Abstract
Ribosome-stimulated hydrolysis of guanosine-5′-triphosphate (GTP) by guanosine triphosphatase (GTPase) translation factors drives protein synthesis by the ribosome. Allosteric coupling of GTP hydrolysis by elongation factor Tu (EF-Tu) at the ribosomal GTPase center to messenger RNA (mRNA) codon:aminoacyl-transfer RNA (aa-tRNA) anticodon recognition at the ribosomal decoding site is essential for accurate and rapid aa-tRNA selection. Here we use single-molecule methods to investigate the mechanism of action of the antibiotic thiostrepton and show that the GTPase center of the ribosome has at least two discrete functions during aa-tRNA selection: binding of EF-Tu(GTP) and stimulation of GTP hydrolysis by the factor. We separate these two functions of the GTPase center and assign each to distinct, conserved structural regions of the ribosome. The data provide a specific model for the coupling between the decoding site and the GTPase center during aa-tRNA selection as well as a general mechanistic model for ribosome-stimulated GTP hydrolysis by GTPase translation factors.
Keywords: FRET, ribosome, single molecule biochemistry, tRNA selection
INTRODUCTION
The ribosome translates an mRNA substrate into a polypeptide chain through repetitive, mRNA-directed binding and incorporation of aa-tRNA substrates. aa-tRNAs are delivered to the two-subunit ribosome in ternary complex with EF-Tu and GTP. Selection of EF-Tu(GTP)aa-tRNA (ternary complex) requires both recognition of cognate codon:anticodon base pairing at the decoding site of the small ribosomal subunit and ribosome-stimulated hydrolysis of GTP by EF-Tu at the GTPase center of the large subunit. aa-tRNA selection is allosterically regulated, with recognition of a correctly base-paired codon:anticodon at the decoding site stimulating the rate of GTP hydrolysis at the GTPase center by 5 × 104 (Rodnina and Wintermeyer 2001; Blanchard et al. 2004a). While relatively well-developed mechanistic models exist for codon:anticodon recognition by the decoding site of the ribosome (Ogle and Ramakrishnan 2005), the mechanisms of GTPase activation and GTP hydrolysis by the GTPase center of the ribosome remain unclear (Rodnina et al. 2000).
Genetic and biochemical studies have identified conserved regions of ribosomal proteins L7/L12, L10, and L11 (Rosendahl and Douthwaite 1993; Rodnina et al. 2000), their 23S ribosomal RNA (rRNA) binding sites (nucleotides 1031–1123) (Rosendahl and Douthwaite 1993; Rodnina et al. 2000), and 23S rRNA nucleotides 2654–2665, known as the sarcin–ricin loop (SRL) (Rodnina et al. 2000), as components of the GTPase center. Cryoelectron microscopy (cryo-EM) has provided a structural model of the interaction of these components of the GTPase center with EF-Tu(GTP)aa-tRNA (Fig. 1; Stark et al. 1997; Valle et al. 2003). While these data provide an excellent biochemical and static structural representation of the GTPase center of the ribosome, dynamic information linking the known structural elements with the biochemical functions of the dynamic GTPase center (Bushuev et al. 1989; Porse et al. 1998; Ban et al. 2000; Seo and Cooperman 2002) is not available.
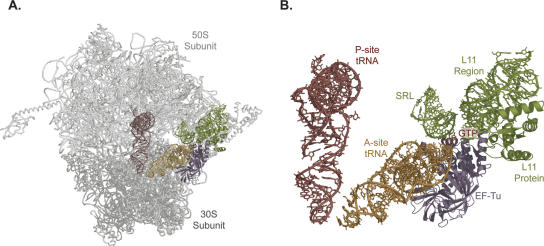
FIGURE 1.
Structural model of ternary complex stalled at the GTPase activation (mid- FRET) state using GDP and the antibiotic kirromycin (Valle et al. 2002, 2003). The model was generated based on previously published fits of the atomic coordinates of EF-Tu(GDP), aa-tRNA, small subunit helix 44, small subunit protein S12, large subunit helix 69, the SRL, and L11 with associated 23S rRNA to the cryo-EM map (Valle et al. 2002, 2003). The coordinates of these initial fits into the cryo-EM map were then used to fit the atomic coordinates of the small and large ribosomal subunits as well as the peptidyl-tRNA into the cryo-EM map. This was done using the small ribosomal subunit from Thermus thermophilus (Wimberly et al. 2000), the large ribosomal subunit from Haloarcula morismortui (Ban et al. 2000), and yeast tRNAPhe which had been previously fitted into the electron density from the 5.5 Å structure of the intact T. thermophilus ribosome. In the model presented here, the large ribosomal subunit is light gray, the small ribosomal subunit is gray, the peptidyl-tRNA is red, and the aa-tRNA is gold with EF-Tu in violet and GTP in red. The SRL and the L11 region of the GTPase center are shown in green. Shown in (A) is the complete model while (B) displays only the relative arrangements of the tRNAs, EF-Tu(GTP), and the vital elements of the GTPase center.
