Abstract
The highly conserved internal stem–loop (ISL) of U6 spliceosomal RNA is unwound for U4/U6 complex formation during spliceosome assembly and reformed upon U4 release during spliceosome activation. The U6 ISL is structurally similar to Domain 5 of group II self-splicing introns, and contains a dynamic bulge that coordinates a Mg++ ion essential for the first catalytic step of splicing. We have analyzed the causes of growth defects resulting from mutations in the Saccharomyces cerevisiae U6 ISL-bulged nucleotide U80 and the adjacent C67–A79 base pair. Intragenic suppressors and enhancers of the cold-sensitive A79G mutation, which replaces the C–A pair with a C–G pair, suggest that it stabilizes the ISL, inhibits U4/U6 assembly, and may also disrupt spliceosome activation. The lethality of mutations C67A and C67G results from disruption of base-pairing potential between U4 and U6, as these mutations are fully suppressed by compensatory mutations in U4 RNA. Strikingly, suppressor analysis shows that the lethality of the U80G mutation is due not only to formation of a stable base pair with C67, as previously proposed, but also another defect. A U6-U80G strain in which mispairing with position 67 is prevented grows poorly and assembles aberrant spliceosomes that retain U1 snRNP and fail to fully unwind the U4/U6 complex at elevated temperatures. Our data suggest that the U6 ISL bulge is important for coupling U1 snRNP release with U4/U6 unwinding during spliceosome activation.
Keywords: Prp24, U4/U6, U6 RNA, pre-mRNA splicing, spliceosome activation
INTRODUCTION
Intron removal from nuclear precursor messenger RNAs (pre-mRNAs) occurs through a two-step transesterification reaction catalyzed by the spliceosome. The spliceosome is a multimegadalton complex consisting primarily of five small nuclear ribonucleoprotein particles, or snRNPs, named U1, U2, U4, U5, and U6 for their respective small nuclear RNA (snRNA) component. These snRNPs bind to an intron sequentially: first U1, then U2, then the U4/U5/U6 tri-snRNP complex. Once fully assembled, the spliceosome is activated through a number of structural rearrangements in the snRNAs, facilitated by protein components (Will and Lührmann 2006). These rearrangements result in the coordinated dissociation of U1 and U4 snRNPs (Kuhn et al. 1999; Staley and Guthrie 1999), during which U6 becomes base paired to the 5′ splice site and U2 RNA. This process requires a complex allosteric cascade of RNA–RNA and RNA–protein interactions (Brow 2002), in which U6 RNA plays a central role.
In the yeast Saccharomyces cerevisiae, most of the U6 RNA exists as a free snRNP, in a two stem–loop conformation bound by its accessory proteins, Prp24 and Lsm2–Lsm8 (Fortner et al. 1994; Ryan and Abelson 2002; Karaduman et al. 2006). While the 5′-terminal stem–loop is thought to be unaltered throughout the splicing cycle, the highly conserved internal stem–loop (ISL; previously called the intramolecular stem–loop or 3′ stem–loop) unwinds during U4/U6 di-snRNP assembly and is reformed during spliceosome activation, when U4 leaves the spliceosome and U6 pairs with U2 RNA (Fig. 1A; Nilsen 1998; Staley and Guthrie 1998). Prp24 has been implicated as an RNA chaperone in both remodeling events (Shannon and Guthrie 1991; Ghetti et al. 1995; Raghunathan and Guthrie 1998a; Vidaver et al. 1999). The U2/U6 RNA complex is thought to catalyze splicing, since a complex consisting solely of fragments of the human U2 and U6 RNAs inefficiently catalyzes a similar reaction (Valadkhan and Manley 2003).
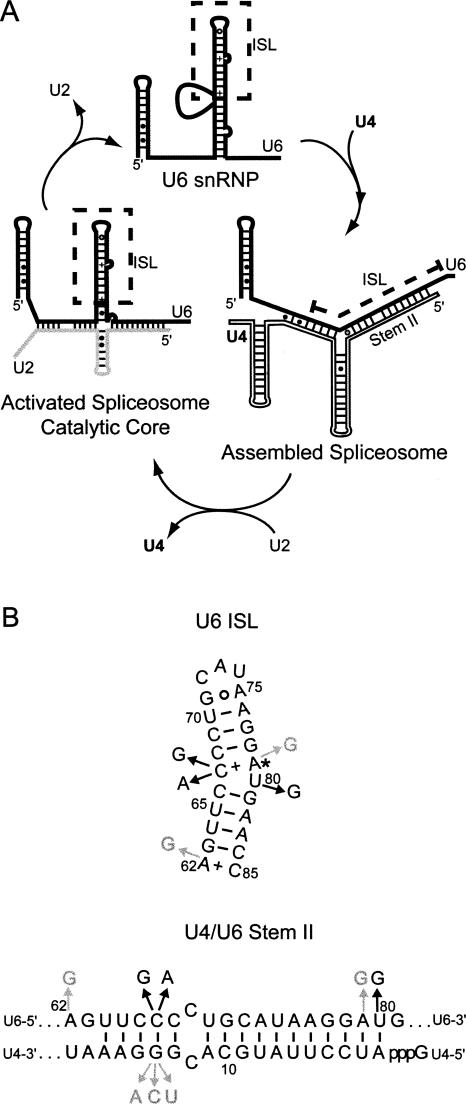
FIGURE 1.
(A) Diagram of structural elements of yeast U6 RNA and their rearrangement during pre-mRNA splicing. The U6 ISL unwinds to form stem II of the U4/U6 complex and is reformed in the U2/U6 complex. (B) Selected lethal (black) and cold-sensitive (gray) substitutions in U6 and U4 RNAs are shown in the U6 ISL (top) and U4/U6 stem II (bottom). (*) A catalytically essential Mg++ binding site.
Consistent with its function as a ribozyme, the U6 ISL has structural similarity to Domain 5 (D5) of group II self-splicing introns, which is essential for catalysis (Sashital et al. 2004; Seetharaman et al. 2006). Indeed, D5 can functionally substitute for the ISL of U6atac in the U12-dependent spliceosome of cultured mammalian cells, at least for one reporter substrate (Shukla and Padgett 2002). The ISL contains two noncanonical A–C base pairs, at residues A62–C85 and C67–A79 (Fig. 1B). A bulged nucleotide at position U80 and the adjacent C67–A79 base pair can act as a dynamic hinge. The C67–A79 pair is stabilized via protonation of A79 (pKa=6.5), which causes U80 to flip out into the major groove. When A79 is in the unprotonated state, U80 stacks inside the ISL, resulting in a 25° bend in the helix (Reiter et al. 2004). Binding of a catalytically necessary Mg++ ion by the U80 pro-Sp phosphate oxygen (Yean et al. 2000) is favored when U80 is stacked inside the helix, suggesting this conformation functions in catalysis (Blad et al. 2005). In addition, residues flanking U80 have been implicated as a binding site for the U6 snRNP protein, Prp24 (Ghetti et al. 1995; Kwan and Brow 2005; Karaduman et al. 2006).
Several conditionally and nonconditionally lethal point mutations have been identified in the ISL bulge region in yeast (Fig. 1B; Madhani et al. 1990; Fortner et al. 1994; McPheeters, 1996). These mutations can alter the base pairing and stability of the ISL, and are known to result in spliceosome assembly defects. Mutations C67A and C67G cause severe growth defects on their own (McPheeters 1996), yet dominantly suppress the cold-sensitive growth defects of U4-G14 mutant strains (Shannon and Guthrie 1991), suggesting that C67 plays an important role in U4/U6 assembly. The cold sensitivity of the A62G and A79G mutations has been postulated to result from ISL stabilization due to the replacement of A–C by G–C base pairs (Fortner et al. 1994). Similarly, the lethal U80G mutation stabilizes the ISL and blocks U4/U6 complex assembly, presumably by inhibiting ISL melting (Madhani et al. 1990; Ryan and Abelson 2002; Sashital et al. 2003).
While it is apparent that ISL bulge mutations can disrupt U4/U6 assembly, it remains unclear if these mutations also affect downstream processes, such as spliceosome activation and/or catalysis. Both in vitro and in vivo evidence from mutational analyses of the human U6 ISL suggest that both base-pairing potential and sequence identity are important for spliceosome assembly and later processes (Wolff and Bindereif 1993; Sun and Manley 1997), and hydroxyl radical probing experiments have placed the human ISL bulge in the vicinity of the U6–5′splice site interaction in activated spliceosomes (Rhode et al. 2006). Mutation of the D5 bulge region of group II self-splicing intron aI5γ was previously found to inhibit either docking of Domain 5 within the intron, splicing catalysis, or both (Schmidt et al. 1996), and a region adjacent to the bulge was shown to interact with the 5′ splice site (Boudvillain et al. 2000). Consistent with these findings, structural studies showed that the D5 bulge from a Pylaiella littoralis group II intron ribozyme construct undergoes dramatic conformational changes upon docking to Domains 1–3 (Gumbs et al. 2006).
We have used two approaches to better understand the phenotypes associated with U6 ISL bulge-region mutations. First, we isolated intragenic suppressors of the cold-sensitive A79G mutation, which substitutes the C67–A79 pair with a C–G pair. Suppressors of A79G map to the upper ISL and telestem regions of U6 and are predicted to destabilize these structures. Several of the selected suppressors of A79G were previously shown to suppress A62G cold sensitivity (Fortner et al. 1994). However, some suppressors of A62G exacerbate the A79G growth defect, suggesting the existence of both shared and antagonistic functions of A62 and A79.
Second, we used directed mutagenesis to test the hypothesis that the lethality of mutations in C67 and U80 is due to formation of a stable base pair between these residues. We find that for neither position is ISL mispairing the sole defect. The lethality of C67A and C67G results from insufficient formation of functional U4/U6 complex and can be fully suppressed by compensatory mutations in U4 RNA. In contrast, the lethal U80G mutation is only weakly suppressed by mutations that eliminate the potential for pairing with position 67 while maintaining pairing potential between U6–67 and U4 RNA. For example, a strain expressing U6-C67A/U80G and U4-G14U has near normal levels of U4/U6 complex, yet is inviable at both high and low temperatures. Cell extracts from this strain exhibit structural alterations in U4/U5/U6 tri-snRNP, assemble aberrant spliceosomes, and are defective for splicing at elevated temperatures. The aberrant spliceosomes fail to release U1 snRNP efficiently, even at permissive temperature, and are additionally impaired for U4/U6 unwinding at restrictive temperature. Our data suggest that sequences in the U6 ISL bulge coordinate U4/U6 unwinding and U1 snRNP release, and therefore have important functions in spliceosome activation, as well as assembly.
