Abstract
Murine leukemia virus (MLV) specifically packages both genomic RNA (FL RNA) and a subgenomic RNA, which we call SD′. SD′ RNA results from alternative splicing of FL RNA. It is reverse-transcribed, and its DNA copy, integrated into the host genome, constitutes a splice donor-associated retroelement. FL and SD′ RNAs share a common 5′-UTR that includes the packaging/dimerization signal (Psi). To investigate whether the mechanism of copackaging of these two RNAs involves RNA heterodimerization, we examined the spontaneous dimerization capacity of the two RNAs as large synthetic RNAs transcribed in vitro. We showed that SD′ RNA not only formed homodimers with similar efficiency as the FL RNA, but that FL and SD′ RNAs also formed FL/SD′ heterodimers via Psi sequences. Comparison of the thermostabilities determined for these different dimeric species and competition experiments with Psi RNA fragments indicate the recruitment of similar dimer-linkage interactions within the Psi region. To validate these results, the dimeric state of the SD′ RNA was analyzed in MLV particles. RNA capture assays performed with the FL RNA as bait revealed that SD′, and not the host packageable U6 or 7SL RNAs, was associated with the FL RNA in virions. Heterodimerization of SD′ RNA with FL RNA may argue for the recent concept of a nuclear dimerization at or near the site of transcription and raises the new hypothesis of RNA dimerization during splicing. Furthermore, FL/SD′ heterodimerization may have leukemogenic consequences by influencing the pool of genomic dimers that will undergo recombinogenic template switching by reverse transcriptase.
Keywords: subgenomic RNA dimerization, encapsidation
INTRODUCTION
The oncogenic murine leukemia virus (MLV) is a gammaretrovirus, prototypic of simple retroviruses. It has been invaluable in our understanding of basic cellular processes such as discovery of oncogenes (Dudley 2003), eukaryotic gene expression, viral pathology (Fan 1997), and therapeutic trials with MLV-based gene transfer vectors (Cavazzana-Calvo and Fischer 2007).
All retroviruses, except the spumaretroviruses, contain two copies of their full-length positive-strand RNA genome (FL RNA) in the form of a dimer. MLV has the particularity to include an additional subgenomic 4.4-kb transcript in virions, called SD′ RNA. SD′ RNA is generated by splicing from an alternative splice donor site (SD′) within the capsid-coding region to the canonical env splice acceptor site (SA) (Fig. 1) leading to synthesis of a polyprotein that harbors Gag and integrase domains. This alternative splicing is required for optimal viral infectivity (Dejardin et al. 2000; Houzet et al. 2003). Unlike other viral spliced RNAs, such as the env mRNA, the SD′ RNA contains the entire 5′-untranslated region (UTR) of the MLV genome including the packaging signal (Psi), which confers specific packaging ability. Indeed, spliced SD′ RNA and unspliced genomic RNA are both efficiently encapsidated into progeny virions. The cellular presence of SD′ as a cDNA copy demonstrates that SD′ RNA is reverse-transcribed after viral entry, and consequently, it is a functional component of infectious MLV virions (Houzet et al. 2003). Thus, it is tempting to consider that the constitution of genomic RNA in virions includes heterodimeric FL/SD′ forms, an issue that we directly address in this study.
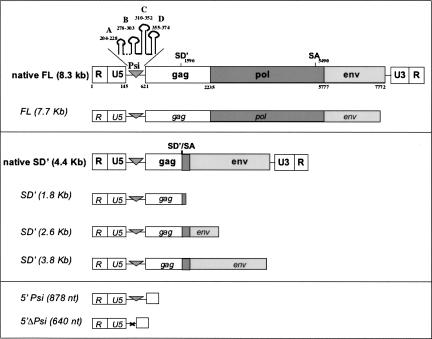
FIGURE 1.
Schematic representation of the MLV RNAs used in this study. Native FL and SD′ RNAs (in boldface) analyzed in virions and synthetic transcripts used in vitro are depicted as large and small boxes, respectively. RNA sizes and features positions are in relation to the numbering from the cap site (+1) of the MLV RNA genome. Non-coding region elements in 5′ and 3′, R U5 and U3 R, respectively, are boxed such as the gag, pol, and env coding regions. The Psi encapsidation signal encompassing stem–loops A, B, C, and D is depicted as a triangle and its deletion by a cross. Donor and acceptor splicing sites, SD′ and SA, are indicated, as well as the SD′/SA junction.
The mechanism of viral RNA dimerization has been studied mainly in vitro using small synthetic RNA fragments encompassing the 5′-UTR. Such fragments dimerize spontaneously at appropriate salt and temperature conditions even though the process is enhanced by nucleocapsid (NC), part of the Gag polyprotein (Prats et al. 1990). Dimerization has been proposed to proceed first through formation of an intermolecular “kissing complex” involving a palindromic loop, called the DIS (dimerization initiation site), of monomeric hairpins followed by conversion to a more stable intermolecular duplex (for review, see Paillart et al. 2004; D'Souza and Summers 2005). Dimerization of the MLV genome appears more complex. In vitro chemical- and enzymatic-accessibility mapping experiments involving the 5′-UTR of the genome upon dimerization indicate that several hairpins participate in the dimerization process (Tounekti et al. 1992; Mougel et al. 1993). In particular, the two pseudo-palindromic loops, DIS-1 (GGCC) and DIS-2 (AGCU), harbored by the two 5′-Psi hairpins, called A (204–228) and B (278–303) (Fig. 1), respectively, can form “kissing dimers” in vitro as well as the two closely spaced C and D hairpins, which are part of the core-encapsidation signal (Prats et al. 1990; Girard et al. 1995; De Tapia et al. 1998; Oroudjev et al. 1999; Kim and Tinoco 2000; Ly and Parslow 2002; Hibbert et al. 2004; Gherghe and Weeks 2006). Although ex vivo mutagenesis studies of the DIS-1 and the DIS-2 structures within vector RNA showed the importance of DIS for RNA packaging (Mougel et al. 1996; Mougel and Barklis 1997; Mikkelsen et al. 2000; Ly and Parslow 2002), their role is less clear in the context of full-length viruses mutated at the DIS regions. This belies the controversial nature of these results (Tchenio and Heidmann 1995; Fisher and Goff 1998; Aagaard et al. 2004). Because of the remarkable functional redundancy of the 5′-UTR and embedded DIS structures (splicing, reverse transcription, translation, dimerization, packaging, transport, and recombination), these results must be interpreted with care.
Genome dimerization is likely to be a prerequisite for genome encapsidation; however, it is still unclear at what point following RNA synthesis, or in what compartment, dimerization occurs. After synthesis, the FL RNA traffics to the translational machinery to encode Gag and Gag/Pol polyproteins, and/or it is captured by the viral Gag proteins to be encapsidated in assembling virus. Similar bifunctionality and traffic apply also to the subgenomic SD′ RNA, which colocalizes with the FL RNA throughout all early and late events of the viral life cycle, providing many opportunities for putative FL/SD′ heterodimerization.
To study the dimerization of the subgenomic SD′ RNA, we examined dimerization properties of synthetic transcripts in vitro, a convenient model system for experimental analysis. We used experimental conditions that allowed for the first time the spontaneous Psi-mediated dimerization of almost full-length SD′ and FL RNAs. Dimerization has been examined in regard to thermostability and dependence of the Psi signal. The validity of the in vitro dimerization results was further tested ex vivo by assaying for FL/SD′ heterodimers isolated from infectious MLV particles. The high frequency of FL/SD′ heterodimerization was contrasted with the capability of different small cellular RNAs, also present in virions, to associate with the FL RNA. Absence of such FL/host RNA associations reinforces the specificity of the FL/SD′ interaction.
