Abstract
Formation of transcription-induced R-loops poses a critical threat to genomic integrity throughout evolution. We have recently shown that the SR protein ASF/SF2 prevents R-loop formation in vertebrates by cotranscriptionally binding to nascent mRNA precursors to prevent their reassociation with template DNA. Here, we identify another RNA binding protein, RNPS1, that when overexpressed strongly suppresses the high molecular weight (HMW) DNA fragmentation, hypermutation, and G2 cell cycle arrest phenotypes of ASF/SF2-depleted cells. Furthermore, ablation of RNPS1 by RNA interference in HeLa cells leads to accumulation of HMW DNA fragments. As ASF/SF2 depletion does not influence RNPS1 expression, and RNPS1 cannot compensate for ASF/SF2 function in splicing, our data suggest that RNPS1 is able to function together with ASF/SF2 to form RNP complexes on nascent transcripts, and thereby prevent formation of transcriptional R-loops.
Keywords: ASF/SF2, RNPS1, genome stability
INTRODUCTION
Alternative splicing factor/splicing factor 2 (ASF/SF2) is a protypical SR protein splicing factor, originally identified by its roles in both constitutive and alternative pre-mRNA splicing (Ge and Manley 1990; Krainer et al. 1990a,b; Ge et al. 1991). While the functions of ASF/SF2 in pre-mRNA splicing have been extensively studied, recent studies have extended its roles into other aspects of mRNA metabolism, including mRNA export (Huang et al. 2003, 2004), mRNA stability (Lemaire et al. 2002), and translation (Sanford et al. 2004). Genetic inactivation experiments have further revealed essential roles of ASF/SF2 in cell viability and animal development (Wang et al. 1996; Xu et al. 2005).
While characterizing how ASF/SF2-depleted chicken DT40 cells die, we found an unexpected but vital function of ASF/SF2 in general cell physiology: protecting chromosomal DNA from the deleterious effects of transcription-induced R-loops formed between nascent RNAs and template DNAs. Defects in this process lead to DNA double-strand breaks (DSBs) and genome instability (Li and Manley 2005). This work, together with studies in bacteria (Drolet et al. 2003; Drolet 2006) and yeast (Huertas and Aguilera 2003), has established an evolutionarily conserved role for cotranscriptional mRNP packaging in preventing the negative effects of nascent transcripts on genomic integrity.
Transcripts synthesized by RNA polymerase II (RNAP II) associate cotranscriptionally with numerous protein factors and exist as mRNPs in the nucleus of eukaryotic cells (Dreyfuss et al. 2002). ASF/SF2, an abundant and multifunctional RNA binding protein, is cotranscriptionally recruited to nascent transcripts and can remain bound to some mRNAs until translation (Sanford et al. 2004). The roles of ASF/SF2 in RNA processing and organization of pre-mRNP particles are entirely consistent with its function in preventing transcription-induced R-loops. However, as numerous other nuclear proteins associate with nascent transcripts and/or function in pre-mRNA splicing, it is important to determine whether any of these proteins function in a similar way to prevent nascent transcripts from hybridizing with template DNAs.
Here, we describe experiments designed to identify other proteins that might function to prevent the formation of R-loops. We first developed a screen for suppressor(s) of ASF/SF2 depletion-induced genome instability in DT40 cells by using a small pool of cDNAs encoding a select group of RNA processing factors and pre-mRNA binding proteins that seemed plausible candidates. This screen identified RNPS1, a nuclear RNA binding protein with multiple roles in mRNA maturation (Badolato et al. 1995). We show that stable overexpression of RNPS1 strongly suppressed the HMW DNA fragmentation, hypermutation, and G2 cell cycle arrest phenotypes that result from ASF/SF2 depletion and subsequent R-loop formation. Depletion of RNPS1 in HeLa cells by siRNA also induced accumulation of HMW DNA fragments, similar to levels observed in ASF/SF2-depleted HeLa cells. As ASF/SF2 depletion does not influence RNPS1 expression, and RNPS1 cannot compensate for ASF/SF2 function in splicing (Mayeda et al. 1999), our data support the view that RNPS1, like ASF/SF2, functions to prevent transcriptional R-looping by forming RNP complexes on nascent transcripts, and that the two proteins function with a considerable degree of redundancy in this process.