RESULTS
Identification of cis-acting suppressors and enhancers of U6-A79G
A number of cis-acting (intragenic) suppressors of the cold-sensitive U6-A62G mutation were previously identified (Fortner et al. 1994). The locations of these mutations in U6 RNA provided clues to the defect of the A62G mutation and resulted in proposal of the telestem (Fig. 2A; Brow and Vidaver 1995). To test if the cold-sensitive A79G mutation acts in a fashion similar to A62G, as previously suggested (Fortner et al. 1994), we subjected the A79G allele to genetic analysis using two approaches: (1) we randomly mutagenized the U6-A79G allele by error-prone PCR and selected cold-resistant alleles, which are expected to include both revertants and cis-acting suppressors, and (2) we tested the effect of selected suppressors of A62G in cis with A79G. The results of these experiments are summarized in Figure 2.
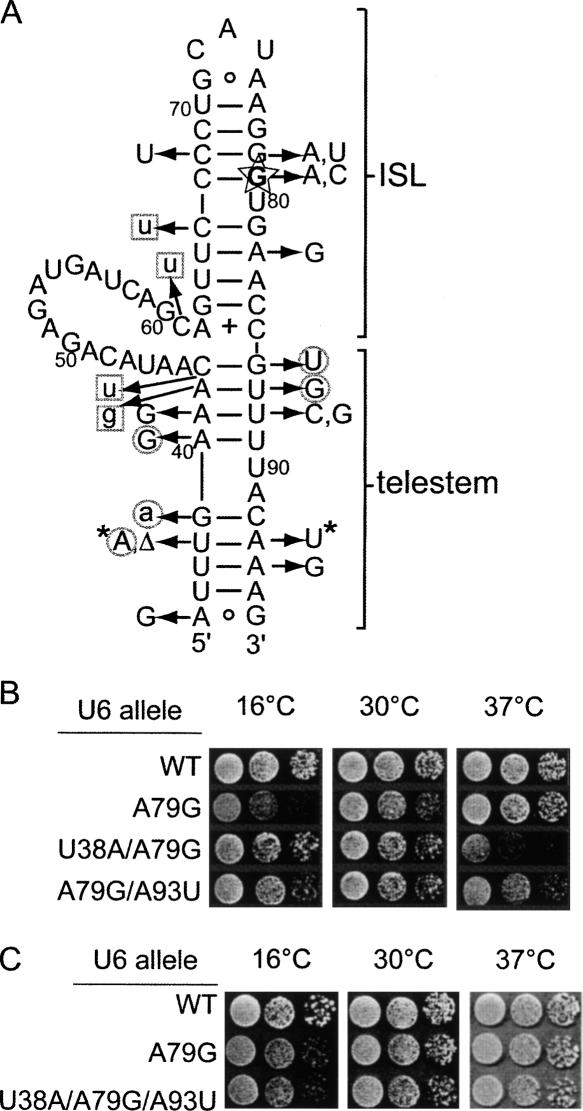
FIGURE 2.
Intragenic suppressors and enhancers of the U6-A79G cold-sensitive mutation. (A) The central region (nucleotides 35–96) of yeast U6 RNA is shown, including the telestem, an intervening loop (nucleotides 44–61), and the ISL. The A79G mutation is outlined with a star and in bold. Arrows point to mutations that were selected as suppressors of A79G (uppercase) or selected as suppressors of A62G (Fortner et al. 1994) and tested with A79G (lowercase). Circled mutations suppress both A62G and A79G, while boxed mutations suppress A62G and enhance (exacerbate) the growth defect of A79G. Four mutations obtained in the A62G selection (A62C, A62U, G63A, and insertion of an A between 62 and 63) neither suppressed or enhanced A79G and are not shown. (Δ) Deletion of a nucleotide. (*) Two mutations that lose suppressor activity when tested in combination. Substitutions selected at position 79, including the true revertant, are also shown. (B) Serial-dilution growth assays on YEPD medium of suppressor mutations U38A and A93U, and (C) a strain with U6-A79G/U38A/A93U show that disruption of the U38-A93 pair is sufficient for suppression of A79G.
The random selection yielded 15 different cis-acting suppressors of A79G, as well as two of the three possible substitutions of G79 (Fig. 2A). The suppressor mutations localize to two structural features: the ISL and the telestem. Most suppressor mutations were isolated only once or twice, but a few were obtained multiple times, including A82G (14), C68U (10), U88C (6), A93U (6), and A35G (5). Four of the selected suppressors of A79G were previously selected as suppressors of A62G (see below). We tested the effect of nine additional suppressors of A62G in combination with A79G. One, G39A, also suppressed A79G, four enhanced (exacerbated) A79G, and four had no genetic interaction with A79G (Fig. 2A).
Interestingly, the telestem is the only region in which suppressors of A79G and A62G colocalize (Fig. 2A, circled suppressors). These suppressor mutations are expected to destabilize the telestem, suggesting that the stability of the telestem contributes to the cold-sensitive growth defect of both the A62G and A79G mutations. This conclusion was previously confirmed for A62G by combining suppressor mutations that together restore pairing of the “lower” (ISL-distal) telestem and observing a loss of suppression (Vidaver et al. 1999). Likewise, combining the U38A and A93U suppressors of A79G (Fig. 2A, marked with asterisks) restores base pairing between residues 38 and 93 in the lower telestem and prevents suppression of A79G (Fig. 2B,C). Furthermore, the heat-sensitive growth exhibited by each suppressor strain is also reverted by combining the mutations. These results support the hypothesis that suppressors of A62G and A79G that map to the lower telestem act by destabilizing this structure. The isolation of the A35G suppressor of A79G five times leads us to propose that the lower telestem is stabilized by a terminal A–G base pair (Elgavish et al. 2001).
The mechanism of suppression by mutations in the upper (ISL-proximal) telestem is less clear. Compensatory combinations of suppressors of A62G in the upper telestem still suppress, suggesting that disruption of base pairing is not the mechanism of suppression (Vidaver et al 1999). Furthermore, two mutations in the upper telestem that suppress A62G, A42G, and C43U (Fig. 2A, boxed), actually enhance A79G, resulting in synthetic lethality at all temperatures tested. Yet mutations in the residues that pair with A42 and C43 in our model of the telestem suppress both A62G and A79G. Thus, the effects of mutations in the upper telestem may be due to disruption of interactions other than intramolecular base pairing (see Discussion).
Four different suppressors of A79G mapped to the ISL, and two of these, C68U and A82G, were the most frequently isolated mutations in the selection. Both suppressors convert Watson–Crick pairs to G–U wobble pairs, and thus, may destabilize the ISL in the vicinity of the C67/G79 pair. In addition to obtaining the true revertant (G79A), we also isolated the G79C mutation. We did not obtain G79U, even though this substitution is not cold sensitive (see Fig. 3), which suggests the selection was not saturated.
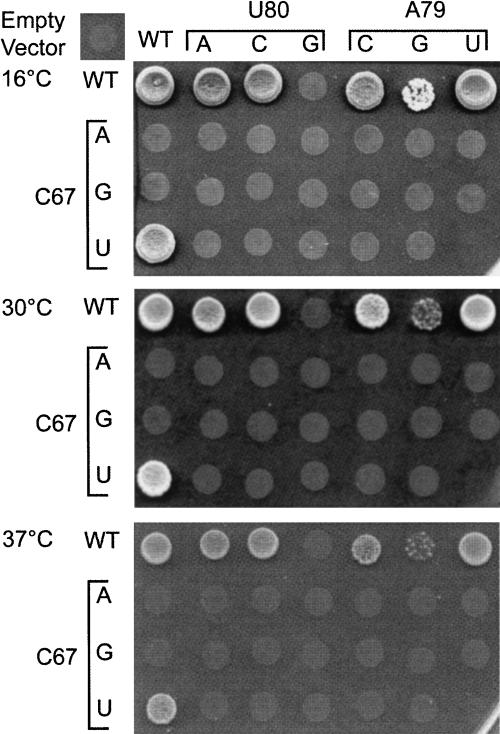
FIGURE 3.
All C67/A79 and C67/U80 double mutants are inviable at 16, 30, and 37°C. Approximately 2×104 cells bearing a wild-type U6 gene on a URA3-marked plasmid and the indicated mutant U6 allele on a TRP1-marked plasmid were plated on medium containing 5-FOA to select for loss of the plasmid bearing the wild-type U6 gene. The phenotypes of all single U6 point mutations except for C67G are the same as previously reported (McPheeters 1996). The apparent heat sensitivity of A79G is not observed in more quantitative assays (cf. Figs. 2 and 4).
Six suppressors of A62G in or adjacent to the ISL were tested with A79G. Four of these, A62C, A62U, G63A, and an insertion of an A between residues 62 and 63, exhibited no genetic interaction with the A79G mutation. The remaining two, C61U and C66U (Fig. 2A, boxed), are synthetic lethal with A79G. The basis of this enhancement of phenotype is not known, but could be explained by disruption of Prp24 binding (see Discussion). Thus, within the ISL, no overlap of suppressors of A62G and A79G was observed.