To investigate real-time EF-Tu(GTP)aa-tRNA conformational dynamics important for allosteric signaling and GTPase center function, we have used single-molecule fluorescence resonance energy transfer (smFRET) between fluorescently labeled tRNAs on the ribosome (Blanchard et al. 2004b). Previous work in our laboratories using this smFRET system has defined a stepwise movement of EF-Tu(GTP)aa-tRNA into the ribosome during aa-tRNA selection and identified three unambiguous EF-Tu(GTP) aa-tRNA conformational states corresponding to initial codon recognition (0.35 FRET), GTPase activation (0.50 FRET), and full accommodation of aa-tRNA into the aa-tRNA binding site (A site) on the ribosome (0.75 FRET). Movement of EF-Tu(GTP)aa-tRNA from initial codon recognition to the GTPase activation state links codon recognition at the small subunit decoding site and GTP hydrolysis at the large subunit GTPase center (Blanchard et al. 2004a).
Here we use a thiazole-containing cyclic-peptide antibiotic, thiostrepton, to probe the function of the GTPase center during aa-tRNA selection. Thiostrepton is best known as an inhibitor of mRNA–tRNA translocation by the GTPase elongation factor G (EF-G) (Pestka 1970; Modolell et al. 1971; Gale et al. 1981; Rodnina et al. 1999), but it also inhibits EF-Tu(GTP)–catalyzed aa-tRNA delivery (Modolell et al. 1971; Gale et al. 1981) as well as the activity of the GTPase initiation factor 2 (IF-2) (Mazumder 1973; Brandi et al. 2004); its mode of action in each of these cases is not known. Mapping of spontaneous ribosome mutations conferring resistance demonstrates that thiostrepton binds the large subunit near L11 and its rRNA binding site near nucleotides 1055–1104 (L11 region) (Fig. 1B; Cameron et al. 2004).
RESULTS AND DISCUSSION
To perform single-molecule fluorescence experiments, Escherichia coli ribosomes carrying 5′-biotinylated mRNA and fluorescent Cy3-labeled fMet-tRNAfMet at the peptidyl-tRNA binding site were tethered to a streptavidin-coated quartz microscope slide as previously described (Blanchard et al. 2004b). Briefly, surface-bound ribosomes were preincubated in 50 μM thiostrepton, and fluorescent Cy5-labeled Phe-tRNAPhe in complex with EF-Tu(GTP) was stopped-flow delivered to the ribosomal A site in the presence of 50 μM thiostrepton. Binding of Cy5-labeled EF-Tu(GTP)Phe-tRNAPhe in the presence of thiostrepton generated a rapidly evolving smFRET signal as observed through a laboratory-built total internal reflection fluorescence microscope. Analysis of data acquired from several such experiments yielded 79 individual ribosomes that were observed to bind EF-Tu(GTP)Phe-tRNAPhe (Table 1) and smFRET versus time traces were plotted for the individual ribosomes (Fig. 2A). Each time trace shows an average of eight attempts to bind EF-Tu(GTP)Phe-tRNAPhe in the presence of thiostrepton (Fig. 2A) for a total of 599 observed binding events (Table 1).
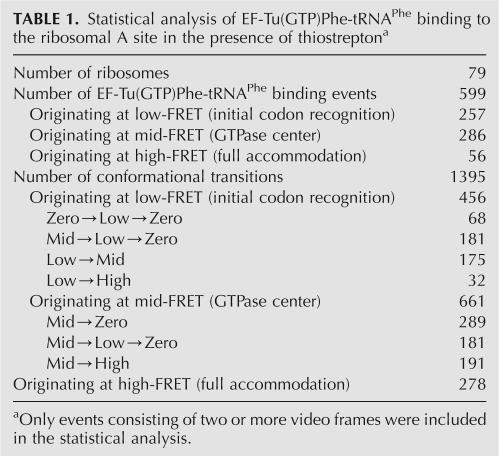
TABLE 1.
Statistical analysis of EF-Tu(GTP)Phe-tRNAPhe binding to the ribosomal A site in the presence of thiostreptona
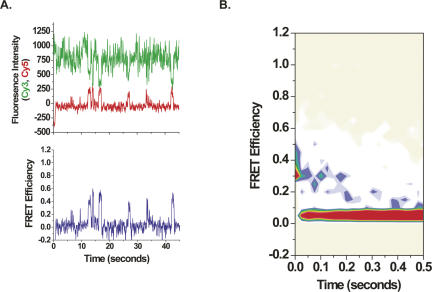
FIGURE 2.