RESULTS
Spontaneous in vitro dimerization
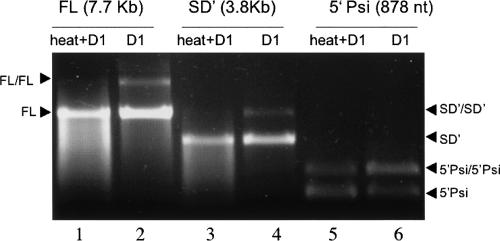
As previously published for short synthetic RNA fragments corresponding to the 5′-end of the MLV RNA, we observed spontaneous dimerization of similar-sized RNAs in vitro (Paoletti et al. 1993). Such spontaneous dimerization reached maximum efficiency when conducted at 58°C (Paoletti et al. 1993; Bonnet-Mathoniere et al. 1996; data not shown). Therefore, we used this to set our experimental conditions for spontaneous dimerization not only of a short 1–878-nt RNA including Psi (called 5′-Psi RNA) but also of much longer fragments, 1–7,706 nt, for the FL RNA (7.7 kb FL) and 1–3,812-nt fragment, encompassing the SD′/SA splice junction, for the SD′ RNA (3.8 kb SD′) (Fig. 1). The standard assay was realized at 58°C with 1 pmol of synthetic RNA (0.1 μM), a similar concentration as previously described (Paoletti et al. 1993). The reaction (R) buffer used to incubate the RNA corresponds to the buffer previously described for resuspension of dimers isolated from MLV virions (Fu and Rein 1993; Flynn and Telesnitsky 2006), and it was chosen in order to keep the same conditions between in vitro and ex vivo investigations. It is currently known that under standard spontaneous dimerization conditions with a prior denaturation step, long RNA fragments were less able to adopt the tertiary structures required for stable dimer formation, and thus, they may be restricted in their ability to dimerize post-synthesis (Prats et al. 1990; Bonnet-Mathoniere et al. 1996; Flynn and Telesnitsky 2006). Indeed, after unfolding by denaturation for 2 min at 90°C, chilling on ice, and refolding by incubation for 10 min at 58°C in R buffer (“heated+D1” conditions), the large 7.7-kb FL and 3.8-kb SD′ RNAs appear as monomeric forms without detectable dimer species (Fig. 2, lanes 1,3). In contrast and as expected for short in vitro transcript, reduced mobility RNA dimer was readily detectable with the 878-nt 5′-Psi fragments under similar heated+D1 conditions (Fig. 2, lane 5). However, because it was recently reported that MLV RNA dimerization can occur more efficiently for longer RNA during in vitro transcription than post-synthesis (Flynn and Telesnitsky 2006), RNA dimerization was analyzed without prior denaturation (“D1” conditions) (Fig. 2, lanes 2,4,6). Under D1 conditions, the two longer 7.7-kb FL and 3.8-kb SD′ RNAs clearly showed dimeric species, detectable by ethidium bromide staining, with 17.5% ± 2.4% and 14% ± 1.3% of dimer for the FL and the SD′ RNAs, respectively. Also, the D1 conditions generated less RNA degradation than the heated+D1 conditions. In contrast, the 5′-Psi RNA dimerization was less sensitive to the absence of denaturation, with an average of 54.4% ± 9.3% of dimers formed under the D1 conditions. These results correlated with previous studies (Roy et al. 1990; Flynn and Telesnitsky 2006) and showed that RNA fragments as long as 7.7 kb can dimerize spontaneously upon in vitro transcription. In addition, these experiments, for the first time, showed that the spliced SD′ RNA is capable of dimerization in vitro.
FIGURE 2.
Conditions of spontaneous dimerization in vitro of FL (7.7 kb), SD′ (3.8 kb), and 5′-Psi (878 nt) RNAs. Comparison between the ability to dimerize with (heat+D1) or without (D1) unfolding step. All experiments were performed with 1 pmol of RNA subjected to dimerization conditions as described in Materials and Methods, analyzed by native agarose gel electrophoresis, and visualized by ethidium bromide staining.
Specificity of the dimer linkage
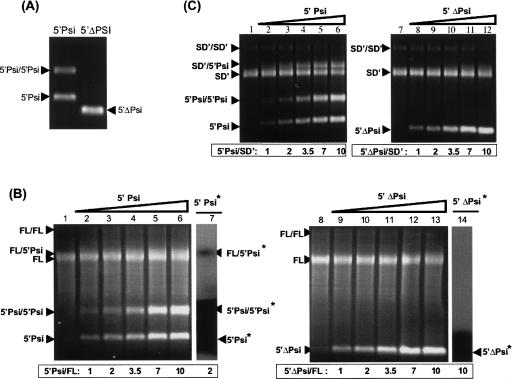
The sequences involved in the genomic RNA dimerization map within the Psi encapsidation element (Prats et al. 1990). In order to determine the specificity of dimer linkage sequences involved in our RNA dimerization assays, we synthesized a 5′-Psi RNA transcript lacking the 168–405-nt segment encompassing the whole Psi signal. We named this the 5′-ΔPsi RNA (Fig. 1). In contrast to the wild-type 5′-Psi RNA, the 5′-ΔPsi RNA was not able to dimerize, indicating that the 5′-Psi/5′-Psi dimer formed under the D1 conditions required the Psi signal (Fig. 3A). These results correlate with previous studies showing that deletion of Psi region impairs spontaneous dimerization of short MLV RNA transcripts (Roy et al. 1990; Girard et al. 1995; Girard et al. 1996; De Tapia et al. 1998; Oroudjev et al. 1999). In order to monitor the direct involvement of the Psi signal within the FL/FL and SD′/SD′ dimers, competition experiments were conducted with either the wild-type (wt) 5′-Psi or the mutant 5′-ΔPsi RNAs. A fixed amount of FL or SD′ RNAs (0.5 pmol) was incubated with increasing amounts (0–5 pmol) of either wt or Psi-deleted 5′-Psi RNA under D1 conditions, and products were analyzed on agarose gels (Fig. 3B,C, respectively). In order to circumvent the problem of gel resolution between FL monomer and FL/5′-ΔPsi dimer bands, experiments were also conducted with radiolabeled 5′-Psi RNA (wt or ΔPsi) (Fig. 3B, lanes 7,14). Fewer FL/FL dimers were detected here compared to the previous experiment (Fig. 2) due to the lower amount of FL RNA (0.5 pmol) used and the longer gel migration. When mixed in equimolecular amounts, FL and 5′-Psi RNAs form FL/FL and 5′-Psi/5′-Psi dimers, respectively. The increasing addition of the 5′-Psi RNA led to FL/5′-Psi dimer formation (Fig. 3B, lanes 3–7). Indeed, adding excess of the wt 5′-Psi transcript, and not the mutant 5′-ΔPsi RNA (lane 14), led to production of a FL/5′-Psi dimer (lane 7). Similar results were obtained with competition experiments conducted in parallel with the SD′ and the 5′-Psi RNAs that showed SD′/5′-Psi RNA dimers (Fig. 3C, lanes 1–6). In contrast, competition with the 5′-ΔPsi RNA did not show 5′-ΔPsi/5′-ΔPsi and SD′/5′-ΔPsi dimers (lanes 7–12). Note that a faint broadening of FL (Fig. 3B, lanes 5–6) and SD′ (Fig. 3C, lanes 11–12) monomers appeared when a high excess of 5′-ΔPsi fragment was added. Such observation likely corresponds to heterodimers with the 5′-ΔPsi, illustrating weak interactions along the 5′-ΔPsi fragment. Altogether, these results indicated that Psi sequences were specifically required for efficient dimerization during transcription of long transcripts such as FL (7.7 kb) and SD′ (3.8 kb) RNAs.