Growth phenotypes of mutations in the U6 ISL bulge region are not suppressed by secondary mutations that disrupt pairing potential
Since the cold-sensitivity of mutations in the A62–C85 pair correlates with base-pairing potential (Fortner et al. 1994) and several of the suppressors of A79G have the potential to destabilize the ISL, we hypothesized that the cold sensitivity of the U6-A79G mutation could be due to the formation of a stable C–G base pair with C67. This hypothesis is consistent with the observation that the A79C and A79U mutations do not cause growth defects (McPheeters 1996), and predicts that mutations in C67 that disrupt pairing should suppress the cold sensitivity of A79G. To test this prediction, we created SNR6 alleles with all possible combinations of bases at positions 67 and 79 (Fig. 3). Contrary to the prediction, all nine C67/A79 double-mutant strains were dead at 16°, 30°, and 37°C. For example, the C67U mutation has no growth phenotype alone, but is lethal in combination with A79G (Fig. 3), even though it is expected to replace a C–G pair with a U–G pair. Likewise, A79C and A79U were synthetic lethal with C67U, even though the resulting base pairs should not be stable. These results indicate that ISL bulge residues function cooperatively, and the growth defects of mutations in positions 67 and 79 are not due simply to the formation of a stable base pair.
One complication for the interpretation of the phenotypes of mutations at position 67 is the existence of an unpaired U residue across the helix at position 80 (Fig. 1B). Thus, mutations at position 67 affect not only its ability to pair with position 79, but also position 80. For example, C67U creates the potential for a stable U–A pair with A79 or an unstable U–U mismatch with U80. This mutation has no growth defect (McPheeters 1996). In contrast, the C67A mutation allows formation of an A–A mismatch with A79 or an A–U pair with U80. This mutation is lethal at all temperatures tested (Fig. 3; McPheeters 1996), suggesting that formation of a stable base pair between positions 67 and 80 is detrimental to U6 RNA function. Interestingly, McPheeters (1996) reported that C67G is lethal at 16°C but not at 30°C, which is consistent with forming a weaker G–U pair with U80. Yet, in our strain background C67G is lethal at 30°C as well as at 16°C (Fig. 3). However, when the U4 RNA gene is on a plasmid and subject to gene amplification, the C67G strain is weakly viable at 30°C, while the C67A strain is not (see Fig. 4). Thus, C67G does appear to have a less-severe phenotype than C67A.
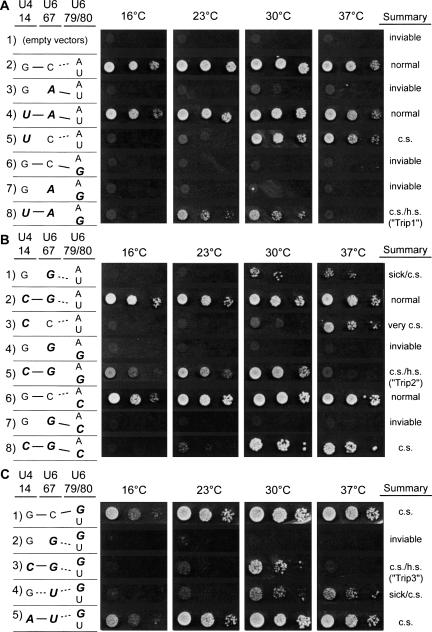
FIGURE 4.
Suppression of U6 single and double mutant alleles by mutations at position 14 in U4 RNA. Strains bearing single and double mutations in the U6 ISL bulge region, at position 14 in U4 RNA, or both, were plated in 10-fold serial dilutions onto medium containing 5-FOA to select for loss of the plasmid bearing wild-type U4 and U6 alleles. Mutated residues are indicated in bold italics. (A) Mutation U6-C67A is fully, while U6-C67A/U80G is only partially suppressed by the compensatory mutation U4-G14U. (B) Mutation U6-C67G is fully, while U6-C67G/U80G and C67G/U80C are only partially suppressed by the compensatory mutation U4-G14C. (C) Strains expressing single and double mutations in the U6 ISL bulge region including A79G are cold sensitive regardless of base-pairing potential. The presence of the C67G/A79G allele confers cold (c.s.) and heat sensitivity (h.s.) when base pairing is restored with U4 RNA.
The phenotypes of single mutations at position 80 also support the hypothesis that growth defects are due to stable base pairing with position 67. U80A and U80C, which are expected to form unstable C–A or C–C pairs, confer no growth defect, while U80G, which could form a C–G pair, is lethal (Fig. 3; Madhani et al. 1990). Furthermore, formation of the C67/G80 pair to the exclusion of the normal C67/A79 pair has been observed in the isolated ISL by NMR (Sashital et al. 2003).
To test whether the formation of a stable base pair at position 67–80 causes lethality, we assessed the effects of double mutations expected to disrupt mispairing. As observed for C67/A79 substitutions, all C67/U80 double-mutant strains are inviable (Fig. 3). These results indicate that mispairing is not the sole cause of the growth phenotypes of mutations at positions 67 and 80, although it does not exclude the possibility that mispairing contributes to the phenotype.
Other possible causes for growth defects due to mutations at positions 67, 79, and 80 of U6 RNA include decreased U6 RNA accumulation and inhibition of U4/U6 complex formation. To assess these potential defects, total cellular RNA was extracted from the mutant strains under conditions that leave existing U4/U6 complex intact. Both U6 RNA and U4/U6 complex levels were assayed by a solution hybridization/native gel analysis procedure (see Fig. 5; Li and Brow 1993). Strains bearing lethal mutations in U6 RNA also contained a “pseudo-wild-type” U6 RNA allele that supports viability but produces U6 RNA and U4/U6 complex with gel mobilities distinguishable from those of full-length U6 RNA. Eighteen of the 21 lethal mutant U6 RNAs were tested and all accumulated to approximately normal levels, but assembled inefficiently into U4/U6 RNA complex (data not shown). These data indicate that the lethality of these mutations is not due to a defect in U6 RNA accumulation, but may be due to a defect in U4/U6 snRNP formation.
FIGURE 5.
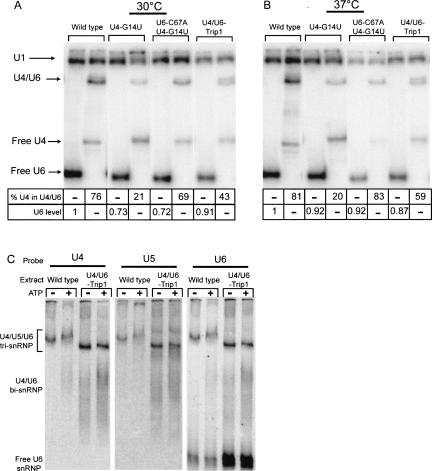
Analysis of snRNA and snRNP complexes in U4 and U6 mutant strains. (A,B) Compensatory mutation of U4 position 14 suppresses the U4/U6 assembly defect of U6-C67A and C67A/U80G. Strains expressing the indicated U4 and U6 alleles were grown at 30°C (A) or shifted to 37°C for 3 h (B) and RNA was prepared under nondenaturing conditions, hybridized to radiolabeled probes complementary to U1 (loading control) and U6 (odd lanes) or U4 (even lanes) RNAs, and separated on a 9% polyacrylamide gel. The percentage of U4 in U4/U6 complex is shown below each mutant. To determine total U6 RNA levels, RNA in odd lanes was heated to 70°C for 5 min prior to hybridization and the U6:U1 ratio was normalized to the ratio in the wild-type strain. (C) U4/U6-Trip1 extracts assemble normal levels of an aberrantly migrating U4/U5/U6 tri-snRNP. Whole-cell extracts prepared from wild-type and U4/U6-Trip1 mutant strains were incubated at room temperature for 30 min in the absence (odd lanes) or presence (even lanes) of ATP and then separated on a 4% polyacrylamide gel, transferred to a nylon membrane, and probed for the indicated snRNAs.
Restoration of U4/U6 pairing in U6 double-mutant strains unmasks suppression of U80 mutations by mutations in C67
Inhibition of U4/U6 complex formation is an expected consequence of hyperstabilization of the U6 ISL, since the ISL competes with U4/U6 pairing (Fortner et al. 1994). However, many of the double mutants are not expected to hyperstabilize the ISL, yet still have a U4/U6 complex accumulation defect. In some of these cases, the defect may be due to destabilization of U4/U6 pairing. Mutations at position 67 are more likely to strongly destabilize U4/U6 base pairing than mutations at positions 79 or 80, because C67 is part of a C–G pair that is flanked by C–G pairs, while A79 and U80 are in A–U pairs at the end of U4/U6 stem II (Fig. 1B). Furthermore, any mutation in C67′s pairing partner in U4 RNA, U4-G14, confers a cold-sensitive growth defect that is dominantly suppressed by compensatory mutations in U6-C67 (Shannon and Guthrie 1991). To test if U4/U6 destabilization contributes to the phenotype of 67/79 and 67/80 double mutations, we combined selected double mutations with mutations in U4-G14.
We first established that compensatory mutations in U4-G14 suppress the recessive phenotypes of single mutations in U6-C67. The lethal U6-C67A single mutation is still lethal at 30°C in combination with U4-G14A and U4-G14C (data not shown), but is fully suppressed at all temperatures in combination with U4-G14U, which alone confers cold-sensitive growth (Fig. 4A, rows 3–5). Likewise, the U6-C67G mutation is not suppressed by U4-G14A, but grows better in combination with U4-G14U (data not shown), and grows normally at all temperatures in combination with U4-G14C (Fig. 4B, rows 1–3). Thus, the growth defects of single mutations in U6-C67 and U4-G14 can be attributed to disruption of U4/U6 pairing.
We next tested the ability of compensatory mutations in U4-G14 to “unmask” suppression of mutations in U6-U80 by mutations in U6-C67. All three mutations at U6-C67 and U4-G14 were combined with U6-U80G. With a U–G pair at U6–67/80, the compensatory U4-G14A mutation did not rescue the lethal growth phenotype (data not shown), possibly because the intramolecular U–G wobble pair still competes too strongly with the intermolecular A–U pair. However, growth of double-mutant strains with either a A–G or G–G pair at 67/80 is rescued in the presence of the compensatory mutation in U4 (Fig. 4A, row 8; B, row 5), indicating that U6–67/80 mispairing contributes to the lethal phenotype of the U80G mutation. These mutations are the first identified suppressors of U6-U80G.