(A) Single-molecule fluorescence intensity and FRET time traces. The figure shows fluorescence data from single molecules after stopped-flow delivery of EF-Tu(GTP)Phe-tRNAPhe(Cy5-acp3U47) in the presence of 50 μM thiostrepton to surface-immobilized ribosome complexes carrying fMet-tRNAfMet(Cy3-s4U8) in the ribosomal peptidyl-tRNA binding site and preincubated with 50 μM thiostrepton. Weak, direct excitation of EF-Tu(GTP)Phe-tRNAPhe(Cy5-acp3U47) in solution by 532-nm illumination led to change in the baseline Cy5 fluorescence upon mixing. The time trajectory of smFRET data provides information about tRNA dynamics on individual ribosome complexes. The top panel displays Cy3 and Cy5 fluorescence intensity, and the bottom panel shows FRET for a representative molecule. Cy3 (green) and Cy5 (red) fluorescence intensity is plotted in arbitrary units (AU) as recorded by the CCD. FRET, characterized by anti-correlated changes in Cy3 and Cy5 fluorescence, is calculated from the fluorescence intensity (I) according to the equation I Cy5/(I Cy3+I Cy5). Each ribosome displays an average of eight individual attempts to bind EF-Tu(GTP)Phe-tRNAPhe(Cy5-acp3U47), and each separate binding event undergoes an average of two transitions between different FRET states. (B) Contour plots of the time evolution of population FRET. The plot was generated by superimposing the individual smFRET time traces obtained in stopped-flow delivery experiments (as in Fig. 2). The time traces were postsynchronized to the first observation of FRET ≥0.25. Contours are plotted from tan (lowest population) to red (highest population). Upon binding to ribosomes, EF-Tu(GTP)Phe-tRNAPhe samples the initial codon recognition state (low-FRET state, ∼0.33) and rapidly moves into the GTPase center (transition to a mid-FRET state, ∼0.43). In the presence of thiostrepton, the majority of EF-Tu(GTP)Phe-tRNAPhe(Cy5-acp3U47) is not allowed to proceed to full accommodation (high-FRET state, ∼0.74) and is rejected from the ribosome (transition to FRET=0). Therefore, the FRET=0 state arises primarily from dissociation of EF-Tu(GTP)Phe-tRNAPhe(Cy5-acp3U47) from the ribosome and photobleaching of Cy3 or Cy5.
To analyze the general trajectory of EF-Tu(GTP)Phe-tRNAPhe binding to the ribosome, a population analysis of smFRET was conducted resulting in a contour plot diagramming the time evolution of population FRET following initial binding (Fig. 2B; Blanchard et al. 2004b). Analysis of the contour plot is consistent with EF-Tu(GTP)Phe-tRNAPhe sampling initial codon recognition (low-FRET state, ∼0.33) followed rapidly by movement of EF-Tu(GTP)Phe-tRNAPhe into the GTPase center (transition to a mid-FRET state, ∼0.43). Unlike the unperturbed delivery of ternary complex (Blanchard et al. 2004b), the majority of EF-Tu(GTP)Phe-tRNAPhe are not allowed to proceed to full accommodation (high-FRET state, ∼0.74), however, and are rejected from the ribosome. The absolute FRET values presented here differ slightly from those reported previously (Blanchard et al. 2004b) due to a quantum yield difference in the Cy5 spectral region of a newly employed charge-coupled device (CCD) (Lee et al. 2007).
Each of the 599 binding events displays an average of two transitions between conformational states of EF-Tu(GTP)Phe-tRNAPhe on the ribosome for a total of 1395 analyzable conformational transitions: 43% of these transitions originate at initial codon recognition, 48% at the GTPase center, and 9% at full accommodation (Table 1). These initial transition populations are fully consistent with the time resolution of the measurement (25 msec per frame) and the observed lifetimes of aa-tRNA at the initial codon recognition state and at the GTPase center in the presence of thiostrepton (discussed below).