FIGURE 3.
Influence of the Psi sequences on dimerization. (A) Dimerization ability of 5′-Psi (878 nt) and 5′-ΔPsi (640 nt) RNAs under standard D1 conditions. (B) FL RNA specifically dimerizes through the Psi sequences. 0.5 pmol of FL RNA (7.7 kb) was incubated alone (lanes 1,8) or with increasing amounts (0.5–5 pmol) of 5′-Psi RNA (lanes 2–7) or 5′-ΔPsi RNA (lanes 9–14) under D1 conditions as described in Materials and Methods. Autoradiographies obtained from similar experiments conducted with radiolabeled wt or mutant Psi RNAs are given (lanes 7 and 14, respectively). (C) SD′ RNA specifically dimerizes through the Psi sequence. The same experiments as in B were achieved with SD′ RNA (3.8 kb). Heterodimerizations were performed with either the 5′-Psi RNA (lanes 1–6) or 5′-ΔPsi RNA (lanes 7–12).
In vitro heterodimerization between the FL and the SD′ RNAs via Psi
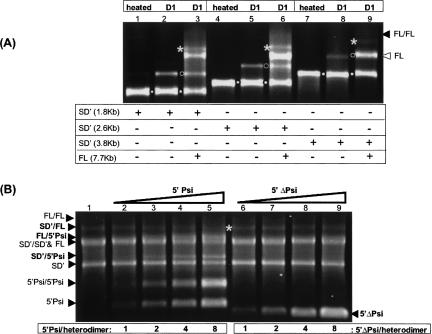
Recently we showed that SD′ RNA acts as a defective retroelement that required the FL genomic RNA for dissemination (Houzet et al. 2003). Therefore, the SD′ RNA should be encapsidated in the presence of the FL RNA to form infectious MLV particles. To address the hypothesis that SD′ and FL RNAs may form heterodimers leading to their copackaging, we examined the heterodimerization between the 7.7-kb FL RNA and various lengths of the SD′ transcripts (1,820; 2,580; and 3,812 nt) (Fig. 1). In these assays, SD′ and FL RNAs were cotranscribed by including the templates for SD′ and 7.7-kb FL RNAs in a single transcription reaction mixture. After synthesis, the RNA mix was incubated under D1 conditions and separated on a native agarose gel. A representative gel is illustrated in Figure 4A. Under these conditions, all SD′ RNAs homodimerized with efficiencies that were inversely proportional with their RNA lengths (lanes 2,5,8). As seen in lanes 3, 6, and 9, when the SD′ RNA was mixed with the FL RNA, a distinct RNA species was visible that migrated between the FL monomer and FL/FL homodimers and was presumed to be SD′/FL heterodimers. In addition, a portion of the cotranscribed RNA was monomeric, and the remaining RNA appeared to homodimerize. The 1.8-, 2.6-, and 3.8-kb SD′ RNAs heterodimerized to roughly similar extents (16% ± 4%). These results indicated that FL and SD′ RNAs are able to form heterodimers spontaneously. Comparison between the different lengths of the SD′ RNA indicated that sequences downstream from the position 1800, corresponding to the integrase coding sequence, are not required for the SD′ RNA dimerization process. To further analyze the FL/SD′ dimer-linkage, competition experiments were conducted as above (Fig. 3) with the wt or mutant 5′-Psi RNAs. As shown in Figure 4B, increasing addition of 5′-Psi RNA lead to decreasing FL/SD′ heterodimerization to the benefit of both SD′/5′-Psi and FL/5′-Psi heterodimer formation. In contrast, addition of 5′-ΔPsi RNA did not change the SD′/FL heterodimerization, indicating that FL and SD′ RNAs specifically formed heterodimers through the Psi sequences.
FIGURE 4.
Analysis of heterodimerization between FL and SD′ RNAs. (A) FL and SD′ RNAs were cotranscribed by mixing the SD′ template cut either with BamHI, BstEII, or ClaI to, respectively, yield 1.8-kb, 2.6-kb, and 3.8-kb SD′ RNA (lanes 3,6,9, respectively) with the FL plasmid template. The cotranscribed RNA mixes were submitted to D1 conditions. Respective FL/SD′ heterodimers are indicated by a star and FL monomer and homodimer by open and black arrowheads, respectively. In parallel, each SD′ RNA was separately submitted to D1 dimerization conditions to form (open circles) SD′/SD′ homodimers (lanes 2,5,8). (Black circles) The different SD′ monomers were obtained by denaturation for 10 min at 85°C (lanes 1,4,7). (B) FL and SD′ RNAs form heterodimers through the Psi sequences. The same amount of FL/SD′ heterodimer (indicated by a star), previously formed by mixing SD′ and FL DNA templates, was incubated with increasing amounts of 5′-Psi (lanes 2–5) or 5′-ΔPsi (lanes 6–9) RNAs for competition experiments.
Thermal stability of the dimers
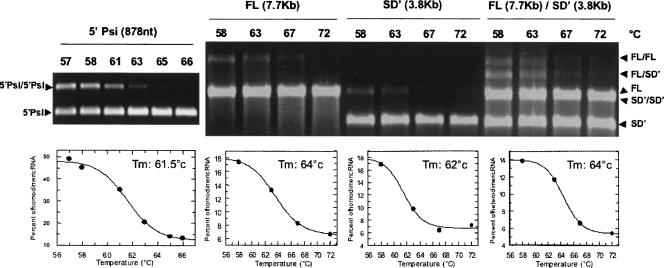
To further analyze the linkage between the SD′ RNA and the genomic RNA, we examined and compared the thermostability of the FL/FL, 5′-Psi/5′-Psi, and SD′/SD′ homodimers with the FL/SD′ heterodimer. Dimers were formed as above and submitted to various temperatures for 10 min. Typical gel electrophoresis results corresponding to a thermal denaturation experiment are shown in Figure 5 together with the denaturation curve corresponding to the quantification of the amount of dimeric RNA. The melting temperatures estimated from these experiments are ∼63°C for all homodimers. However, it cannot be excluded that the modest increased stability, if significant, of the FL RNA compared to the two shorter RNAs might illustrate additional contacts within the FL/FL dimers.
FIGURE 5.
Thermal stability of the dimers. A representative experiment is shown with the corresponding T m plots. Temperature-induced dissociation of RNA dimers were done as described in Materials and Methods with an RNA concentration of 0.1 μM. The percentage of dimer was estimated for each RNA species at different temperatures, and curves were fitted to calculate the melting temperature, T m. Results of at least three independent experiments gave the following T m values: 61.3° ± 1.5°C for 5′/5′; 64.5° ± 0.5°C for FL/FL; 62.0° ± 0.4°C for SD′/SD′, and 64.3° ± 0.8°C for FL/SD′.
These experiments revealed that the FL/SD′ heterodimer displays similar melting temperature (64.3° ± 0.8°C) as the homodimers and definitively not a lower stability. These results correlate with the thermal stabilities previously determined by electron microscopy for genomic RNA (8332 nt) dimers purified from wild-type Moloney or Friend MLV virions (T m: 60°C) (Bender et al. 1978). Also, the T m we estimated here was only 1.1-fold higher than the T m reported for mature dimers extracted from Moloney MLV virions (Fu and Rein 1993; Fu et al. 2006).