Strikingly, the triple-mutant strains have severe conditional growth defects, indicating that U6–67/80 pairing is not the only defect of the U80G mutation. In particular, the U4-G14U, U6-C67A/U80G triple mutant (hereafter, U4/U6-Trip1) (Fig. 4A, row 8) has tight heat- and cold-sensitive growth defects that are not exhibited by the U6-C67A, U4-G14U double-mutant strain (Fig. 4A, row 4), and, therefore, are not simply due to altered U4/U6 pairing. Likewise, the U4-G14C, U6-C67G/U80G triple mutant (U4/U6-Trip2) (Fig. 4B, row 5) has heat- and cold-sensitive growth defects that are not exhibited by the U6-C67G, U4-G14C double-mutant strain (Fig. 4B, row 2). When grown on rich medium, U4/U6-Trip1 is less cold sensitive, but remains highly heat sensitive (data not shown). Thus, the U to G transversion at U6 position 80 interferes with U6 RNA function, particularly at elevated temperature, regardless of the potential for mispairing between positions 67 and 80. In contrast, the inviability of a U6-C67G/U80C double mutant strain (Fig. 4B, row 7) appears to be due primarily to an imbalance between ISL stability and U4/U6 stability, since only residual cold sensitivity is observed when the U4/U6 pairing defect is corrected by the U4-G14C mutation (Fig. 4B, row 8).
We also analyzed the ability of mutations in U6-C67 to suppress the cold sensitivity of U6-A79G in the presence of compensatory U4-G14 mutations. U6-A79G is only weakly cold sensitive when the U4 RNA gene is carried on a centromere plasmid (Fig. 4C, row 1), presumably due to increased U4 gene dosage resulting from plasmid amplification. Furthermore, the C67U/A79G double mutant is viable when the U4 gene is present on a plasmid instead of in the genome, although it is strongly cold sensitive (Fig. 4C, row 4). This cold-sensitive growth defect is suppressed by the compensatory U4-G14A mutation (Fig. 4C, row 5), but is exacerbated by the noncompensatory G14C or G14U (data not shown) mutations. In contrast, the C67G/A79G double mutant is inviable even when the U4 gene is on a plasmid (Fig. 4C, row 2), and the compensatory U4-G14C mutation only partially rescues the inviability, leaving strong cold- and heat-sensitive growth defects (U4/U6-Trip3) (Fig. 4C, row 3). Given that the G14C/C67G double mutant is viable at all temperatures tested (Fig. 4B, row 2), the strong growth defects of U4/U6-Trip3 must be due to the combination of C67G and A79G.
We analyzed cellular U4/U6 levels to confirm that the partial suppression of mutations in the ISL bulge region by complementary mutations at U4-G14 is due to restoration of bi-snRNP complex levels. As was the case for U6-C67A, we found that U4-G14U has low levels of U4/U6 complex. As predicted, the combination of U6-C67A with U4-G14U restored U4/U6 complex to nearly wild-type levels (Fig. 5A). The U4/U6-Trip1 strain has an increased level of U4/U6 compared with U4-G14U, but complex levels were not restored to that of wild type. This result suggested that the growth defects seen in the triple-mutant strain may be due in part to a U4/U6 assembly defect. To further test this possibility, we repeated the analysis after incubating the strains at 37°C for 3 h prior to harvesting cells. The shift to the nonpermissive temperature actually increased rather than decreased the amount of U4/U6 complex in the U4/U6-Trip1 strain (Fig. 5B), suggesting that the growth defect is not due to inadequate U4/U6 complex formation. Similar results were obtained using the U4/U6-Trip2 strain (data not shown). U4/U6 complex from the Trip1 strain appears as a doublet, which could reflect conformational heterogeneity in the mutant complex. However, similar amounts of slow migrating U4/U6 are sometimes observed in RNA prepared from other strains, including wild type.
U4/U6-Trip1 extracts assemble altered tri-snRNPs and spliceosomes, and are inactive for splicing at elevated temperature
To better define the molecular defects of the U4/U6-Trip1 mutation, we analyzed whole-cell extracts made from a U4/U6-Trip 1 strain. Native gel electrophoresis and Northern blot analysis were used to compare the snRNP composition of the mutant extract with that of a wild-type strain (Fig. 5C). Although U4/U6-Trip1 extracts contain the U4/U5/U6 tri-snRNP at levels similar to wild type, they also show an increased amount of free U6 snRNP. This result suggests that U4/U6 complex formation is somewhat impaired in the mutant extracts, consistent with our RNA analysis. Strikingly, tri-snRNPs from U4/U6-Trip1 extracts have an increased native gel mobility compared with those from wild-type extracts, raising the possibility that these tri-snRNPs have altered composition or conformation. Incubation of the cell extracts with ATP, which stimulates turnover of the tri-snRNP pool (Raghunathan and Guthrie 1998a), did not reveal an obvious defect in tri-snRNP assembly in vitro (Fig. 5C).
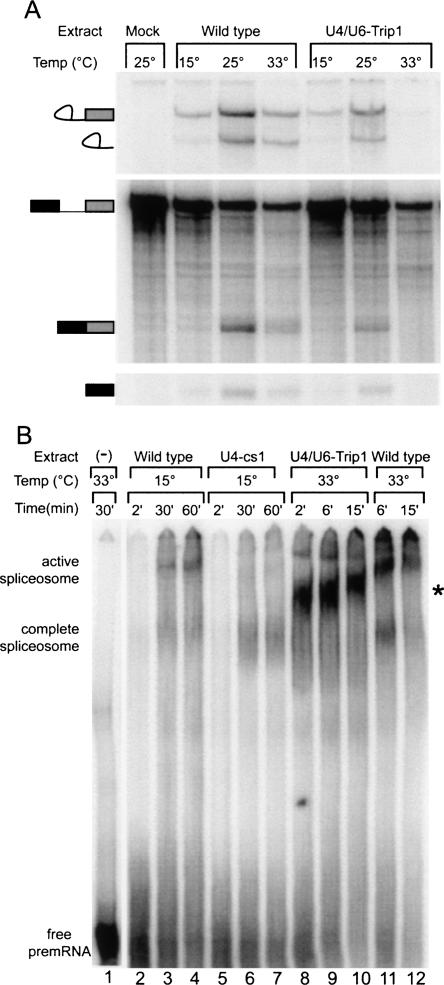
We next tested the U4/U6-Trip1-mutant extract's ability to catalyze splicing in vitro. Wild-type extracts are most active for splicing at 25°C, but exhibit significant activity at 15°C and 33°C (Fig. 6A). U4/U6-Trip1 extracts display a similar pattern, except that splicing is almost completely blocked prior to the first catalytic step at 33°C. Impaired splicing at 33°C was observed for three independent U4/U6-Trip1 extract preparations. Thus, the activity of U4/U6-Trip1 extract parallels the strain's heat-sensitive growth defect. The lack of a cold-sensitive in vitro splicing defect may reflect the weaker nature of the cold-sensitive growth defect when U4/U6-Trip1 is grown on YEPD, as it was for extract preparation.
FIGURE 6.
U4/U6-Trip1 extracts are deficient for splicing at high temperature and assemble aberrant spliceosomes. (A) Whole-cell extracts were incubated in the presence of a radiolabeled actin pre-mRNA for 25 min at the temperatures shown. Splicing products were separated from pre-mRNA on a 6% denaturing polyacrylamide gel. (B) Native gel analysis of U4/U6-Trip1 splicing intermediates. Splicing complexes were assembled onto radiolabeled pre-mRNA at 15° (lanes 2–7), and 33°C (lanes 1,8–12) for the times shown. The complexes were treated with heparin and separated on a 4% polyacrylamide gel. Extracts from a strain expressing the U4-cs1 mutation (Kuhn et al. 1999) were used as a control for complete spliceosomes. (*) The position of aberrant U4/U6-Trip1 complex.
A block prior to the first step of splicing could result from a defect in spliceosome assembly or activation. In order to discriminate between these two possibilities, we first analyzed U4/U6-Trip1 spliceosome formation on native polyacrylamide gels. Spliceosomes from U4-cs1 extracts arrest after assembly and prior to activation when incubated at low temperature (Kuhn et al. 1999) and were used as a control for the complete spliceosome, which contains all five snRNPs. U4-cs1 extracts exhibit a single major complex with pre-mRNA at 15°C (Fig. 6B, lanes 5–7), which we interpret to be the fully assembled but inactive spliceosome. Extracts from a wild-type strain accumulated two complexes, the complete and activated forms of the spliceosome, at 15°, 24°, and 33°C (Fig. 6B, lanes 2–4,11,12; data not shown). Extracts from U4/U6-Trip1 rapidly assemble pre-mRNA complexes at 15°, 24°, and 33°C (Fig. 6B, lanes 8–10; data not shown). Intriguingly, at all three temperatures, the major U4/U6-Trip1 complex has a gel mobility slower than fully assembled spliceosomes, but faster than activated spliceosomes.
One possible explanation for the aberrant gel migration of U4/U6-Trip1 pre-mRNA complexes is that they are stalled intermediates in spliceosome activation. If this is the case, the complexes may retain U1 snRNP, U4 snRNP, or both. To determine the snRNP composition of the aberrant complex formed by U4/U6-Trip1 extracts, we affinity purified spliceosomes using biotinylated pre-mRNA and streptavidin-agarose beads, then analyzed their snRNA content by solution hybridization (Kuhn et al. 1999).
Spliceosomes purified from U4-cs1 control extracts at 15°C are arrested prior to activation, and thus contain both U1 RNA and U4/U6 complex, but not “free” U6 RNA (Kuhn et al. 1999) (Fig. 7A, lane 1). Purification of spliceosomes is dependent on the presence of biotin label, as negligible amounts of complex are recovered from the no-biotin control (Fig. 7A, lane 2). In contrast to the U4-cs1 control, wild-type extracts contain both complete and activated forms of the spliceosome at both 24° and 34°C, as judged by the presence of both U4/U6 complex and free U6 (Fig. 7A, lanes 3,4). U4/U6-Trip1 extracts also assembled complexes containing U4/U6 and free U6 at 24° and 34°C (Fig. 7A, lanes 7,8). In order to test for the presence of free U4 RNA, wild-type and U4/U6-Trip1 complexes assembled at 34°C were affinity purified and hybridized with probes for U1, U2, and U4 RNAs (Fig. 7A, lanes 5,9). As a marker for free U4 RNA, duplicate samples were incubated at 65°C to denaturate any U4/U6 complex. No accumulation of free U4 was detected in either wild-type or U4/U6-Trip1-purified complexes.