RESULTS
Identification of suppressor(s) for ASF/SF2 depletion-induced HMW DNA fragmentation
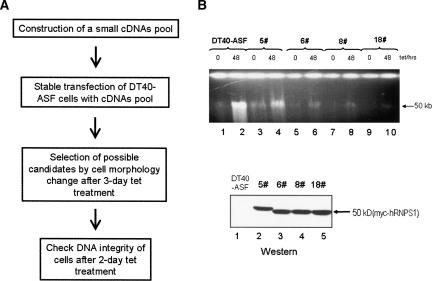
To gain more insights into the mechanism by which cells protect their genomes from the destabilizing effects of nascent transcripts, we set out to establish a screen for potential suppressor(s) of ASF/SF2 depletion-induced genome instability. To this end, we made a small pool of cDNAs encoding nine myc-tagged proteins (UAP56, TAP, Y14, SRM160, RNPS1, SRP20, 9G8, U2AF35, and SC35). These proteins include several SR proteins and non-snRNP splicing factors, components of the exon junction complex (EJC), a splicing-dependent mRNP complex (Le Hir et al. 2000; Reichert et al. 2002; Tange et al. 2004), and components of the TREX complex, which has been demonstrated to function in preventing transcriptional R-looping in yeast (Huertas and Aguilera 2003). DT40-ASF cells were transfected with this cDNA pool and distributed into 96-well plates with zeocin selective medium. Zeocin-resistant transfectants were expanded, and aliquots of each colony were subjected to tet treatment (Fig. 1A). Notably, while parental DT40-ASF cells began to show signs of cell death (see Materials and Methods), such as cytoplasmic shrinkage and membrane blebbing, after 2 d of tet treatment, several of the stable transfectants retained their normal round shape even after 3 d of tet treatment.
FIGURE 1.
Identification of suppressor(s) of ASF/SF2 depletion-induced HMW DNA fragmentation. (A) Strategy of isolating suppressor(s) of ASF/SF2 depletion-induced HMW DNA fragmentation. (B) ASF/SF2 depletion-induced HMW DNA fragmentation was partially suppressed in the selected stable transfectants. Selected colonies were expanded and grown for 48 h in the presence (+) or absence (−) of tet. Cell extracts were analyzed for the integrity of chromosomal DNA by 0.5% agarose (top panel) gel electrophoresis and for myc-tagged protein(s) by Western blot (bottom panel). The colony number is indicated.
R-loop-mediated DNA damage leads to genomic instability, G2 phase cell cycle arrest, and contributes to cell death in ASF/SF2-depleted cells (Li and Manley 2005). We therefore reasoned that the improvement in cell morphology of the stable transfectants reflected to some degree alleviation of R-loop-mediated DNA damage. To investigate this possibility, we examined the formation of HMW DNA fragments in the stable transfectants after 48 h of tet treatment. Most of the cells indeed showed a significant decrease in HMW DNA fragmentation compared with parental DT40-ASF cells (Fig. 1B; data not shown). Remarkably, cells showing the most significant decreases in HMW DNA fragmentation (Fig. 1B, top panel, lanes 6,8,10) always expressed a ∼50-kD myc-tagged protein, which most closely approximates the size of myc-tagged RNPS1 (Fig. 1B, bottom panel, lanes 3–5). These data suggest a potential role of RNPS1 in suppressing ASF/SF2 depletion-induced HMW DNA fragmentation.