Capture of FL/SD′ heterodimer in MLV virions
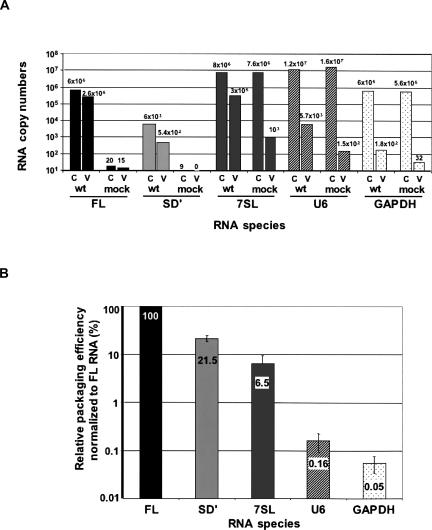
To address the issue of heterodimerization in virus particles, we assayed the presence of FL/SD′ heterodimers in virions released from 293T cells producing replication-competent MLV. Previous studies by Northern blot analysis showed that SD′ RNA was faintly detectable in MLV-infected cells (Dejardin et al. 2000). Therefore, we used RT-QPCR to monitor the SD′ and FL abundance in cells and virions as described previously (Houzet et al. 2003). The copy numbers of SD′ and FL RNAs in assays and mock controls were measured in 50 ng of cellular RNA samples (C) and in 1/1600 of the total released virions (V); a representative experiment is shown in Figure 6A. To determine encapsidation efficiencies, copy numbers measured in cells and in virions were reported to the total input of cellular and virions RNA samples, and virus-to-cell RNA ratios [(V/C) × 100] were calculated and normalized to the FL level (Fig. 6B). As previously reported with the murine Dunni cell system (Houzet et al. 2003), the SD′ RNA was specifically incorporated, although 4.6-fold less than the FL RNA, in virions released from human 293T cells transfected with MLV.
FIGURE 6.
Ex vivo analysis of natural RNAs. (A) RNA levels in cell and virion samples of a representative experiment. Host and viral RNAs were quantitated by RT-QPCR in both (C) transfected cells and (V) virus. Values correspond to copy numbers measured in 50 ng of cellular RNA samples and in 1/1600 of total released virions. (B) Relative efficiencies of RNA encapsidation in MLV particles. Encapsidation efficiencies were determined [(V/C) × 100] and normalized to the FL level. Results represent the mean±standard deviations of at least three independent experiments.
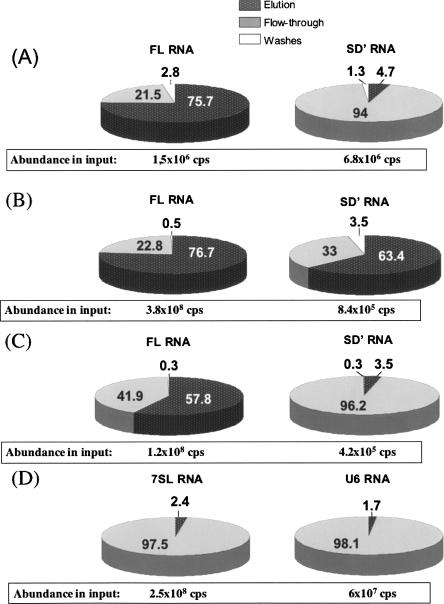
Next, to determine the state of the encapsidated SD′ RNA, the presence of FL/SD′ heterodimers was investigated by an RNA capture assay (RCA). The RCA was developed by Telesnitsky and coworkers to precisely capture an RNA able to form heterodimers with the MLV FL RNA (Flynn et al. 2004). We used the same 3′-biotinylated oligonucleotide, complementary to a sequence within MLV pol as previously used by Flynn et al. (2004). For these experiments, all previously described steps were followed scrupulously (Flynn et al. 2004), except that the initial RNase protection assay used to monitor RNA was substituted by more sensitive and quantitative RT-QPCR analysis. FL and SD′ RNAs were quantitated for inputs, the flowthroughs, the elutions, and wash samples in order to monitor the proportion of RNA retained on the beads. To control that the anti-pol oligonucleotide allows specific capture of FL RNA and was not specific to the SD′ RNA, RCA experiments were realized with FL and SD′ synthetic transcripts either alone (data not shown) or mixed (Fig. 7A). The FL and SD′ RNAs, transcribed in vitro separately, were mixed and kept at 4°C prior to the RCA procedure to prevent their dimerization. The SD′ synthetic transcript, alone or mixed with the FL transcript, was not retained in the elution fraction (∼5%), whereas a large majority of the FL transcript (76%) was captured (Fig. 7A). Then, the FL/SD′ heterodimers were sought in virions, and RCA experiments were conducted with RNA samples extracted from released virions (Fig. 7B). As expected, the FL RNA was mainly present in the elution sample (77% ± 17%). Indeed, it has been generally found that released wild-type retroviral particles contain genomic RNA exclusively in dimeric form (Housset et al. 1993; Buxton et al. 2005; Fu et al. 2006; for review, see Paillart et al. 2004). The significant level of the unretained FL RNA fraction (23%) probably indicated the limit of the system with RNAs that did not bind to the biotinylated oligonucleotide. Like the genomic RNA, a large part of the SD′ RNA (63% ± 18%) was found in the elution. Since the SD′ RNA could not directly anneal to the biotinylated oligonucleotide and bind the streptavidin beads (Fig. 7A), eluted SD′ RNA must directly represent the proportion of SD′ RNA linked to the FL RNA, presumably as FL/SD′ heterodimers. In the case of SD′, the remaining proportion (33%) of unretained SD′ molecules, measured in the flowthrough and the washes, could correspond to the limit of the system, as seen for the FL RNA capture, or to SD′ homodimers.
FIGURE 7.
RNA capture assays. Abundances of SD′ and FL RNAs were quantified by RT-QPCR, and RNA copy numbers (cps) in input were given in boxes. To monitor the specificity of the capture, repartitions of FL and SD′ RNAs were determined in the different fractions collected during the RCA procedure (see Materials and Methods). (A) Specificity of the biotinylated oligonucleotide allowing the FL RNA capture. Control RNA capture experiments were conducted with the synthetic FL (7.7 kb) and SD′ (3.8 kb) RNAs transcribed separately in vitro, mixed, and kept at 4°C prior to the RCA to prevent dimerization. (B) Heterodimerization of SD′ RNA in MLV particles by the RNA capture assay. The RNA copy numbers measured in the input correspond to 30 mL of virus-containing supernatant. Average frequencies of SD′/FL and FL/FL associations were 63.4% ± 18% and 77.0% ± 17%, respectively, and were obtained from triplicate experiments. (C) The SD′ capture depends on specific FL RNA association in virions. For this control, an additional step was added before the standard RCA procedure used in B. RNA samples were heated for 10 min at 70°C and chilled on ice prior to annealing to the biotinylated oligonucleotide. (D) Heterodimerization of the host RNAs. The 7SL and U6 RNAs were quantified in the same input (number of copies in boxes) and RCA fractions as in C for viral RNA analysis. Triplicate experiments gave an average proportion of unretained RNA of 97.5% ± 0.9% and 98% ± 0.7% for 7SL and U6 RNAs, respectively.
To demonstrate that the SD′ RNA is captured through its FL RNA association, control RCA experiments were conducted with native viral RNA extracted from virions, and heat-denaturated before annealing to the biotinylated oligonucleotide, in order to dissociate the RNA dimers (Fig. 7C). These results showed that the SD′ viral RNA was not captured (3.5%), whereas the FL genomic RNA was still retained (58%), presumably as monomers. Indeed, the high level of unbound FL genomic RNA in the flowthrough fraction (42%) likely corresponds to heat-denaturated FL/FL dimers (Fig. 7C).
Altogether, these results indicate that SD′ capture depends on specific FL RNA association and suggest that encapsidated SD′ RNA is more frequently in a FL/SD′ heterodimeric state than in a SD′/SD′ homodimeric form in MLV particles.