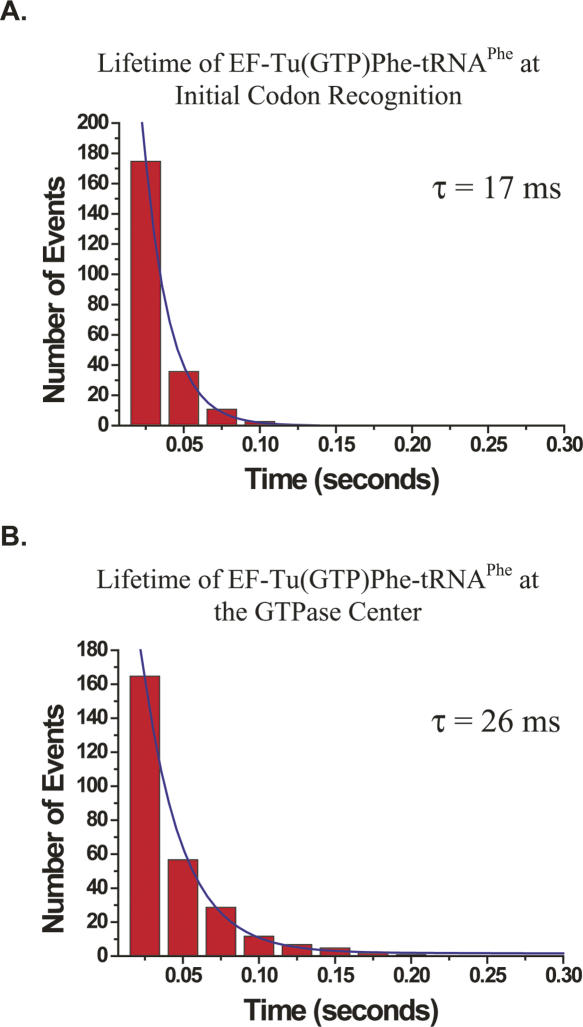
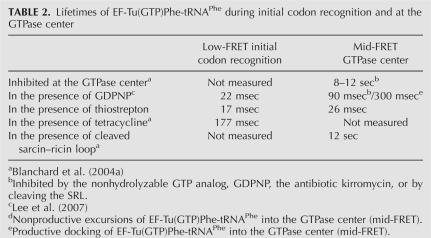
Analysis of conformational transitions originating at initial codon recognition reveals that thiostrepton does not affect the codon-recognition state of EF-Tu(GTP)Phe-tRNAPhe delivery. The total lifetime of initial codon recognition before it undergoes a conformational transition is 17 ms (Fig. 3A; Table 2). This is in close agreement with the uninhibited initial codon recognition lifetime measured for the delivery of EF-Tu(GDPNP)Phe-tRNAPhe in the presence of the nonhydrolyzable GTP analog, GDPNP (22 msec) (Table 2; Lee et al. 2007), consistent with a model in which thiostrepton does not directly inhibit initial codon recognition or movement of EF-Tu(GTP)Phe-tRNAPhe into the GTPase center of the ribosome.
FIGURE 3.
Lifetime analysis of EF-Tu(GTP)Phe-tRNAPhe at the (A) initial codon-recognition state and (B) GTPase center in the presence of 50-μM thiostrepton. Cy5-labeled EF-Tu(GTP)Phe-tRNAPhe was delivered to surface-immobilized ribosomes carrying Cy3-labeled fMet-tRNAfMet in the peptidyl-tRNA ribosomal binding site. In (A), the total lifetime of EF-Tu(GTP)Phe-tRNAPhe at the initial codon recognition includes transitions from low-FRET to zero-FRET (assigned as dissociation from the ribosome) and to mid- or high-FRET (assigned as rotation into the GTPase center and full accommodation, respectively). In this analysis, the threshold between low- and zero-FRET states was set to 0.23 and that between low- and mid- or high-FRET states was set to 0.34. In (B), the lifetime of EF-Tu(GTP)Phe-tRNAPhe at the GTPase center before a transition to low- or zero-FRET occurs includes transitions from mid-FRET to low- or zero-FRET (assigned as reverse rotation into the initial codon-recognition state and dissociation from the ribosome, respectively). In this analysis, the threshold between mid- and low- or zero-FRET states was set to 0.34, and mid-FRET events were defined as those events occurring between 0.34 and 0.60. These threshold values were determined as in Lee et al. 2007). Histograms of the times spent in (A) the initial codon recognition state or (B) the GTPase center were then fit to a single exponential decay function using a nonlinear least-squares fitting algorithm (Origin, Microcal). Only events consisting of two or more video frames were included in the data fits.
TABLE 2.
Lifetimes of EF-Tu(GTP)Phe-tRNAPhe during initial codon recognition and at the GTPase center
The majority of transitions (55%) that originate at the initial codon recognition state (low-FRET) correspond to rejection and dissociation of EF-Tu(GTP)Phe-tRNAPhe from the ribosome (transition from low-FRET to zero-FRET, Table 1). Strikingly, 73% of the these rejection events derive from EF-Tu(GTP)Phe-tRNAPhe that has reversed its trajectory, moving from the GTPase center back into the initial codon recognition state before dissociating from the ribosome (Table 1). EF-Tu(GTP)Phe-tRNAPhe thus does not dissociate directly from the GTPase center of the ribosome but follows similar paths into and out of the ribosome.
The majority (71%) of EF-Tu(GTP)Phe-tRNAPhe that arrive at the GTPase center (mid-FRET) are rapidly rejected from the ribosome (mid-FRET to low- or zero-FRET, Table 1). Of these rejection events, 39% correspond to rejection of EF-Tu(GTP)Phe-tRNAPhe via reverse rotation through the initial codon recognition state while 61% correspond to EF-Tu(GTP)Phe-tRNAPhe, apparently dissociating directly from the GTPase center (Table 1). This apparent direct dissociation from the GTPase center, however, may represent events in which residence in the low-FRET state was missed at the 25 msec time resolution.