Overexpression of RNPS1 suppresses ASF/SF2 depletion-induced HMW DNA fragmentation, G2 cell cycle arrest, and hypermutation phenotypes
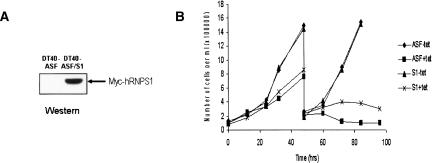
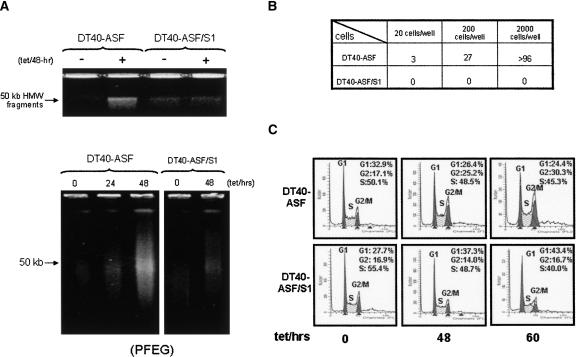
We next wished to obtain direct evidence that RNPS1 functions in preventing ASF/SF2 depletion-induced DNA damage. For this, we constructed a stable derivative of DT40-ASF cells, DT40-ASF/S1, that expresses myc-tagged human RNPS1 (myc-RNPS1) (Fig. 2A). We first asked whether expression of myc-RNPS1 affected cell viability in the presence of tet. Although DT40-ASF/S1 cells still underwent cell death upon ASF/SF2 depletion, viability of these cells was extended ∼1–2 d (Fig. 2B). We next examined whether HMW DNA fragmentation was observed in DT40-ASF/S1 cells upon ASF/SF2 depletion. As predicted from the initial screening experiments, both conventional and pulse-field agarose gel electrophoresis revealed that DT40-ASF/S1 cells indeed showed a striking decrease in HMW DNA fragmentation, as compared with the parental DT40-ASF cells, after 48 h of tet treatment (Fig. 3A).
FIGURE 2.
RNPS1 overexpression delays cell death induced by ASF/SF2 depletion. (A) Expression of myc-hRNPS1 in DT40-ASF/S1 cells. Myc-hRNPS1 expression in DT40-ASF/S1 cells was detected by Western blot using an anti-myc-antibody. (B) Growth curve of DT40-ASF and DT40-ASF/S1 cells in the presence or absence of tet.
FIGURE 3.
RNPS1 overexpression suppresses HMW DNA fragmentation, G2 cell cycle arrest, and hypermutation phenotypes in ASF/SF2-depleted cells. (A) Overexpression of RNPS1 reduces HMW DNA fragmentation in ASF/SF2-depleted cells. DT40-ASF and DT40-ASF/S1 cells were grown in the absence (−) or presence (+) of 1 μg/mL tet for the indicated times. The integrity of chromosomal DNA was analyzed by 0.5% agarose gel electrophoresis (upper panel) and pulse-field gel electrophoresis (PFEG) (lower panel). (B) Reversion of cell death in DT40-ASF cells and DT40-ASF/S1 cells after ASF/SF2 depletion. After a 2-d tet treatment, DT40-ASF and DT40-ASF/S1 cells were assayed for tet resistance, and the number of wells with surviving cells was determined. (C) Cell cycle phenotype of ASF/SF2-depleted cells. DT40-ASF and DT40-ASF/S1 cells, grown with or without 1 μg/mL tet for the indicated times, were stained with propidium iodide and analyzed by FACS. Raw data were quantitated and plotted, and the fraction of cells in G1, S, and G2/M phases are indicated.
Our initial indication that ASF/SF2 is essential for the maintenance of genomic stability was the observation that depletion of ASF/SF2 from DT40-ASF cells resulted in a very high reversion frequency of the tet-sensitive phenotype (Li and Manley 2005). This led to the finding that ASF/SF2 depletion caused HMW DNA fragmentation, and the hypothesis that the reversion arose from DNA rearrangements. If this is indeed the case, then the suppression of fragmentation observed in DT40-ASF/S1 cells should be correlated with a reduction in reversion of tet sensitivity. To test this, DT40-ASF and DT40-ASF/S1 cells were grown in the presence of tet for >10 d and the number of revertants determined (Fig. 3B). Strikingly, while DT40-ASF cells displayed the same high reversion frequency observed previously, no revertants were obtained from DT40-ASF/S1 cells in this assay. As the revertants were counted after an extended tet treatment when the majority of cells were dead, the suppression of tet reversion suggests that DNA damage and HMW fragmentation were suppressed and not simply delayed in DT40-S1 cells.
ASF/SF2 depletion also results in G2 cell-cycle arrest, likely reflecting a DNA damage-induced checkpoint (Li and Manley 2005). We next asked whether the suppression of DNA damage by RNPS1 overexpression also repressed the cell-cycle arrest phenotype in ASF/SF2-depleted DT40-ASF/S1 cells. To test this, FACS analysis was performed to determine the cell-cycle profile of DT40-ASF/S1 cells at different time points after tet addition (Fig. 3C). In contrast to parental DT40-ASF cells, no change in the G2/M phase population was observed in DT40-ASF/S1 cells upon 48-h or 60-h tet treatment.