Specificity of the heterodimerization in the virions
Retroviruses also package significant amounts of cellular RNAs, but the encapsidation process of these RNAs remains unknown. We and others have reported specific packaging of the 7SL RNA and U6 snRNA in retroviral particles (Giles et al. 2004; Onafuwa-Nuga et al. 2005, 2006; Houzet et al. 2007). Thus, we examined encapsidation of these cellular RNAs as above for the SD′ RNA (Fig. 6). As a marker of random packaging (Onafuwa-Nuga et al. 2005), we also analyzed incorporation of a housekeeping GAPDH mRNA. Similarly, we measured copy numbers of these three host RNAs in both transfected or mock-transfected cells and in the corresponding supernatants (Fig. 6A). The U6 and 7SL RNA amounts in the supernatant of transfected cells were, respectively, 36- and 300-fold higher than those measured in mock, whereas GAPDH displays a 5.5-fold increased level in MLV particles compared to mock. To assess the significance of the presence of those host RNAs in virions, the percentages of host RNA leakage were calculated as [(total cps in V)/(total cps in C)] × 100 of mock samples, where cps means copy numbers. Leakage was <0.002% for the three host RNAs and could have resulted from association with pelleted exosomes as already reported (Valadi et al. 2007; Zeilfelder et al. 2007). Leakages were substracted from assays, and encapsidation efficiencies of host RNAs were determined as above for viral RNAs (Fig. 6B). These results show that all host RNAs were not encapsidated with the same efficiency. The 7SL RNA (6.5%) and to a lesser extent the U6 snRNA (0.16%) appear specifically enriched in MLV particles released by MLV-producing transfected 293T cells.
We examined whether encapsidation of the 7SL and U6 host RNAs might also result from association with the FL RNA, as determined above for the SD′ RNA (Fig. 7B). We searched for the presence of heterodimers formed between FL and these cellular RNAs by RCA. Then, the host 7SL and the U6 RNAs were quantified by specific RT-QPCR in parallel to FL and SD′ RNA within all RNA samples collected throughout the RCA procedure (input, flowthrough, washes, and elution) as in Figure 7B. The results, reported in Figure 7D, showed that 7SL and U6 RNAs were found mainly in the flowthrough samples (97.5% ± 0.9%, 98% ± 0.7%, respectively). These results not only confirm the specificity of the RCA approach but also reveal that these two host RNAs were not captured by the FL RNA. Consequently, neither 7SL nor U6 RNAs appeared to be significantly associated with FL RNA in virions, at least not stably enough to be detected under these RCA conditions.
DISCUSSION
In vitro spontaneous dimerization of the SD′ RNA was examined, and we showed that the SD′ RNA forms stable dimers, implying Psi-directed dimerization. The results described herein not only indicate that synthetic large fragments like the 3.8-kb SD′ RNA and the 7.7-kb FL RNA dimerize spontaneously during the in vitro transcription, but also that FL and SD′ RNAs can also form FL/SD′ heterodimers as stable as genomic FL/FL homodimers. The similar thermostability of these homo- and heterodimers suggests that the nature of the interactions within the FL/SD′ heterodimer is comparable to that of the genomic FL/FL homodimer and that they likely occur through similar dimer linkage sequences as characterized for the FL/FL dimerization.
Since the SD′ RNA is selectively packaged in wild-type MLV particles (Houzet et al. 2003; this study), we tested whether the mechanism of SD′ RNA encapsidation involves SD′ RNA dimerization. Ex vivo investigations of SD′ RNA dimerization were performed to analyze the state of SD′ RNA in MLV particles. By using the RCA approach, a tool designated to examine heterodimerization between two distinct RNAs, we showed that SD′ RNA was mainly associated with the genomic RNA in virions as heterodimers (FL/SD′), since only a minor portion of the total virion SD′ RNA content appeared unlinked to the FL RNA. The presence of other multimeric forms (e.g., FL/FL/SD′ or FL/SD′/SD′), however, cannot be excluded. This propensity to associate with the genomic RNA indicates a mechanism for SD′ RNA encapsidation directly dependent on genomic RNA packaging, and it favors the concept that dimerization governs viral RNA packaging (Sakuragi et al. 2002). Furthermore, we examined whether this may be extended to the cellular RNAs found to be enriched in retroviral particles.
The presence and abundance of cellular RNAs in retroviral particles vary greatly from one RNA species to another as evident from the specific packaging of 7SL RNA and U6 snRNA and the random packaging of GAPDH mRNA into MLV virions. Their role in viral assembly and the mechanism of their selective incorporation into particles are poorly understood. To provide clues regarding this mechanism, we examined the possibility that the 7SL RNA and the U6 snRNA associate with the genomic FL RNA in MLV particles. RNA capture assays showed that the majority (98%) of these RNAs were not physically linked with the genomic RNA in virions. These findings correlate with previous work reporting a weak association, detectable as a faint signal by Northern blot experiments, between 7SL and genomic RNA in MLV virions (Rulli et al. 2007). The absence of stable associations between host RNAs and genomic RNA in virions does not support the hypothesis that prior association with genomic RNA is essential for their selection and packaging, unlike SD′ RNA. These results also correlate with previous studies demonstrating that encapsidation of host RNAs is mediated by a mechanism independent of the viral genomic RNA (Onafuwa-Nuga et al. 2005; Houzet et al. 2007; Rulli et al. 2007). Attempts at identifying common determinants between cellular and viral encapsidated RNAs by sequence alignments showed no significant homologies or complementary between the host RNAs and the MLV genome. They do not have any obvious sequences in common that may mediate their recruitment (Onafuwa-Nuga et al. 2005; Rulli et al. 2007). Altogether these results enforce the specificity of the FL/SD′ dimerization and reveal the SD′ RNA as a specific partner of the FL RNA in MLV particles. By definition, dimerization involves colocalization of the two RNAs at some step during the packaging process. Recent studies report evidence for preferential copackaging of MLV genomic RNAs transcribed at the same chromosomal locus (Kharytonchyk et al. 2005; Flynn and Telesnitsky 2006). A direct linkage may exist between the degree of RNA copackaging and nuclear RNA colocalization, suggesting that dimer-partner selection occurs in the nucleus in the vicinity of the transcription site (Rasmussen and Pedersen 2006). Since transcription and splicing are spatially and functionally two linked steps in the RNA life (Szentirmay and Sawadogo 2000), the SD′ production by splicing of the genomic RNA also provides the close proximity required to allow the FL/SD′ heterodimerization. Importantly, the SD′ RNA is also generated by direct transcription of its SD′ DNA copy. Indeed, as a retroelement, SD′ cDNA integrates into the host genome by the viral integrase (Houzet et al. 2003). Both, FL and SD′ cDNAs may enter the nucleus through a common preintegration complex, yielding tandem integration. Consequently, the spatial position of their proviral transcription sites may favor the heterodimerization observed between the two RNAs. Experiments designed to discriminate between these two possibilities for SD′/FL heterodimerization, while not trivial, are in progress.
Heterodimerization between the genomic and the SD′ RNAs could influence recombination by template switching during viral reverse transcription and may have consequences in MLV pathogenesis (Muriaux and Rein 2003; Moore et al. 2007). Indeed, the propensity of the SD′ RNA to form heterodimers with the genome might restrict the association of genomic RNA with other defective endogenous retroviruses or virus-like elements. Such recombination may generate recombinant transforming viruses like the Harvey sarcoma virus carrying the rasH oncogene and derived from recombinations with both VL30 retrotransposon and Mo-MLV sequences (Makris et al. 1993; Torrent et al. 1994) or like the mink cell focus-forming viruses (MCFs) associated with the T-lymphomagenesis (for review, see Fan 1997). Previously, we showed that a mutant Mo-MLV, lacking the SD′ RNA, had the propensity to induce lymphoid disease with MCFs production (Ramirez et al. 2004). This earlier study can be seen as evidence that in the absence of SD′ RNA, more recombinant MCF viruses were generated. This could explain the remarkable extension of leukemic properties harbored by this mutant Mo-MLV lacking the SD′ RNA (Audit et al. 1999; Ramirez et al. 2004). Interestingly, Sorensen et al. (2007) have recently reported the broadened oncogenic potential of a B-lymphomagenic Akv MLV with an impaired SD′ splice site.