FIGURE 7.
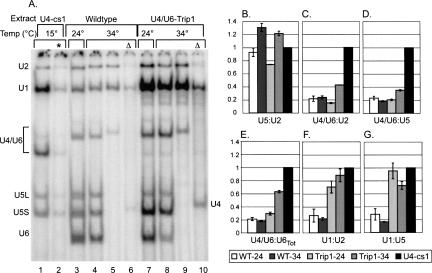
U4/U6-Trip1 aberrant spliceosomes are defective for U1 release and U4/U6 unwinding. (A) Affinity purification of U4/U6-Trip1 splicing complexes. Splicing complexes were assembled onto biotinylated pre-mRNA at the temperatures shown for 30 (lanes 1,2) or 15 (lanes 3–10) min. Complexes were affinity purified using streptavidin-agarose beads. After washing the beads, splicing complexes were digested with proteinase K and their associated snRNAs were analyzed by solution hybridization using probes for U1, U2, U5, and U6 (lanes 1–4,7,8) or for U1, U2, and U4 (lanes 5,6,9,10). Yeast U5 is processed into long (U5L) and short (U5S) forms. U4-cs1/U6 complex has an aberrant (fast) mobility (Li and Brow 1996). Nonbiotinylated pre-mRNA was used as a control for nonspecific binding (*, lane 2). To determine the position of free U4 RNA on the gel, samples were heated at 65°C before electrophoresis (Δ). (B–G) Quantification of affinity purified complexes. The U5:U2 (B), U4/U6:U2 (C), U4/U6:U5 (D), U4/U6:U6total (E), U1:U2 (F), and U1:U5 (G) ratios were normalized to ratios from cold-arrested U4cs1 complexes. Data shown are an average of duplicate experiments, with bars indicating the range.
The spliceosomes assembled in U4/U6-Trip1 extract appeared to contain a deficit of free U6 RNA at 34°C, and an excess of U1 RNA at both 24° and 34°C. To confirm these observations, we quantified the relative snRNA levels in the affinity-purified complexes. Because U4-cs1 extract predominantly accumulates complete spliceosomes at low temperature (Kuhn et al. 1999), the signal from U4-cs1 affinity-purified complexes represents ∼1:1 stoichiometries for the snRNAs and was used as a normalization control. In order to assess the efficiency of tri-snRNP addition, we first analyzed the U5:U2 ratio. Complexes from both wild-type and U4/U6-Trip1 extracts had U5:U2 ratios similar to a 1:1 stoichiometry (Fig. 7B), suggesting that the majority of purified complexes were either assembled or activated spliceosomes, and not pre-spliceosomes (which lack U4/U5/U6 tri-snRNP). However, U4/U6-Trip1 complexes purified from extracts incubated at 24°C did have a somewhat reduced U5:U2 ratio (∼0.75), implying that a small fraction of these complexes are pre-spliceosomes. This finding suggests a mild tri-snRNP addition defect at 24°C.
The U4/U6:U2 ratio was used to investigate the activation status of purified complexes. Wild-type complexes assembled at 24° and 34°C, and U4/U6-Trip1 complexes assembled at 24°C had equally low U4/U6:U2 ratios of about 0.2 (Fig. 7C), consistent with efficient activation of assembled spliceosomes. In contrast, U4/U6-Trip1 complexes assembled at 34°C had an approximately twofold higher U4/U6:U2 ratio (Fig. 7C). Similar results were obtained by calculating the U4/U6:U5 ratio (Fig. 7D) and the fraction of total U6 signal found in the U4/U6 band (Fig. 7E). Thus, U4/U6 unwinding in U4/U6-Trip1 spliceosomes is impaired at 34°C.
To determine the U1 RNA stoichiometry, we analyzed U1:U2 and U1:U5 ratios. Regardless of assembly temperature, wild-type complexes had U1:U2 ratios approximately equal to their U4/U6:U2 ratios (Fig. 7, cf. C and F), as would be expected for activated spliceosomes that have released both U1 and U4 snRNPs in a coordinated manner. Strikingly, U4/U6-Trip1 complexes assembled at 24° and 34°C had much higher U1:U2 ratios (Fig. 7F), consistent with a failure of activated spliceosomes to release U1 snRNP. Measurement of the U1:U5 ratios confirms the U1 retention defect, although this value is slightly inflated in the 24°C complexes due to the mild tri-snRNP addition defect (Fig. 7G). Therefore, U4/U6-Trip1 spliceosomes are impaired for U1 release even at splicing permissive temperature and have reduced U4/U6 unwinding at nonpermissive temperature.
DISCUSSION
Here we report an analysis of the U6 ISL bulge region mutations C67A, C67G, A79G, and U80G. Our data show that the lethality of C67A and C67G is due to loss of base pairing with residue G14 of U4 RNA, as compensatory mutations of U4-G14 completely suppress the C67A and C67G growth defects. The more severe growth defects associated with U6-C67A and C67G compared with U4-G14 mutations is likely due to mispairing between C67 and U80. Similarly, our results suggest that mispairing in the bulge region contributes to the A79G and U80G growth defects. However, suppressor analysis of U6-A79G and U80G suggests that these mutations also disrupt other interactions. Below, we discuss the possible functions of U6-A79 and U80 in spliceosome activation.
Both cross-ISL pairing and at least one other defect contribute to A79G and U80G growth phenotypes
ISL/telestem stabilization by A79G is likely a major contributor to its cold-sensitive phenotype, since destabilizing mutations in both helices were selected as suppressors. However, three mutations predicted to disrupt either the ISL (C66U) or telestem (A42G and C43U) enhance the A79G growth defect. Furthermore, we found that A79G greatly enhances the otherwise viable C67G/U4-G14C mutation (Fig. 4), even though A79G does not have the potential to form a stable base pair with C67G. These results suggest that the cold-sensitive growth of the A79G strain is due both to stabilization of the ISL and at least one other defect. We suggest that this defect is disruption of an intermolecular rather than intramolelcular interaction, possibly involving the intron 5′ splice site and/or Prp24 (see below).
We found that all 18 U6-C67/A79 and C67/U80 double mutations confer lethality, including many that do not have the potential to form stable intramolecular base pairs. Since all tested lethal double mutations showed low levels of U4/U6 complex, their phenotypes may be due, at least in part, to a bi-snRNP assembly defect. Given that binding of Prp24 to U6 in vitro is weakened by both the A79G and U80G mutations (Kwan and Brow 2005; S. Kwan and D. Brow, unpubl.), the assembly defect may result from the combined effects of reduced U4/U6 stability and disruption of Prp24 binding.
We describe the first identified suppressors of U6-U80G. Disruption of C67/G80 pairing by either the C67A or C67G mutation (in the presence of the compensatory U4 position 14 mutation) partially suppresses U80G lethality, but both heat-and cold-sensitive growth defects persist in the U4/U6-Trip strains (Fig. 4). The modest U4/U6 complex assembly defect observed in the U4/U6-Trip1 and Trip2 strains does not correlate with the heat-sensitive growth defect, indicating that inhibition of U4/U6 formation is not a major contributor to the growth defect. These results suggest that the splicing block in U4/U6 Trip strains occurs after U4/U6 assembly.
Function of the ISL bulge in coordination of U4/U6 unwinding and U1 snRNP release
Our analysis of whole-cell extracts from the U4/U6-Trip1 strain revealed altered native gel mobility of the U4/U5/U6 tri-snRNP, which could reflect a difference in composition, conformation, or both. Our results cannot discriminate between these possibilities; however, the snRNPs are functional for splicing at room temperature, so it is unlikely that they are missing essential factors. One possibility is that U4/U6-Trip1 tri-snRNPs are less stable than their wild-type counterparts and lose weakly associated factors during gel electrophoresis. Alternatively or additionally, these tri-snRNPs may have altered conformation due to changes in U4/U6 stem II base pairing. Either alteration could account for the weak tri-snRNP addition defect we observed with U4/U6-Trip1 extracts.
Extracts from the U4/U6-Trip1 mutant form pre-mRNA complexes that migrate aberrantly on native gels. Strikingly, spliceosomes affinity purified from U4/U6-Trip1 extract maintain high levels of U1, despite having reduced amounts of U4/U6. This observation suggests a previously unknown function for the ISL; coordinating U4/U6 unwinding and U1 snRNP release. Although they are defective for U1 release, U4/U6-Trip1 spliceosomes are catalytically active at room temperature. There are two possible explanations for this observation, (1) a small fraction of U4/U6-Trip1 spliceosomes release U1 snRNP entirely and only this subpopulation is competent for splicing, or (2) U4/U6-Trip1 spliceosomes that retain U1 snRNP are still able to splice. We cannot yet discriminate between these possibilities. At elevated temperatures, U4/U6-Trip1 extracts are impaired for both U1 snRNP release and U4/U6 unwinding. However, U4/U6 unwinding is not completely blocked, as a small amount of free U6 was observed. The fact that splicing was almost completely inhibited even after significant U4/U6 unwinding suggests that the Trip1 mutation may also block splicing catalysis, perhaps due to loss of Mg++ binding or disruption of other interactions in the active site.