In the presence of thiostrepton, the lifetimes of EF-Tu(GTP)Phe-tRNAPhe at the GTPase center (26 msec) Fig. 3B; Table 2) and in the initial codon recognition state (17 msec) are similar. This finding provides unambiguous evidence that, upon movement of EF-Tu(GTP)Phe-tRNAPhe into the GTPase center, thiostrepton inhibits the formation of additional stabilizing contacts between EF-Tu(GTP)Phe-tRNAPhe and the ribosome over and above those already made during initial codon recognition.
The single-molecule fluorescence experiments described here and elsewhere (Blanchard et al. 2004a,b) allow real-time observation of aa-tRNA selection. Cognate EF-Tu(GTP)Phe-tRNA undergoes initial codon recognition (low-FRET), moves from the initial codon recognition state to the GTPase activation state (low- to mid-FRET transition), and binds stably to the GTPase center (mid-FRET state). Binding of a correctly base-paired, cognate EF-Tu(GTP)aa-tRNA to the decoding site on the small ribosomal subunit leads to a conformational change of the decoding site (Yoshizawa et al. 1999; Ogle and Ramakrishnan 2005) that is propagated via movement (Blanchard et al. 2004a) and stable binding of the EF-Tu(GTP)aa-tRNA substrate into the catalytic GTPase center on the large ribosomal subunit. Near-cognate EF-Tu(GTP)aa-tRNAs form incomplete interactions in the decoding site (Ogle and Ramakrishnan 2005), have shorter lifetimes in the initial codon recognition state (Blanchard et al. 2004a), and make primarily unsuccessful excursions to the GTPase center (Blanchard et al. 2004a).
Antibiotics are powerful tools to probe ribosome mechanisms. Comparison of the effect of tetracycline and thiostrepton provides important insight into the nature of allosteric signaling in aa-tRNA selection. Tetracycline binds at the decoding site where it was shown to interfere directly with initial codon recognition, increasing the lifetime of initial codon recognition from 50 msec to 177 msec and inhibiting the movement of ternary complex into the GTPase center of the ribosome, causing the majority of EF-Tu(GTP)aa-tRNA to be rejected during initial codon recognition (Blanchard et al. 2004a). In contrast, thiostrepton binds at the GTPase center where it blocks stable binding of ternary complex, causing the majority of EF-Tu(GTP)aa-tRNA to be rejected from the GTPase center. Tetracycline and thiostrepton, although chemically and functionally distinct, bind to two different, but coupled, active sites on the ribosome and have the same overall effect on protein synthesis: rejection of EF-Tu(GTP)aa-tRNA from the ribosome.
During uninhibited delivery of EF-Tu(GTP)aa-tRNA to the A site, stable docking of ternary complex into the GTPase center leads to formation of the full complement of stabilizing contacts between EF-Tu(GTP)aa-tRNA and components of the GTPase center. Inhibition of EF-Tu(GTP)Phe-tRNAPhe delivery to the A site with a non-hydrolyzable GTP analog, the antibiotic kirromycin, or cleavage of the SRL, all of which act after docking of EF-Tu(GTP)Phe-tRNAPhe at the GTPase center, leads to a stably bound ternary complex at the GTPase center with a lifetime of 8–12 sec (Blanchard et al. 2004a). Also observed during uninhibited delivery of EF-Tu(GTP)Phe-tRNAPhe is a small population of ternary complexes that move into the GTPase center but dissociate rather than progress to the fully accommodated (i.e., high-FRET) state (Lee et al., in press). In these nonproductive movements into the GTPase center, only a partial subset of the stabilizing contacts are made and EF-Tu(GTP)Phe-tRNAPhe is less stably bound, with a lifetime of 90 msec (Lee et al. 2007) at the GTPase center. EF-Tu(GTP)Phe-tRNAPhe delivery in the presence of thiostrepton represents the extreme case, with very few, if any, stabilizing contacts made between the ternary complex and the GTPase center. This leads to an even less stably bound ternary complex and a lifetime of EF-Tu(GTP)Phe-tRNAPhe at the GTPase center comparable to that observed for the initial codon recognition state.