MATERIALS AND METHODS
Plasmid constructions
Plasmids were constructed by using common cloning techniques (Sambrook and Russell 2001) and propagated in the E. coli STBL2 strain at 30°C to prevent any recombination. The plasmid pIS1 (Roy et al. 1990) allows T7 transcription starting at position +1 of the Mo-MLV genome without any additional sequence derived from the plasmid. To produce FL (+1 to +7706) and SD′ (+1 to +3812) RNAs, p57(2LTR) and p57cDNASD′ constructs (Houzet et al. 2003) corresponding to the entire molecular clone of Friend-MLV and Friend-SD′ retroelement, respectively, were cloned into the PIS1 construct to produce pT7F57FL and pT7F57-SD′ plasmids, respectively. To obtain these constructs, KpnI fragments of either the p57(2LTR) corresponding to position 30–8327 or the pT7F57-SD′ (position 30–4433) were introduced into pIS1 opened by KpnI. Deletion of the Psi sequences (positions 168–405) was made by standard PCR mutagenesis approach (details on request) to produce the pT7F57–5′ΔPsi construct. All constructs used in this study as template for in vitro transcription harbored a unique substitution (A to T) at position +17.
RNA synthesis
Both the pT7F57-FL and pT7F57-SD′ plasmids were cut by ClaI prior to transcription to yield FL (7.7 kb) and SD′ (3.8 kb) RNAs, respectively. The pT7F57-SD′ plasmid was cut with either BamHI or BstEII to serve as template for transcription of 1.8-kb or 2.6-kb SD′ RNAs, respectively. The short 5′-RNAs, wt or ΔPsi, were transcribed from of pT7F57-FL or pT7F57–5′ΔPsi, respectively, cut with HindIII. The digested templates were extracted with phenol–chloroform and chloroform, ethanol-precipitated, and dissolved in sterile MilliQ water. RNAs were produced by incubating template DNA for 2 h at 37°C with 75 U of T7 RNA polymerase (Invitrogen); 1 mM ATP, GTP, CTP, and UTP (Invitrogen); 10 mM DTT; 1× transcription buffer (Invitrogen); and 40 U of RNasin (Invitrogen) in 25 μL of final reaction volume; then ethanol-precipitated. After DNase treatment with 3 U of RQ1 DNase (Promega) for 20 min at 37°C, the RNA was phenol–chloroform- and chloroform-extracted and precipitated with ethanol. After washing, the precipitate was dissolved in sterile Milli-Q (Millipore) water. Internally labeled RNAs were synthesized as previously described (De Tapia et al. 1998).
Spontaneous in vitro dimerization
In a standard experiment, 0.5 or 1 pmol of RNA in Milli-Q water was adjusted to 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 100 mM NaCl, and 1% SDS (R buffer) in a final volume of 10 μL. For radioactive experiments, 4 × 104 cpm of 32P-labeled transcript was added into samples before resuspension in R buffer. The samples were heated or not at indicated temperatures for 10 min then chilled on ice, mixed with 2 μL of 50% (mass/vol) glycerol, 1% Na2EDTA, and 0.4% (mass/vol) bromophenol blue and submitted to 0.8% agarose gel electrophoresis at 4 V/cm in 50 mM Tris/borate (pH 8), 1 mM EDTA, and 0.1 μg/mL ethidium bromide. The quantification of each conformer to estimate the percentage of dimer was performed with ImageQuant software (Molecular Dynamics). Grafit software was used to fit a curve corresponding to % of dimer=f(temperature) to estimate the temperature of melting (T m).
Virus production and RNA encapsidation
Human embryonic kidney 293 (HEK 293T) cells were maintained at 37°C in Dulbecco's modified Eagle's medium (DMEM) supplemented with glutamine (2 mM), penicillin, streptomycin, and 10% (v/v) heat-inactivated fetal calf serum. Transfections of 293T cells were performed with 3 × 106 cells seeded the day before in 100-mm dishes in 8 mL of medium by calcium phosphate precipitation with 10 μg of Mo-MLV molecular clone (pBSKeco; a kind gift from F.L. Cosset) (Shinnick et al. 1981). The cells were washed with PBS to eliminate plasmid excess and split into three 100-mm dishes 24 h after transfection. The virus-containing medium was harvested at 72 h post-transfection corresponding to 48 h of virions releasing. Residual cells were removed by filtration through 0.45-μm filters, and virus-containing medium was stored at −80°C prior to use.
RNA encapsidation efficiency was determined by RT-QPCR as previously described (Houzet et al. 2003). The RNA copy numbers measured in cells and in virions were reported to the total input of cell and virion RNA samples, and virus-to-cell RNA ratios [(V/C) × 100] were calculated and normalized to the FL RNA level.
Isolation of dimeric viral RNA
Approximately 30 mL of virus-containing medium was centrifuged at 125,000g for 90 min at 4°C. Pelleted virus was suspended in 400 μL of lysis buffer (50 mM Tris at pH 7.5, 10 mM EDTA, 1% SDS, 100 mM NaCl, 50 μg/mL tRNA, 100 μg/mL proteinase K) and incubated for 30 min at 37°C, extracted with phenol–chloroform, chloroform, and ethanol-precipitated. For DNase treatment, RNA was suspended in 50 μL of TE buffer and incubated with 3 U of RQ1 DNase (Promega) for 20 min at 37°C. After phenol–chloroform, chloroform extraction, and ethanol precipitation, the RNA was dissolved in 15 μL of R buffer.
RNA capture assay
RNA capture
The RNA capture assay protocol (RCA) was previously described in detail (Flynn et al. 2004). Briefly, virions were harvested from the media, and virion RNAs were isolated under conditions that maintained dimer linkages. An aliquot (1/5) of the RNA sample extracted from released virions was save to generate the input sample, whereas the rest (4/5) of the RNA sample was subjected to the capture assay by using the 3′-biotinylated anti-MLV pol oligonucleotide (5′-CAGTCTCTGTATGTGGGGCTTG-3′). Annealing to virion RNAs was achieved under reaction conditions determined to maintain dimer linkages. The oligonucleotide-bound FL RNA, as well as their dimer partners, were recovered by magnetic streptavidin-coated beads (Promega) by using a magnetic stand, and the supernatant was saved for quantification (flowthrough sample). After several washes (washes sample), the bound RNA was eluted by heating for 5 min at 85°C in water (elution sample). All samples—input, flowthrough, washes, and elution—were ethanol-precipitated with 15 μg of carrier tRNA, and RNA analysis was performed in all samples by specific RT-QPCR.
RNA analysis
Reverse transcription (RT) was performed as previously described with an oligo(dT) primer for viral RNA quantifications with 1 μg of each RNA sample and with specific internal primers for U6 and 7SL RNA analysis with 0.5 μg of each RNA sample (Smagulova et al. 2005). Quantitative PCR was achieved with 2.5% of the RT-d(T), 0.125% of RT-U6, and 0.05% of RT-7SL reactions with the SensiMix kit (Quantace) and the RotorGene (Corbett) system. The primers used for amplifications of FL, SD′, U6, and 7SL RNAs are described in Houzet et al. (2003, 2007) and Smagulova et al. (2005). A standard curve was generated from 50 to 500,000 copies of pBSKeco plasmid.