The heat sensitivity of the U4/U6 unwinding defect is unusual, since mutations that block U4/U6 unwinding typically result in cold-sensitive growth defects (e.g., brr2–1 and U4-cs1). However, mutations have been reported in the U5 snRNP protein Snu114 that cause heat-sensitive U4/U6 unwinding defects, presumably by disrupting physical interactions with other factors (Bartels et al. 2003; Brenner and Guthrie 2005). The heat sensitivity of U4/U6 unwinding in the U4/U6-Trip1 strain may result from similar disruption of U6 ISL interactions important for spliceosome activation. We propose that sequences in the U6 ISL bulge region make important contacts that promote both U1 snRNP release and U4/U6 unwinding,
Possible direct and indirect mechanisms for promotion of U1 snRNP release and U4/U6 unwinding by the U6 ISL bulge region
The assembled spliceosome contains a network of RNA:RNA interactions, in which U1 is base paired to the 5′splice site of the intron, U2 is base paired to the branchpoint region, and U4 and U6 RNAs are base paired together. Spliceosome activation involves several RNA base-pairing rearrangements that are policed by multiple proteins (Brow 2002). GTP-bound Snu114 derepresses the helicase Brr2, which unwinds U4/U6 (Bartels et al. 2002; Small et al. 2006). The helicase Prp28 also becomes activated and catalyzes the transfer of the 5′ splice site from base pairing with U1 to base pairing with U6 (Staley and Guthrie 1999). Importantly, U1 release normally requires U4/U6 unwinding (Kuhn et al. 1999) and vice versa (Staley and Guthrie 1999). The mechanism by which these processes are coupled is not well understood. Here, we report the remarkable uncoupling of U1 release and U4/U6 unwinding in U4/U6-Trip1 spliceosomes. The simplest interpretation of our data is that recognition of sequences in the U6 ISL bulge region during U4/U6 unwinding signals U1 release.
RNA internal loops and bulges often function as recognition sites for proteins, metal ions, and other RNAs (Hermann and Patel 2000). It is possible that the U6 ISL bulge functions directly by modulating interactions between U6 and the 5′splice site. Early in activation, the ACA box at U6 nucleotides 42–44 base pairs with the highly conserved UGU sequence found in the intron at positions 4–6 (Sawa and Abelson 1992; Li and Brow 1996; Johnson and Abelson 2001; Chan et al. 2003). Upon complete unwinding of U4/U6, this pairing interaction is switched to the U6 ACA box at positions 47–49. Hydroxyl-radical cleavage experiments have shown that sequences in the human U6 ISL bulge region are in close proximity to the U6–5′ splice-site helix in activated spliceosomes (Rhode et al. 2006). In group II self-splicing introns, the λ–λ′ interaction between Domains 1 and 5 brings the D5 bulge region into close proximity to the ε–ε′ interaction (Boudvillain et al. 2000), the functional homolog of the U6–5′ splice-site pairing (Jacquier and Michel 1990). Additionally, Gumbs et al. (2006) recently reported that the Domain 5 bulge region undergoes conformational changes upon docking into the active site of a group II intron. Combined with the data reported herein, these results are consistent with a model in which sequences in the U6 ISL bulge region function in spliceosome activation by stabilizing the U6–5′ splice-site interaction through tertiary contacts similar to λ–λ′ (Fig. 8; Boudvillain et al. 2000; Yean et al. 2000). Disruption of such stabilization by the A79G mutation could explain the enhancement of this mutation by mutations in A42 and C43, which pair with the 5′ splice site.
FIGURE 8.
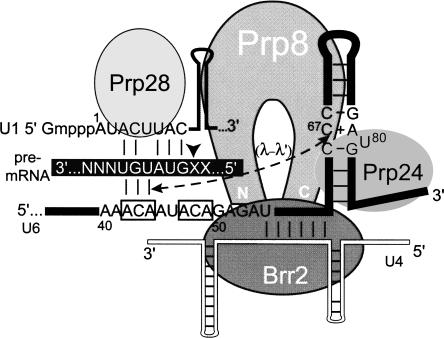
Model of U6 ISL bulge function during spliceosome activation. The pre-mRNA is highlighted in black with an arrowhead designating the 5′ splice site. “N” and “X” represent nonconserved intron and exon sequences, respectively. The bulge region might promote U1 release directly (dashed arrow), by stabilizing interactions between U6 ACA boxes (nucleotides 42–44 and/or 47–49), analogous to the λ–λ′ tertiary interaction of group II self-splicing introns, or indirectly, through recognition by Prp24, Prp8, or other splicing factors. The N and C termini of Prp8 are labeled. See text for descriptions of the other factors.
Alternatively, sequences in the bulge region could indirectly promote U1 release. The U6 ISL may function in spliceosome activation through interactions with Prp24 or another splicing protein. Prp24 has been proposed to promote spliceosome activation because loss-of-function Prp24 alleles enhance the growth defects of mutations in other activation factors. The prp24-1 allele results in synthetic lethality with a mutation in PRP28 (Strauss and Guthrie 1991). In addition, the prp24-RRM3sub mutation enhances the growth defect of the snu114-60 allele (Brenner and Guthrie 2005). Similarly, substitutions in Prp24 RRM 2 (R158S) and RRM 3 (F257I) synthetically enhance the cold-sensitive growth defect of U4-cs1 (Kuhn and Brow 2000).
Prp24 also has genetic and biochemical interactions with U6-C67, U6-A79, and U6-U80. Mutations in Prp24 RRMs 2 and 3 that are synthetically lethal with U6-A79G (Vidaver 1999) fully suppress C67A, but not C67A/U80G (our unpublished results). In addition, the intragenic enhancers of A79G found in this study, A42G, C43U, C61U, and C66U, lie in regions that have been implicated as Prp24-binding sites (Shannon and Guthrie 1991; Ghetti et al. 1995; Jandrositz and Guthrie 1995; Kwan and Brow 2005; Karaduman et al. 2006). Notably, A79G and U80G decrease the affinity of Prp24 for U6 in vitro (Kwan and Brow 2005; S. Kwan and D. Brow, unpubl.). Therefore U6-A79G and U80G may block spliceosome activation by altering Prp24/U6 binding interactions in such a way that Prp28 is not activated to release U1 snRNP.
It has been proposed that the U5 snRNP protein Prp8 regulates the activities of the putative U4/U6 helicase Brr2, and the putative U1/5′ splice-site helicase Prp28, because mutations in Prp8 suppress the cold-sensitive prp28-1 and brr2-1 alleles (Kuhn et al. 2002). The results of two hybrid experiments suggest that both the N- and C-terminal regions of Prp8 bind to Brr2 (Dix et al. 1998; Grainger and Beggs 2005). Two-hybrid interactions were also observed between the Prp8 N-terminal region and the U1 snRNP proteins Prp39, Prp40, and Snp1 (Abovich and Rosbash 1997; Awasthi et al. 2001; van Nues and Beggs 2001). It is conceivable that both the U4/U6 unwinding defect and the U1 snRNP release defect observed in U4/U6-Trip1 extracts are due to the failure of U6-U80G to allosterically induce a conformational change in Prp8 that normally results in Brr2 activation and displacement of U1 snRNP proteins.
In summary, we provide evidence that mutations in the U6 ISL bulge region can alter at least four processes in the splicing cycle: U4/U6 complex formation, tri-snRNP assembly, U1 snRNP release, and U4/U6 unwinding. Our data suggest recognition of sequences in the U6 ISL bulge region promotes coupled U4/U6 unwinding and U1 release (Fig. 8). This recognition could occur directly through interaction with U6:5′ splice site helix, or indirectly through the function of protein factors involved in activation. The identities of factors involved in recognizing the ISL bulge remain unknown; however, Prp24 and Prp8 are likely candidates. Further experiments will be necessary to identify the exact contacts made by the ISL bulge during activation.
MATERIALS AND METHODS
Plasmids and construction of alleles
snr6-A79C, A79U, U80A, U80C, and U80G alleles carried on the yeast-Escherichia coli shuttle vector pSX6T (CEN4, ARS1, TRP1) were kind gifts from David McPheeters (Case Western Reserve University) (McPheeters 1996). SNR6-C67A, C67G, and C67U were constructed via site-directed mutagenesis of pRS314-544H6 (CEN6, ARSH4, TRP1) (Kaiser and Brow 1995). The snr6-A79G allele in pSE358 was described previously (Fortner et al. 1994). snr6 C67H/A79B and C67H/U80V alleles were constructed by EcoRI/EcoNI digestion of the single-mutant plasmids and ligation of the C67H fragments into the A79B and U80V-containing vectors. snr6-C61U, 61A62, G63A, C66U, C61U/A79G, 61A62/A79G, A62C/A79G, A62U/A79G G63A/A79G, and C66U/A79G were constructed by ligation of synthetic DNAs with BclI/EcoNI compatible ends into BclI/EcoNI-cut pRS314-539H6 and pRS314-539H6-A79G. pRS316-U4wt-U6mini (CEN6, ARSH4, URA3) was constructed by ligation of a PCR-amplified region (from −48 to +260) of SNR6 into the BamHI digestion product of pRS316-U4wt (Kuhn et al. 2002). SNR14 mutations G14A and G14U were created by Quikchange mutagenesis (Stratagene) of pRS313-U4wt; pRS313-snr14-G14C was described previously (Kuhn et al. 1999). pJPS149, a Bluescript plasmid containing the actin pre-mRNA construct, was a kind gift from Jon Staley (University of Chicago) (Mayas et al. 2006).
Yeast strains
MWK027 (MATα, trp1-1, ura3-1, leu2-3,112, his3-11,15, ade2-1, can1-100, GAL2+, met2-1, lys2-2, Δsnr6∷LEU2, [YCp50-ψwt]) was previously described (Kaiser and Brow 1995). MWK027-A79G is a version of MWK027 containing YCp50-snr6-A79G (URA3) in place of YCp50-ψwt. CJM000 (MATa, snr6∷LEU2, snr14∷trp1∷ADE2, trp1, ura3, lys2, his3, ade2, [pRS316-U4wt-U6mini]) was derived from ANK640 (Kuhn et al. 2002) by the following manipulations. The TRP1 marker of ANK640 was converted to an ADE2 marker by transformation with the BamHI digestion product of plasmid M3938 (Voth et al. 2003), selection on –Ade media, and verification of tryptophan auxotrophy. The resulting strain was transformed with pRS314-544H6 and loss of YCp50-39D6 was selected by plating onto media containing 5-fluoroorotic acid (5-FOA). The subsequent strain was transformed with pRS316-U4wt-U6mini. Loss of plasmids pRS317-U4-wt and pRS314–544H6 was monitored by replica plating onto –lys and –trp media.