Together, these results provide strong evidence that RNPS1 overexpression is sufficient to suppress HMW DNA fragmentation, G2 cell arrest, and hypermutation phenotypes in ASF/SF2-depleted cells.
Overexpression of other nuclear RNA binding proteins suppresses ASF/SF2 depletion-induced genomic instability weakly or not at all
The results of our initial screen pointed to RNPS1 but none of the other proteins tested as potential suppressors of ASF/SF2 depletion-induced genomic instability. To investigate the specificity of the RNPS1 effect further, we analyzed DT40-ASF cells transformed with expression vectors encoding several of the proteins referred to above.
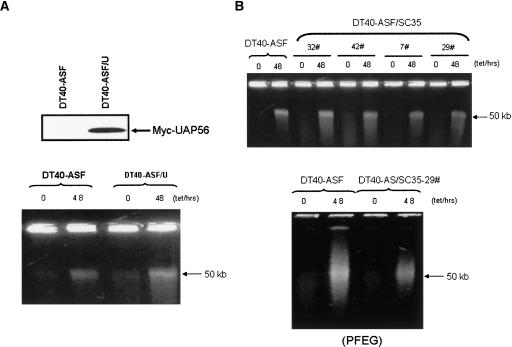
We first analyzed in more detail the effect of UAP56 overexpression in DT40-ASF cells. UAP56 is especially interesting because its yeast counterpart, Sub2, has been shown to play an important role in preventing transcription-coupled R-loop formation in yeast (Fan et al. 2001). We therefore stably transfected DT40-ASF cells with a UAP56 expression vector. No obvious effects on the cell morphology or viability changes observed following ASF/SF2 depletion were detected in any of the UAP56-expressing clones isolated (results not shown). Furthermore, analysis of one of the clones expressing myc-UAP56 (Fig. 4A) revealed no significant effect on the suppression of HMW DNA fragmentation resulting from ASF/SF2 depletion (Fig. 4B).
FIGURE 4.
Effects of UAP56 and SC35 overexpression on ASF/SF2 depletion-induced HMW DNA fragmentation. (A) Overexpression of UAP56 has no effect on HMW DNA fragmentation in ASF/SF2-depleted cells. Myc-hUAP56 expression in DT40-ASF/U cells was detected by Western blot using an anti-myc-antibody (upper panel). DT40-ASF and DT40-ASF/U cells were grown in the absence (−) or presence (+) of 1 μg/mL tet for the indicated times. The integrity of chromosomal DNA was analyzed by 0.5% agarose gel electrophoresis (lower panel). (B) DT40-ASF/SC35 cells display weak suppression on HMW DNA fragmentation induced by ASF/SF2 depletion. DT40-ASF cells and several DT40-ASF/SC35 colonies were grown in the absence (−) or presence (+) of 1 μg/mL tet for the indicated times. The integrity of chromosomal DNA was analyzed by 0.5% agarose gel electrophoresis (upper panel) and PFEG (lower panel).
We also stably transfected constructs expressing the SR proteins 9G8, SRp20, or SC35 into DT40-ASF cells separately. We examined 96 stable transfectants for each. No obvious changes were observed in cell morphology or viability induced by ASF/SF2 depletion (results not shown). We further examined HMW fragmentation in 16 SC35 stable transfectants. Fifteen of the colonies tested did not display reduced HMW fragmentation following ASF/SF2 depletion (Fig. 4B, upper panel; results not shown). However, a decrease in HMW fragmentation after ASF/SF2 depletion was observed in one SC35 stable transfectant, although the suppression was significantly weaker than observed in DT40-ASF/S1 cells (Fig. 4B).
