydration and recognition of methylated CpG steps in DNA. EMBO J. 1998;17:2709–2718. doi: 10.1093/emboj/17.9.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFail-Isom, L., Sines, C.C., Williams, L.D. DNA structure: Cations in charge? Curr. Opin. Struct. Biol. 1999;9:298–304. doi: 10.1016/S0959-440X(99)80040-2. [DOI] [PubMed] [Google Scholar]
- Murray, J.B., Seyhan, A.A., Walter, N.G., Burke, J.M., Scott, W.G. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- Pan, B., Xiong, Y., Shi, K., Deng, J., Sundaralingam, M. Crystal structure of an RNA purine-rich tetraplex containing adenine tetrads: Implications for specific binding in RNA tetraplexes. Structure. 2003;11:815–823. doi: 10.1016/s0969-2126(03)00107-2. [DOI] [PubMed] [Google Scholar]
- Pyle, A.M. Metal ions in the structure and function of RNA. J. Biol. Inorg. Chem. 2002;7:679–690. doi: 10.1007/s00775-002-0387-6. [DOI] [PubMed] [Google Scholar]
- Rao, S.T., Sundaralingam, M. Stereochemistry of nucleic acids and their constituents. V. The crystal and molecular structure of a hydrated monosodium inosine 5′-phosphate. A commonly occurring unusual nucleotide in the anticodons of tRNA. J. Am. Chem. Soc. 1969;91:1210–1217. doi: 10.1021/ja01033a031. [DOI] [PubMed] [Google Scholar]
- Rosenberg, J.M., Seeman, N., Day, R.O., Rich, A. RNA double-helical fragments at atomic resolution: II. The crystal structure of sodium guanylyl-3′,5′-cytidine nonahydrate. J. Mol. Biol. 1976;104:145–167. doi: 10.1016/0022-2836(76)90006-1. [DOI] [PubMed] [Google Scholar]
- Safiranos, S.G., Das, K., Tantillo, C., Clark, A.D., Ding, J., Whitcomb, J.M., Boyer, P.L., Hughes, S.H., Arnold, E. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 2001;20:1449–1461. doi: 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, W.G., Finch, J.T., Klug, A. The crystal structure of an all-RNA hammerhead ribozyme: A proposed mechanism for RNA catalytic cleavage. Cell. 1995;81:991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
- Stefan, L.R., Zhang, R., Levitan, A.G., Hendrix, D., Brenner, S.E., Holbrook, S. MeRNA: A database of metal ion binding sites in RNA structures. Nucleic Acids Res. 2006;34:D131–D134. doi: 10.1093/nar/gkj058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesnitsky, A., Goff, S.P. Reverse transcriptase and the generation of retroviral DNA. In: Coffin J.M., et al., editors. Retroviruses. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. pp. 121–160. [PubMed] [Google Scholar]
- Tereshko, V., Wilds, C.J., Minasov, G., Prakash, T.P., Maier, M.A., Howard, A., Wawrzak, Z., Manoharan, M., Egli, M. Detection of alkali metal ions in DNA crystals using the state-of-the-art X-ray diffraction experiments. Nucleic Acids Res. 2001;29:1208–1215. doi: 10.1093/nar/29.5.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Várnai, P., Zakrzewska, C. DNA and its counterions: A molecular dynamics study. Nucleic Acids Res. 2004;32:4269–4280. doi: 10.1093/nar/gkh765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H.-J., Hakoshima, T., van der Marel, G., van Boom, J.H., Rich, A. AT base pairs are less stable than GC base pairs in Z-DNA: The crystal structure of d(m5CGTAm5CG) Cell. 1984;37:321–331. doi: 10.1016/0092-8674(84)90328-3. [DOI] [PubMed] [Google Scholar]