ACKNOWLEDGMENTS
We thank Z. Morichaud, J.C. Paillart, and A. Onafuwa-Nuga for technical assistance and helpful discussions. This work was supported by the Ministère de la Recherche (ACI) and the CNRS grants to M.M. and by NIH grant CA 069300 to A.T. S.M. was supported by ACI and Wood-Whelan fellowships, and L.H. by a fellowship from SIDACTION.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.713807.
REFERENCES
- Aagaard, L., Rasmussen, S.V., Mikkelsen, J.G., Pedersen, F.S. Efficient replication of full-length murine leukemia viruses modified at the dimer initiation site regions. Virology. 2004;318:360–370. doi: 10.1016/j.virol.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Audit, M., Dejardin, J., Hohl, B., Sidobre, C., Hope, T.J., Mougel, M., Sitbon, M. Introduction of a cis-acting mutation in the capsid-coding gene of Moloney murine leukemia virus extends its leukemogenic properties. J. Virol. 1999;73:10472–10479. doi: 10.1128/jvi.73.12.10472-10479.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender, W., Chien, Y.H., Chattopadhyay, S., Vogt, P.K., Gardner, M.B., Davidson, N. High-molecular-weight RNAs of AKR, NZB, and wild mouse viruses and avian reticuloendotheliosis virus all have similar dimer structures. J. Virol. 1978;25:888–896. doi: 10.1128/jvi.25.3.888-896.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet-Mathoniere, B., Girard, P.M., Muriaux, D., Paoletti, J. Nucleocapsid protein 10 activates dimerization of the RNA of Moloney murine leukaemia virus in vitro. Eur. J. Biochem. 1996;238:129–135. doi: 10.1111/j.1432-1033.1996.0129q.x. [DOI] [PubMed] [Google Scholar]
- Buxton, P., Tachedjian, G., Mak, J. Analysis of the contribution of reverse transcriptase and integrase proteins to retroviral RNA dimer conformation. J. Virol. 2005;79:6338–6348. doi: 10.1128/JVI.79.10.6338-6348.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavazzana-Calvo, M., Fischer, A. Gene therapy for severe combined immunodeficiency: are we there yet? J. Clin. Invest. 2007;117:1456–1465. doi: 10.1172/JCI30953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejardin, J., Bompard-Marechal, G., Audit, M., Hope, T.J., Sitbon, M., Mougel, M. A novel subgenomic murine leukemia virus RNA transcript results from alternative splicing. J. Virol. 2000;74:3709–3714. doi: 10.1128/jvi.74.8.3709-3714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Tapia, M., Metzler, V., Mougel, M., Ehresmann, B., Ehresmann, C. Dimerization of MoMuLV genomic RNA: Redefinition of the role of the palindromic stem–loop H1 (278–303) and new roles for stem–loops H2 (310–352) and H3 (355–374) Biochemistry. 1998;37:6077–6085. doi: 10.1021/bi9800303. [DOI] [PubMed] [Google Scholar]
- D'Souza, V., Summers, M.F. How retroviruses select their genomes. Nat. Rev. Microbiol. 2005;3:643–655. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- Dudley, J.P. Tag, you're hit: Retroviral insertions identify genes involved in cancer. Trends Mol. Med. 2003;9:43–45. doi: 10.1016/s1471-4914(03)00003-0. [DOI] [PubMed] [Google Scholar]
- Fan, H. Leukemogenesis by Moloney murine leukemia virus: A multistep process. Trends Microbiol. 1997;5:74–82. doi: 10.1016/S0966-842X(96)10076-7. [DOI] [PubMed] [Google Scholar]
- Fisher, J.A., Goff, S.P. Mutational analysis of stem–loops in the RNA packaging signal of the Moloney murine leukemia virus. Virology. 1998;244:133–145. doi: 10.1006/viro.1998.9090. [DOI] [PubMed] [Google Scholar]
- Flynn, J.A., Telesnitsky, A. Two distinct Moloney murine leukemia virus RNAs produced from a single locus dimerize at random. Virology. 2006;344:391–400. doi: 10.1016/j.virol.2005.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn, J.A., An, W., King, S.R., Telesnitsky, A. Nonrandom dimerization of murine leukemia virus genomic RNAs. J. Virol. 2004;78:12129–12139. doi: 10.1128/JVI.78.22.12129-12139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, W., Rein, A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 1993;67:5443–5449. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, W., Dang, Q., Nagashima, K., Freed, E.O., Pathak, V.K., Hu, W.S. Effects of Gag mutation and processing on retroviral dimeric RNA maturation. J. Virol. 2006;80:1242–1249. doi: 10.1128/JVI.80.3.1242-1249.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherghe, C., Weeks, K.M. The SL1–SL2 (stem–loop) domain is the primary determinant for stability of the gammaretroviral genomic RNA dimer. J. Biol. Chem. 2006;281:37952–37961. doi: 10.1074/jbc.M607380200. [DOI] [PubMed] [Google Scholar]
- Giles, K.E., Caputi, M., Beemon, K.L. Packaging and reverse transcription of snRNAs by retroviruses may generate pseudogenes. RNA. 2004;10:299–307. doi: 10.1261/rna.2150604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard, P.M., Bonnet-Mathoniere, B., Muriaux, D., Paoletti, J. A short autocomplementary sequence in the 5′ leader region is responsible for dimerization of MoMuLV genomic RNA. Biochemistry. 1995;34:9785–9794. doi: 10.1021/bi00030a016. [DOI] [PubMed] [Google Scholar]
- Girard, P.M., de Rocquigny, H., Roques, B.P., Paoletti, J. A model of PSI dimerization: Destabilization of the C278–G303 stem–loop by the nucleocapsid protein (NCp10) of MoMuLV. Biochemistry. 1996;35:8705–8714. doi: 10.1021/bi952454s. [DOI] [PubMed] [Google Scholar]
- Hibbert, C.S., Mirro, J., Rein, A. mRNA molecules containing murine leukemia virus packaging signals are encapsidated as dimers. J. Virol. 2004;78:10927–10938. doi: 10.1128/JVI.78.20.10927-10938.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housset, V., De Rocquigny, H., Roques, B.P., Darlix, J.L. Basic amino acids flanking the zinc finger of Moloney murine leukemia virus nucleocapsid protein NCp10 are critical for virus infectivity. J. Virol. 1993;67:2537–2545. doi: 10.1128/jvi.67.5.2537-2545.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houzet, L., Battini, J., Bernard, E., Thibert, V., Mougel, M. A new retroelement constituted by a natural alternatively spliced RNA of murine replication-competent retroviruses. EMBO J. 2003;22:4866–4875. doi: 10.1093/emboj/cdg450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houzet, H., Paillart, J., Smagulova, F., Maurel, S., Morichaud, Z., Marquet, R., Mougel, M. HIV controls the selective packaging of genomic, spliced viral, and cellular RNAs into virions through different mechanisms. Nucleic Acids Res. 2007;35:2695–2704. doi: 10.1093/nar/gkm153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharytonchyk, S.A., Kireyeva, A.I., Osipovich, A.B., Fomin, I.K. Evidence for preferential copackaging of Moloney murine leukemia virus genomic RNAs transcribed in the same chromosomal site. Retrovirology. 2005;2:3. doi: 10.1186/1742-4690-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C.H., Tinoco I., Jr A retroviral RNA kissing complex containing only two G•C base pairs. Proc. Natl. Acad. Sci. 2000;97:9396–9401. doi: 10.1073/pnas.170283697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly, H., Parslow, T.G. Bipartite signal for genomic RNA dimerization in Moloney murine leukemia virus. J. Virol. 2002;76:3135–3144. doi: 10.1128/JVI.76.7.3135-3144.