Growth assays
Plasmids containing the SNR6 alleles described above were transformed into MWK027. Single-colony transformants were selected from plates containing media lacking tryptophan (−trp) and grown overnight in liquid –trp media before being plated onto (−trp) media containing 5-FOA to select against the plasmid containing the pseudo-wild-type SNR6. Plasmids bearing SNR14 and SNR6 alleles were simultaneously transformed into CJM000, and transformants were selected on plates lacking both tryptophan and lysine, grown overnight in liquid –trp, −lys medium, and plated onto (−trp) (−lys) medium containing 5-FOA to select for the loss of pRS316−U4wt-U6mini.
Genetic suppression analysis
In order to generate random mutations in snr6-A79G, we amplified the −799 to +233 region of pRS314-539H6-A79G in the presence of 40–100 μM MnCl2 and a threefold increase in individual dNTP concentrations. MWK027-A79G was cotransformed with the large fragment of EcoRI/EcoNI-digested pRS314–539H6-A79G and the mutagenized PCR product, as previously described (Muhlrad et al. 1992). Plasmid repair was selected by plating onto –trp medium and incubation at 30°C. Loss of plasmid YCp50-SNR6-A79G was selected by replica plating onto –trp 5-FOA. Suppression was determined by replica plating onto YEPD at 16°C. Robustly growing colonies were picked and their genomic DNA was prepared as described (Hoffman and Winston 1987). The SNR6 coding region was PCR amplified and sequenced to identify suppressor mutations. Candidate suppressor mutations were subcloned from recovered plasmids into pRS314-539H6-A79G and transformed into MWK027. Transformants were cured of YCp50-ψwt by plating on 5-FOA and were tested for heat and cold sensitivity by serial dilution plating onto YEPD.
In vitro splicing
Splicing extracts were prepared using the liquid nitrogen method, and pre-mRNA splicing was performed essentially as previously described (Umen and Guthrie 1995), with the exception that U4/U6-Trip1 extracts were prepared by resuspending washed cells in 0.2 vol lysis buffer (as opposed to 0.4 vol) to increase total protein concentration and stabilize the extracts. Splicing was performed as described (Lin et al. 1985). Pre-mRNA was transcribed using plasmid pJPS149 as described (Mayas et al. 2006). Native gel analysis of splicing complexes was performed as described (Cheng and Abelson 1987), with the exception that gels (4% polyacrylamide 80:1) were run in 50 mM TBE buffer at 10V/cm for 17 h at 4°C.
Affinity purification of splicing complexes was adapted from Kuhn et al. (1999). Biotinylated pre-mRNA was transcribed in vitro in the presence of 1:20 Bio11-UTP (Ambion):UTP. Streptavidin-agarose beads were prepared by washing twice with 10 vol “washing buffer” (60 mM Hepes at pH 7.6, 150 mM NaCl, 3 mM MgCl2, 0.5 mM DTT, 15% glycerol, 0.05% NP-40), blocked in 10 vol “general buffer” (40% Buffer D, 60 mM KPO4 at pH 7, 2.5 mM MgCl2, 3% PEG 8000, 20 mM EDTA, 0.2 mM DTT) with 0.1 mg/mL BSA, 0.1 mg/mL tRNA, and 0.1 mg/mL glycogen for 1 h at 4°C, and then resuspended in 2 vol “general buffer” and kept on ice. Splicing reactions (20 μL) were stopped by the addition of 75 μL ice-cold “general buffer” containing 25 μL prepared streptavidin-agarose beads (Pierce). Splicing complexes were bound to the beads for 1 h at 4°C, washed three times with 20 vol “washing buffer,” and eluted in 50 μL solution hybridization buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% SDS) containing 1 mg/mL proteinase K for 30 min at 37°C. After proteinase K digestion, snRNAs were detected by solution hybridization using oligos U1-SH, U2-SH (Kuhn et al. 1999), U6-6D, U5B, and U4-14B (Li and Brow 1993).
snRNA and snRNP analysis
U4/U6 complex assembly was assayed by solution hybridization as previously described (Li and Brow 1993). Briefly, total cellular RNA was prepared under nondenaturing conditions (Treco 1989) from strains grown in media lacking tryptophan and histidine to select for maintenance of both the U6 and U4 alleles, respectively. A total of 1–5 μg of total RNA were resuspended in 3 μL of DEPC-treated ddH2O, added to 2 μL annealing buffer (150 mM NaCl, 50 mM Tris-Cl at pH 7.5, 1 mM EDTA final), and 5′ 32P-labeled oligonucleotide probe (0.2 pmoles U1-SH and U6-D or U4–14B), and incubated for 15 min at 37°C. Assays were electrophoresed at 150 V/cm for ∼4 h at 4°C on a 9% polyacrylamide gel.
SnRNP levels were analyzed as described (Raghunathan and Guthrie 1998b). Splicing reactions (5 μL; +/− ATP) lacking pre-mRNA were incubated 30 min at room temperature and loaded on a 4% polyacrylamide (80:1) gel. Gels were run at 13 V/cm for 5 h at 4°C in 50 mM Tris/50 mM Glycine/2 mM MgCl2 and electro-transferred onto Hybond-N membranes overnight in 23 mM TBE. Blots were probed for U1, U2, U4, and U5 using radiolabeled DNA probes prepared according to Davis and Ares (2006) under the hybridization conditions described by Ruby (1999). The U6 snRNA was detected using a kinased oligo U6-dep (5′-ATCTCTGTATTGTTTCAAATTGACCAA-3′).
ACKNOWLEDGMENTS
We thank David McPheeters and David Stillman for kindly sharing plasmids; Hyunsic Choi for initial subcloning of some U6 double-mutant plasmids; Rabiah Mayas and Jon Staley for in vitro splicing protocols, materials, and helpful advice; members of the Brow, Dahlberg, and Butcher laboratories for helpful advice and discussions; and Sharon Kwan, Janina Görnemann, Jon Staley, and anonymous reviewers for critical reading and comments on the manuscript. This work was supported by grants GM54018 (D.A.B.) and GM65166 (S.E.B.) from the National Institutes of Health (NIH). C.J.M. was supported by NIH predoctoral training grant T32 GM7215.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.699907.
REFERENCES
- Abovich, N., Rosbash, M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- Awasthi, S., Palmer, R., Castro, M., Mobarak, C.D., Ruby, S.W. New roles for the Snp1 and Exo84 proteins in yeast pre-mRNA splicing. J. Biol. Chem. 2001;276:31004–31015. doi: 10.1074/jbc.M100022200. [DOI] [PubMed] [Google Scholar]
- Bartels, C., Klatt, C., Lührmann, R., Fabrizio, P. The ribosomal translocase homologue Snu114p is involved in unwinding U4/U6 RNA during activation of the spliceosome. EMBO Rep. 2002;3:875–880. doi: 10.1093/embo-reports/kvf172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels, C., Urlaub, H., Lührmann, R., Fabrizio, P. Mutagenesis suggests several roles of Snu114p in pre-mRNA splicing. J. Biol. Chem. 2003;278:28324–28334. doi: 10.1074/jbc.M303043200. [DOI] [PubMed] [Google Scholar]
- Blad, H., Reiter, N.J., Abildgaard, F., Markley, J.L., Butcher, S.E. Dynamics and metal ion binding in the U6 RNA intramolecular stem–loop as analyzed by NMR. J. Mol. Biol. 2005;353:540–555. doi: 10.1016/j.jmb.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Boudvillain, M., de Lencastre, A., Pyle, A.M. A tertiary interaction that links active-site domains to the 5′ splice site of a group II intron. Nature. 2000;406:315–318. doi: 10.1038/35018589. [DOI] [PubMed] [Google Scholar]
- Brenner, T.J., Guthrie, C. Genetic analysis reveals a role for the C terminus of the Saccharomyces cerevisiae GTPase Snu114 during spliceosome activation. Genetics. 2005;170:1063–1080. doi: 10.1534/genetics.105.042044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow, D.A. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- Brow, D.A., Vidaver, R.M. An element in human U6 RNA destabilizes the U4/U6 spliceosomal RNA complex. RNA. 1995;1:122–131. [PMC free article] [PubMed] [Google Scholar]
- Chan, S.P., Kao, D.I., Tsai, W.Y., Cheng, S.C. The Prp19p-associated complex in spliceosome activation. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- Cheng, S.C., Abelson, J. Spliceosome assembly in yeast. Genes & Dev. 1987;1:1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- Davis, C.A., Ares M., Jr Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. 2006;103:3262–3267. doi: 10.1073/pnas.0507783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix, I., Russell, C.S., O'Keefe, R.T., Newman, A.J., Beggs, J.D. Protein-RNA interactions in the U5 snRNP of Saccharomyces cerevisiae . RNA. 1998;4:1675–1686. doi: 10.1017/s1355838298412998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgavish, T., Cannone, J.J., Lee, J.C., Harvey, S.C., Gutell, R.R. AA.AG@helix.ends: A:A and A:G base-pairs at the ends of 16S and 23S rRNA helices. J. Mol. Biol. 2001;310:735–753. doi: 10.1006/jmbi.2001.4807. [DOI] [PubMed] [Google Scholar]
- Fortner, D.M., Troy, R.G., Brow, D.A. A stem/loop in U6 RNA defines a conformational switch required for pre-mRNA splicing. Genes & Dev. 1994;8:221–233. doi: 10.1101/gad.8.2.221. [DOI] [PubMed] [Google Scholar]
- Ghetti, A., Company, M., Abelson, J. Specificity of Prp24 binding to RNA: A role for Prp24 in the dynamic interaction of U4 and U6 snRNAs. RNA. 1995;1:132–145. [PMC free article] [PubMed] [Google Scholar]
- Grainger, R.J., Beggs, J.D. Prp8 protein: At the heart of the spliceosome. RNA. 2005;11:533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbs, O.H., Padgett, R.A., Dayie, K.T. Fluorescence and solution NMR study of the active site of a 160-kDa group II intron ribozyme. RNA. 2006;12:1693–1707. doi: 10.1261/rna.137006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann, T., Patel, D.J. RNA bulges as architectural and recognition motifs. Structure. 2000;8:R47–R54. doi: 10.1016/s0969-2126(00)00110-6. [DOI] [PubMed] [Google Scholar]