Thiostrepton therefore blocks formation of stabilizing contacts between EF-Tu(GTP)aa-tRNA and the GTPase center that occur even in nonproductive excursions of EF-Tu(GTP)aa-tRNA into the GTPase center. The L11 region of the GTPase center, which forms the thiostrepton binding site, regulates stable binding of EF-Tu(GTP)aa-tRNA at the GTPase center. This is achieved either by thiostrepton (1) blocking the direct interaction between the L11 region and EF-Tu(GTP)aa-tRNA or (2) blocking a conformational change in which the L11 region participates and is important for other components of the GTPase center to bind and stabilize EF-Tu(GTP)aa-tRNA. In fact, recent X-ray crystallographic structures of the E. coli ribosome in two different conformations support the latter; the structures display a large-scale conformational change of the L11 region that is controlled by the rRNA helix that forms part of the L11 region and the thiostrepton binding site (Schuwirth et al. 2005). In either case, the L11 region of the GTPase center serves to control the events that lead to stable binding of EF-Tu(GTP)aa-tRNA at the GTPase center.
The means by which an intersubunit allosteric signal is propagated from the decoding site to the GTPase center has been the subject of much speculation, with proposals for both propagation via conformational changes of the aa-tRNA (Pape et al. 1999; Piepenburg et al. 2000) or the subunit interface of the ribosome (Powers and Noller 1994; Yusupov et al. 2001). The data presented here and elsewhere (Blanchard et al. 2004a) establish that movement and stable binding of EF-Tu(GTP)aa-tRNA at the GTPase center allosterically link the decoding site to the GTPase center. This model does not preclude conformational changes of the subunit interface, which may be coupled to movement and stable binding of EF-Tu(GTP)aa-tRNA at the GTPase center.
Despite contradictory reports on the effect of thiostrepton on GTP hydrolysis by EF-Tu (Modolell et al. 1971; Takahashi et al. 1986), the results presented here are consistent with a model in which stable binding of EF-Tu(GTP)aa-tRNA at the GTPase center is a prerequisite for GTPase activation and GTP hydrolysis. Additional components of the GTPase center, such as the SRL (Valle et al. 2003; Blanchard et al. 2004a), may then stimulate GTP hydrolysis. In addition to this potential role in stimulating GTP hydrolysis, our smFRET experiments suggest that the SRL may also be involved in allosteric signaling of a post-GTP hydrolysis EF-Tu conformational change to the peptidyltransferase center of the ribosome (Blanchard et al. 2004a). Cleavage of the SRL by the cytotoxin restrictocin does not perturb binding of EF-Tu(GTP)aa-tRNA to the GTPase center (Table 2), in contrast to inhibition by thiostrepton. These data strongly support a model in which the L11 region mediates stable binding of EF-Tu(GTP)aa-tRNA at the GTPase center and the SRL has a fundamental role in stimulating the GTPase activity of EF-Tu. This mechanism may be general for other GTPase translation factors.
The results presented here demonstrate the power of single-molecule fluorescence to link structure, dynamics, and biochemical function to elucidate molecular mechanisms of allosteric coupling between multiple active sites in the translating ribosome. In addition, this approach provides direct observation of antibiotic action on the conformational dynamics of the translating ribosome.
MATERIALS AND METHODS
Reagent preparation and purification
Ribosomes, translation initiation and elongation factors, methionyl-tRNA synthetase and formylmethionyl-tRNA formyltransferase, from E. coli were prepared and purified using standard protocols (Blanchard et al. 2004b). tRNAfMet (Sigma) labeled with Cy3-maleimide (Amersham Biosciences) at the s4U8 position and tRNAPhe (Sigma) labeled with Cy5-N-hydroxysuccinimidyl ester (Amersham Biosciences) at the acp3U47 position were prepared and purified as previously described (Blanchard et al. 2004b). The formyl donor, 10-formyltetrahydrofolate, was prepared as described (Blanchard et al. 2004b). Aminoacylation and formylation of tRNAfMet(Cy3-s4U8) were achieved as reported (Blanchard et al. 2004b), and aminoacylation of tRNAPhe(Cy5-acp3U47) was achieved following standard protocols (Blanchard et al. 2004b). The 5′-biotin mRNA used in all experiments was derived from T4 gene product 32 and was in vitro transcribed and purified as reported previously (Blanchard et al. 2004b). All experiments were performed at room temperature (22°C) in Tris-polymix buffer which was composed of 50 mM Tris-OAc (pH25°C=7.5), 100 mM KCl, 5 mM NH4OAc, 0.5 mM Ca(OAc)2, 15 mM Mg(OAc)2, 6 mM BME, 5 mM putrescine, and 1 mM spermidine. 70S complexes were initiated on gene32-derived mRNA in vitro in Tris-polymix buffer at 5 mM Mg(OAc)2 as reported (Blanchard et al. 2004b), and initiation complexes were purified by sucrose density ultracentrifugation in Tris-polymix buffer at 20 mM Mg(OAc)2. Phe-tRNAPhe(Cy5-acp3U) was complexed with EF-Tu(GTP) at a final concentration of 2 μM as previously reported (Blanchard et al. 2004b). Quartz microscope slides and glass coverslips for use in total internal reflection fluorescence microscopy were prepared and passivated as described (Blanchard et al. 2004b). Initiated ribosomal complexes were immobilized onto the passivated quartz microscope slides as previously described (Blanchard et al. 2004b).