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makris, A., Patriotis, C., Bear, S.E., Tsichlis, P.N. Structure of a Moloney murine leukemia virus–virus-like 30 recombinant: Implications for transduction of the c-Ha-ras proto-oncogene. J. Virol. 1993;67:1286–1291. doi: 10.1128/jvi.67.3.1286-1291.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen, J.G., Lund, A.H., Duch, M., Pedersen, F.S. Mutations of the kissing-loop dimerization sequence influence the site specificity of murine leukemia virus recombination in vivo. J. Virol. 2000;74:600–610. doi: 10.1128/jvi.74.2.600-610.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, M.D., Fu, W., Nikolaitchik, O., Chen, J., Ptak, R.G., Hu, W.S. Dimer initiation signal of human immunodeficiency virus type 1: Its role in partner selection during RNA copackaging and its effects on recombination. J. Virol. 2007;81:4002–4011. doi: 10.1128/JVI.02589-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougel, M., Barklis, E. A role for two hairpin structures as a core RNA encapsidation signal in murine leukemia virus virions. J. Virol. 1997;71:8061–8065. doi: 10.1128/jvi.71.10.8061-8065.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougel, M., Tounekti, N., Darlix, J.L., Paoletti, J., Ehresmann, B., Ehresmann, C. Conformational analysis of the 5′ leader and the gag initiation site of Mo-MuLV RNA and allosteric transitions induced by dimerization. Nucleic Acids Res. 1993;21:4677–4684. doi: 10.1093/nar/21.20.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougel, M., Zhang, Y., Barklis, E. Cis-active structural motifs involved in specific encapsidation of Moloney murine leukemia virus RNA. J. Virol. 1996;70:5043–5050. doi: 10.1128/jvi.70.8.5043-5050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriaux, D., Rein, A. Encapsidation and transduction of cellular genes by retroviruses. Front. Biosci. 2003;8:D135–D142. doi: 10.2741/950. [DOI] [PubMed] [Google Scholar]
- Onafuwa-Nuga, A.A., King, S.R., Telesnitsky, A. Nonrandom packaging of host RNAs in Moloney murine leukemia virus. J. Virol. 2005;79:13528–13537. doi: 10.1128/JVI.79.21.13528-13537.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onafuwa-Nuga, A.A., Telesnitsky, A., King, S.R. 7SL RNA, but not the 54-kD signal recognition particle protein, is an abundant component of both infectious HIV-1 and minimal virus-like particles. RNA. 2006;12:542–546. doi: 10.1261/rna.2306306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oroudjev, E.M., Kang, P.C., Kohlstaedt, L.A. An additional dimer linkage structure in Moloney murine leukemia virus RNA. J. Mol. Biol. 1999;291:603–613. doi: 10.1006/jmbi.1999.2984. [DOI] [PubMed] [Google Scholar]
- Paillart, J.C., Shehu-Xhilaga, M., Marquet, R., Mak, J. Dimerization of retroviral RNA genomes: An inseparable pair. Nat. Rev. Microbiol. 2004;2:461–472. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- Paoletti, J., Mougel, M., Tounekti, N., Girard, P.M., Ehresmann, C., Ehresmann, B. Spontaneous dimerization of retroviral MoMuLV RNA. Biochimie. 1993;75:681–686. doi: 10.1016/0300-9084(93)90099-e. [DOI] [PubMed] [Google Scholar]
- Prats, A.C., Roy, C., Wang, P., Erard, M., Housset, V., Gabus, C., Paoletti, C., Darlix, J.L. cis elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. J. Virol. 1990;64:774–783. doi: 10.1128/jvi.64.2.774-783.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez, J.M., Houzet, L., Koller, R., Bies, J., Wolff, L., Mougel, M. Activation of c-myb by 5′ retrovirus promoter insertion in myeloid neoplasms is dependent upon an intact alternative splice donor site (SD′) in gag . Virology. 2004;330:398–407. doi: 10.1016/j.virol.2004.09.038. [DOI] [PubMed] [Google Scholar]
- Rasmussen, S.V., Pedersen, F.S. Co-localization of γ retroviral RNAs at their transcription site favours co-packaging. J. Gen. Virol. 2006;87:2279–2289. doi: 10.1099/vir.0.81759-0. [DOI] [PubMed] [Google Scholar]
- Roy, C., Tounekti, N., Mougel, M., Darlix, J.L., Paoletti, C., Ehresmann, C., Ehresmann, B., Paoletti, J. An analytical study of the dimerization of in vitro generated RNA of Moloney murine leukemia virus MoMuLV. Nucleic Acids Res. 1990;18:7287–7292. doi: 10.1093/nar/18.24.7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulli S.J., Jr, Hibbert, C.S., Mirro, J., Pederson, T., Biswal, S., Rein, A. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J. Virol. 2007;81:6623–6631. doi: 10.1128/JVI.02833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuragi, J., Iwamoto, A., Shioda, T. Dissociation of genome dimerization from packaging functions and virion maturation of human immunodeficiency virus type 1. J. Virol. 2002;76:959–967. doi: 10.1128/JVI.76.3.959-967.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Russell, D.X. Molecular cloning: A laboratory manual. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Shinnick, T.M., Lerner, R.A., Sutcliffe, J.G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981;293:543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Smagulova, F., Maurel, S., Morichaud, Z., Devaux, C., Mougel, M., Houzet, L. The highly structured encapsidation signal of MuLV RNA is involved in the nuclear export of its unspliced RNA. J. Mol. Biol. 2005;354:1118–1128. doi: 10.1016/j.jmb.2005.10.021. [DOI] [PubMed] [Google Scholar]
- Sorensen, A.B., Lund, A.H., Kunder, S., Quintanilla-Martinez, L., Schmidt, J., Wang, B., Wabl, M., Pedersen, F.S. Impairment of alternative splice sites defining a novel γ retroviral exon within gag modifies the oncogenic properties of Akv murine leukemia virus. Retrovirology. 2007;4:46. doi: 10.1186/1742-4690-4-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szentirmay, M.N., Sawadogo, M. Survey and summary: Spatial organization of RNA polymerase II transcription in the nucleus. Nucleic Acids Res. 2000;28:2019–2025. doi: 10.1093/nar/28.10.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchenio, T., Heidmann, T. The dimerization/packaging sequence is dispensable for both the formation of high-molecular-weight RNA complexes within retroviral particles and the synthesis of proviruses of normal structure. J. Virol. 1995;69:1079–1084. doi: 10.1128/jvi.69.2.1079-1084.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrent, C., Bordet, T., Darlix, J.L. Analytical study of rat retrotransposon VL30 RNA dimerization in vitro and packaging in murine leukemia virus. J. Mol. Biol. 1994;240:434–444. doi: 10.1006/jmbi.1994.1459. [DOI] [PubMed] [Google Scholar]
- Tounekti, N., Mougel, M., Roy, C., Marquet, R., Darlix, J.L., Paoletti, J., Ehresmann, B., Ehresmann, C. Effect of dimerization on the conformation of the encapsidation Psi domain of Moloney murine leukemia virus RNA. J. Mol. Biol. 1992;223:205–220. doi: 10.1016/0022-2836(92)90726-z. [DOI] [PubMed] [Google Scholar]
- Valadi, H., Ekstrom, K., Bossios, A., Sjostrand, M., Lee, J.J., Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Zeilfelder, U., Frank, O., Sparacio, S., Schon, U., Bosch, V., Seifarth, W., Leib-Mosch, C. The potential of retroviral vectors to cotransfer human endogenous retroviruses (HERVs) from human packaging cell lines. Gene. 2007;390:175–179. doi: 10.1016/j.gene.2006.08.019. [DOI] [PubMed] [Google Scholar]