- Hoffman, C.S., Winston, F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli . Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Jacquier, A., Michel, F. Base-pairing interactions involving the 5′ and 3′-terminal nucleotides of group II self-splicing introns. J. Mol. Biol. 1990;213:437–447. doi: 10.1016/S0022-2836(05)80206-2. [DOI] [PubMed] [Google Scholar]
- Jandrositz, A., Guthrie, C. Evidence for a Prp24 binding site in U6 snRNA and in a putative intermediate in the annealing of U6 and U4 snRNAs. EMBO J. 1995;14:820–832. doi: 10.1002/j.1460-2075.1995.tb07060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, T.L., Abelson, J. Characterization of U4 and U6 interactions with the 5′ splice site using a S. cerevisiae in vitro trans-splicing system. Genes & Dev. 2001;15:1957–1970. doi: 10.1101/gad.895601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, M.W., Brow, D.A. Lethal mutations in a yeast U6 RNA gene B block promoter element identify essential contacts with transcription factor-IIIC. J. Biol. Chem. 1995;270:11398–11405. doi: 10.1074/jbc.270.19.11398. [DOI] [PubMed] [Google Scholar]
- Karaduman, R., Fabrizio, P., Hartmuth, K., Urlaub, H., Lührmann, R. RNA structure and RNA-protein interactions in purified yeast U6 snRNPs. J. Mol. Biol. 2006;356:1248–1262. doi: 10.1016/j.jmb.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Kuhn, A.N., Brow, D.A. Suppressors of a cold-sensitive mutation in yeast U4 RNA define five domains in the splicing factor Prp8 that influence spliceosome activation. Genetics. 2000;155:1667–1682. doi: 10.1093/genetics/155.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, A.N., Li, Z., Brow, D.A. Splicing factor Prp8 governs U4/U6 RNA unwinding during activation of the spliceosome. Mol. Cell. 1999;3:65–75. doi: 10.1016/s1097-2765(00)80175-6. [DOI] [PubMed] [Google Scholar]
- Kuhn, A.N., Reichl, E.M., Brow, D.A. Distinct domains of splicing factor Prp8 mediate different aspects of spliceosome activation. Proc. Natl. Acad. Sci. 2002;99:9145–9149. doi: 10.1073/pnas.102304299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan, S.S., Brow, D.A. The N- and C-terminal RNA recognition motifs of splicing factor Prp24 have distinct functions in U6 RNA binding. RNA. 2005;11:808–820. doi: 10.1261/rna.2010905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Brow, D.A. A rapid assay for quantitative detection of specific RNAs. Nucleic Acids Res. 1993;21:4645–4646. doi: 10.1093/nar/21.19.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Brow, D.A. A spontaneous duplication in U6 spliceosomal RNA uncouples the early and late functions of the ACAGA element in vivo. RNA. 1996;2:879–894. [PMC free article] [PubMed] [Google Scholar]
- Lin, R.J., Newman, A.J., Cheng, S.C., Abelson, J. Yeast mRNA splicing in vitro. J. Biol. Chem. 1985;260:14780–14792. [PubMed] [Google Scholar]
- Madhani, H.D., Bordonne, R., Guthrie, C. Multiple roles for U6 snRNA in the splicing pathway. Genes & Dev. 1990;4:2264–2277. doi: 10.1101/gad.4.12b.2264. [DOI] [PubMed] [Google Scholar]
- Mayas, R.M., Maita, H., Staley, J.P. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat. Struct. Mol. Biol. 2006;13:482–490. doi: 10.1038/nsmb1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPheeters, D.S. Interactions of the yeast U6 RNA with the pre-mRNA branch site. RNA. 1996;2:1110–1123. [PMC free article] [PubMed] [Google Scholar]
- Muhlrad, D., Hunter, R., Parker, R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- Nilsen, T.W. RNA–RNA interactions in nuclear pre-mRNA splicing. In: Simons R., Grunberg-Marago M., editors. RNA structure and function. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1998. pp. 279–307. [Google Scholar]
- Raghunathan, P.L., Guthrie, C. A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science. 1998a;279:857–860. doi: 10.1126/science.279.5352.857. [DOI] [PubMed] [Google Scholar]
- Raghunathan, P.L., Guthrie, C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol. 1998b;8:847–855. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- Reiter, N.J., Blad, H., Abildgaard, F., Butcher, S.E. Dynamics in the U6 RNA intramolecular stem–loop: A base flipping conformational change. Biochemistry. 2004;43:13739–13747. doi: 10.1021/bi048815y. [DOI] [PubMed] [Google Scholar]
- Rhode, B.M., Hartmuth, K., Westhof, E., Lührmann, R. Proximity of conserved U6 and U2 snRNA elements to the 5′ splice site region in activated spliceosomes. EMBO J. 2006;25:2475–2486. doi: 10.1038/sj.emboj.7601134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby, S.W. A yeast spliceosome assay. Methods Mol. Biol. 1999;118:365–390. doi: 10.1385/1-59259-676-2:365. [DOI] [PubMed] [Google Scholar]
- Ryan, D.E., Abelson, J. The conserved central domain of yeast U6 snRNA: Importance of U2-U6 helix Ia in spliceosome assembly. RNA. 2002;8:997–1010. doi: 10.1017/s1355838202025013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital, D.G., Allmann, A.M., Van Doren, S.R., Butcher, S.E. Structural basis for a lethal mutation in U6 RNA. Biochemistry. 2003;42:1470–1477. doi: 10.1021/bi027137h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital, D.G., Cornilescu, G., McManus, C.J., Brow, D.A., Butcher, S.E. U2-U6 RNA folding reveals a group II intron-like domain and a four-helix junction. Nat. Struct. Mol. Biol. 2004;11:1237–1242. doi: 10.1038/nsmb863. [DOI] [PubMed] [Google Scholar]
- Sawa, H., Abelson, J. Evidence for a base-pairing interaction between U6 small nuclear RNA and 5′ splice site during the splicing reaction in yeast. Proc. Natl. Acad. Sci. 1992;89:11269–11273. doi: 10.1073/pnas.89.23.11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, U., Podar, M., Stahl, U., Perlman, P.S. Mutations of the two-nucleotide bulge of D5 of a group II intron block splicing in vitro and in vivo: Phenotypes and suppressor mutations. RNA. 1996;2:1161–1172. [PMC free article] [PubMed] [Google Scholar]
- Seetharaman, M., Eldho, N.V., Padgett, R.A., Dayie, K.T. Structure of a self-splicing group II intron catalytic effector domain 5: Parallels with spliceosomal U6 RNA. RNA. 2006;12:235–247. doi: 10.1261/rna.2237806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, K.W., Guthrie, C. Suppressors of a U4 snRNA mutation define a novel U6 snRNP protein with RNA-binding motifs. Genes & Dev. 1991;5:773–785. doi: 10.1101/gad.5.5.773. [DOI] [PubMed] [Google Scholar]
- Shukla, G.C., Padgett, R.A. A catalytically active group II intron Domain 5 can function in the U12-dependent spliceosome. Mol. Cell. 2002;9:1145–1150. doi: 10.1016/s1097-2765(02)00505-1. [DOI] [PubMed] [Google Scholar]
- Small, E.C., Leggett, S.R., Winans, A.A., Staley, J.P. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol. Cell. 2006;23:389–399. doi: 10.1016/j.molcel.2006.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley, J.P., Guthrie, C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Staley, J.P., Guthrie, C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- Strauss, E.J., Guthrie, C. A cold-sensitive mRNA splicing mutant is a member of the RNA helicase gene family. Genes & Dev. 1991;5:629–641. doi: 10.1101/gad.5.4.629. [DOI] [PubMed] [Google Scholar]
- Sun, J.S., Manley, J.L. The human U6 snRNA intramolecular helix: Structural constraints and lack of sequence specificity. RNA. 1997;3:514–526. [PMC free article] [PubMed] [Google Scholar]
- Treco, D. Preparation of yeast RNA. Saccharomyces cerevisiae, current protocols in molecular biology . In: Ausubel F.M., et al., editors. Greene Publishing Associates and Wiley-Interscience; New York: 1989. pp. 13.12.11–13.12.13. [Google Scholar]
- Umen, J.G., Guthrie, C. A novel role for a U5 snRNP protein in 3′ splice site selection. Genes & Dev. 1995;9:855–868. doi: 10.1101/gad.9.7.855. [DOI] [PubMed] [Google Scholar]
- Valadkhan, S., Manley, J.L. Characterization of the catalytic activity of U2 and U6 snRNAs. RNA. 2003;9:892–904. doi: 10.1261/rna.5440303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nues, R.W., Beggs, J.D. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae . Genetics. 2001;157:1451–1467. doi: 10.1093/genetics/157.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidaver, R.M. Cellular and molecular biology. University of Wisconsin-Madison; Madison, WI: 1999. Mechanisms of U6 RNA conformational changes during pre-mRNA splicing; p. 125. [Google Scholar]
- Vidaver, R.M., Fortner, D.M., Loos-Austin, L.S., Brow, D.A. Multiple functions of Saccharomyces cerevisiae splicing protein Prp24 in U6 RNA structural rearrangements. Genetics. 1999;153:1205–1218. doi: 10.1093/genetics/153.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth, W.P., Jiang, Y.W., Stillman, D.J. New “marker swap” plasmids for converting selectable markers on budding yeast gene disruptions and plasmids. Yeast. 2003;20:985–993. doi: 10.1002/yea.1018. [DOI] [PubMed] [Google Scholar]
- Will, C.L., Lührmann, R. Spliceosome structure and function. In: Gesteland R.F., et al., editors. The RNA world. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 369–400. [Google Scholar]
- Wolff, T., Bindereif, A. Conformational changes of U6 RNA during the spliceosome cycle: An intramolecular helix is essential both for initiating the U4-U6 interaction and for the first step of splicing. Genes & Dev. 1993;7:1377–1389. doi: 10.1101/gad.7.7b.1377. [DOI] [PubMed] [Google Scholar]
- Yean, S.L., Wuenschell, G., Termini, J., Lin, R.J. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature. 2000;408:881–884. doi: 10.1038/35048617. [DOI] [PMC free article] [PubMed] [Google Scholar]