Single molecule experiments
A laboratory-built, prism-based total internal reflection fluorescence (TIRF) apparatus, based on an inverted microscope, was used for all experiments. Cy3-labeled molecules were excited using a diode-pumped 532 nm laser (CrystaLaser). Fluorescence emission was collected by a 1.2 NA/60× water-immersion objective (PlanApo) and imaged onto a cooled, back-illuminated CCD camera (Cascade, Roper Scientific) with 9-pixel binning at 25 ms exposure time. EF-Tu(GTP)tRNAPhe(Cy5-acp3U) at a concentration of 2 μM was diluted with Tris-polymix buffer to a final concentration of 7.5 nM and stopped-flow delivered to immobilized initiated ribosomal complexes. Stopped-flow delivery was achieved using a custom-built, motor driven syringe injection system. The deadtime following stopped-flow delivery of substrates is estimated at ∼500 ms. To extend the lifetimes and reduce the noise of Cy3 and Cy5 fluorescence for fluorescence microscopy, an oxygen scavenging system composed of 1% β-D-glucose, 25 U/mL glucose oxidase, and 250 U/mL catalase was added to all samples. In addition, 1,3,5,7-cyclooctatetraene (Aldrich) and p-nitrobenzyl alcohol (Fluka) were added to all buffers in order to quench a long-lived nonfluorescent triplet state sampled by the Cy5 fluorescent dye (Blanchard et al. 2004b).
ACKNOWLEDGMENTS
We thank Tae-Hee Lee for helpful discussions and assistance with the data analysis. R.L.G. is supported by the American Cancer Society and by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund. This work was supported by grants to J.D.P. from the U.S. National Institutes of Health (GM51266) and the David and Lucille Packard Foundation; to S.C. from the U.S. National Science Foundation, the U.S. National Aeronautics and Space Administration, and the U.S. Air Force Office of Scientific Research; and to S.C. and J.D.P. from the David and Lucille Packard Foundation Interdisciplinary Science Program (grant no. 2000-01671).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.499407.
REFERENCES
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., Steitz, T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Blanchard, S.C., Gonzalez R.L., Jr, Kim, H.D., Chu, S., Puglisi, J.D. tRNA selection and kinetic proofreading in translation. Nat. Struct. Mol. Biol. 2004a;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- Blanchard, S.C., Kim, H.D., Gonzalez R.L., Jr, Puglisi, J.D., Chu, S. tRNA dynamics on the ribosome during translation. Proc. Natl. Acad. Sci. 2004b;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandi, L., Marzi, S., Fabbretti, A., Fleischer, C., Hill, W.E., Gualerzi, C.O., Lodmell, J.S. The translation initiation functions of IF2: Targets for thiostrepton inhibition. J. Mol. Biol. 2004;335:881–894. doi: 10.1016/j.jmb.2003.10.067. [DOI] [PubMed] [Google Scholar]
- Bushuev, V.N., Gudkov, A.T., Liljas, A., Sepetov, N.F. The flexible region of protein L12 from bacterial ribosomes studied by proton nuclear magnetic resonance. J. Biol. Chem. 1989;264:4498–4505. [PubMed] [Google Scholar]
- Cameron, D.M., Thompson, J., Gregory, S.T., March, P.E., Dahlberg, A.E. Thiostrepton-resistant mutants of Thermus thermophilus . Nucleic Acids Res. 2004;32:3220–3227. doi: 10.1093/nar/gkh644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, E.F., Cundliffe, E., Reynolds, P.E., Richmond, M.H., Waring, M.J. The molecular basis of antibiotic action. John Wiley & Sons; London: 1981. [Google Scholar]
- Lee, T.-H., Blanchard, S.C., Kim, H.D., Puglisi, J.D., Chu, S. Direct observations of induced fit during initial selection of aminoacyl-tRNA on the ribosome. Proc. Natl. Acad. Sci. 2007;104:13661–13665. doi: 10.1073/pnas.0705988104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder, R. Effect of thiostrepton on recycling of Escherichia coli initiation factor 2. Proc. Natl. Acad. Sci. 1973;70:1939–1942. doi: 10.1073/pnas.70.7.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modolell, J., Cabrer, B., Parmeggiani, A., Vazquez, D. Inhibition by siomycin and thiostrepton of both aminoacyl-tRNA and factor-G binging to ribosomes. Proc. Natl. Acad. Sci. 1971;68:1796–1800. doi: 10.1073/pnas.68.8.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle, J.M., Ramakrishnan, V. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- Pape, T., Wintermeyer, W., Rodina, M.W. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 1999;18:3800–3807. doi: 10.1093/emboj/18.13.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka, S. Thiostrepton: A ribosomal inhibitor of translocation. Biochem. Biophys. Res. Commun. 1970;40:667–674. doi: 10.1016/0006-291x(70)90956-3. [DOI] [PubMed] [Google Scholar]
- Piepenburg, O., Pape, T., Pleiss, J.A., Wintermeyer, W., Uhlenbeck, O.C., Rodnina, M.V. Intact aminoacyl-tRNA is required to trigger GTP hydrolysis by elongation factor Tu on the ribosome. Biochemistry. 2000;39:1734–1738. doi: 10.1021/bi992331y. [DOI] [PubMed] [Google Scholar]
- Porse, B.T., Leviev, I., Mankin, A.S., Garrett, R.A. The antibiotic thiostrepton inhibits a functional transition within protein L11 at the ribosomal GTPase centre. J. Mol. Biol. 1998;276:391–404. doi: 10.1006/jmbi.1997.1541. [DOI] [PubMed] [Google Scholar]
- Powers, T., Noller, H.F. The 530 loop of 16S rRNA: A signal to EF-Tu? Trends Genet. 1994;10:27–31. doi: 10.1016/0168-9525(94)90016-7. [DOI] [PubMed] [Google Scholar]
- Rodnina, M.V., Wintermeyer, W. Fidelity of aminoacyl-tRNA selection on the ribosome: Kinetic and structural mechanisms. Annu. Rev. Biochem. 2001;70:415–435. doi: 10.1146/annurev.biochem.70.1.415. [DOI] [PubMed] [Google Scholar]
- Rodnina, M.V., Savelsbergh, A., Matassova, N.B., Katunin, V.I., Semenkov, Y.P., Wintermeyer, W. Thiostrepton inhibits the turnover but not the GTPase of elongation factor G on the ribosome. Proc. Natl. Acad. Sci. 1999;96:9586–9590. doi: 10.1073/pnas.96.17.9586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina, M.V., Stark, H., Savelsbergh, A., Wieden, H.J., Mohr, D., Matassova, N.B., Peske, F., Daviter, T., Gualerzi, C.O., Wintermeyer, W. GTPases mechanisms and functions of translation factors on the ribosome. Biol. Chem. 2000;381:377–387. doi: 10.1515/BC.2000.050. [DOI] [PubMed] [Google Scholar]
- Rosendahl, G., Douthwaite, S. Ribosomal proteins L11 and L10.(L12)(4) and the antibiotic thiostrepton interact with overlapping regions of 23S ribosomal RNA backbone in the ribosomal GTPase center. J. Mol. Biol. 1993;234:1013–1020. doi: 10.1006/jmbi.1993.1655. [DOI] [PubMed] [Google Scholar]
- Schuwirth, B.S., Borovinskaya, M.A., Hau, C.W., Zhang, W., Vila-Sanjurjo, A., Holton, J.M., Cate, J.H.D. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Seo, H.S., Cooperman, B.S. Large-scale motions within ribosomal 50S subunits as demonstrated using photolabile oligonucleotides. Bioorg. Chem. 2002;30:163–187. doi: 10.1006/bioo.2002.1255. [DOI] [PubMed] [Google Scholar]
- Stark, H., Rodnina, M.V., Rinke-Appel, J., Brimacombe, R., Wintermeyer, W., van Heel, M. Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature. 1997;389:403–406. doi: 10.1038/38770. [DOI] [PubMed] [Google Scholar]
- Takahashi, K., Ghag, S., Chladek, S. Aminoacyl-tRNA–elongation factor Tu–ribosome interaction leading to hydrolysis of guanosine 5′-triphosphate. Biochemistry. 1986;25:8330–8336. doi: 10.1021/bi00373a030. [DOI] [PubMed] [Google Scholar]
- Valle, M., Sengupta, J., Swami, N.K., Grassucci, R.A., Burkhardt, N., Nierhaus, K.H., Agrawal, R.K., Frank, J. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J. 2002;21:3557–3567. doi: 10.1093/emboj/cdf326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle, M., Zavialov, A., Li, W., Stagg, S.M., Sengupta, J., Nielsen, R.C., Nissen, P., Harvey, S.C., Ehrenberg, M., Frank, J. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat. Struct. Biol. 2003;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- Wimberly, B.T., Brodersen, D.E., Clemons W.M., Jr, Morgan-Warren, R.J., Carter, A.P., Vonrhein, C., Hartsch, T., Ramakrishnan, V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- Yoshizawa, S., Fourmy, D., Puglisi, J.D. Recognition of the codon–anticodon helix by ribosomal RNA. Science. 1999;285:1722–1725. doi: 10.1126/science.285.5434.1722. [DOI] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H., Noller, H.F. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]