ASF/SF2 depletion does not influence RNPS1 expression
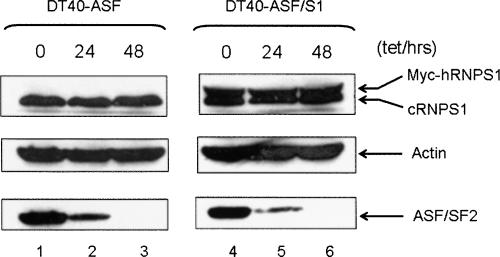
The above results indicate that expression of exogenous RNPS1 is able to alleviate the deleterious effects of ASF/SF2 depletion on genome stability. A reasonable explanation for this is that RNPS1 can substitute for ASF/SF2 in forming RNP particles on nascent transcripts, thereby preventing the formation of the R-loops that lead to DNA strand breaks and rearrangements. But another possibility is that ASF/SF2 depletion decreases RNPS1 levels in cells, and this actually underlies the R-loop-mediated DNA damage in ASF/SF2-depleted cells. To test this, we analyzed lysates of DT40-ASF cells treated with tet for various time points for RNPS1 protein levels by Western blotting. No significant differences in RNPS1 levels were detected after 48 h of tet treatment (Fig. 5, cf. lanes 1–3). Overexpression of RNPS1 also did not influence depletion of ASF/SF2 following tet treatment (Fig. 5, cf. lanes 1–3 and 4–6). These results indicate that the ability of RNPS1 overexpression to suppress ASF/SF2 depletion-induced DNA damage is not attributable to down-regulation of endogenous RNPS1 expression or to a stabilizing effect of RNPS1 overexpression on ASF/SF2 levels after tet treatment.
FIGURE 5.
ASF/SF2 depletion does not influence RNPS1 expression. DT40-ASF and DT40-ASF/S1 cells were transferred into medium with or without 1 μg/mL tet. After incubation for the indicated times, cell lysates were examined for levels of myc-hRNPS1 (top panel), cRNPS1 (top panel), actin (middle panel), and ASF/SF2 (bottom panel) by Western blot.
HMW DNA fragmentation occurs in RNPS1-depleted HeLa cells
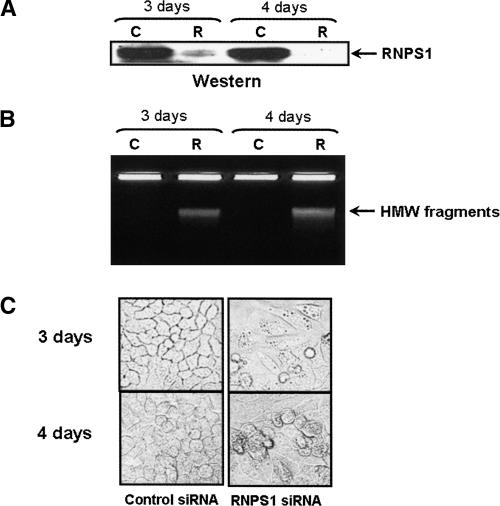
The above data reveal that RNPS1 overexpression is very effective in suppressing ASF/SF2 depletion-induced DNA damage. To extend these results, we next wished to ask whether depletion of RNPS1 can also induce formation of HMW DNA fragments. To this end, we utilized RNAi to deplete RNPS1 in HeLa cells. HeLa cells were transfected with either an RNPS1 or control siRNA duplex, and analyzed for both RNPS1 protein levels and the appearance of HMW DNA fragments 3 or 4 d following transfection. As compared with the control siRNA, the RNPS1 siRNA depleted RNPS1 by >90% (Fig. 6A). Concomitant with depletion of RNPS1, cells transfected with RNPS1 siRNA but not the control siRNA showed accumulation of HMW DNA fragments (Fig. 6B). Indeed, depletion of RNPS1 was at least as effective as depletion of ASF/SF2 in inducing DNA fragmentation (Li and Manley 2005). Also, depletion of RNPS1 resulted in significant changes in cell morphology and viability (Fig. 6C). In contrast to control siRNA treatment, RNPS1 siRNA-treated cells displayed a spindle-shaped morphology, and significant cell death was observed.
FIGURE 6.
Depletion of RNPS1 induces HMW DNA fragmentation in HeLa cells. HeLa cells were treated with control (C) or RNPS1 (R) siRNA for 3 or 4 d. The integrity of chromosomal DNA was analyzed by 0.5% agarose gel electrophoresis (A), and expression of RNPS1 was determined by Western blot (B). Cell morphology was examined by microscopy at 100× magnification (C).
DISCUSSION
The SR protein splicing factor ASF/SF2 functions in the maintenance of genomic stability by preventing nascent transcripts from forming mutagenic R-loop structures during transcription. In this study, we provide evidence that another nuclear RNA binding protein also participates in protecting chromosomal DNA from the deleterious effects of nascent transcripts. We found that overexpression of a single RNA binding protein, RNPS1, is able to suppress the effects of ASF/SF2 depletion on genome integrity. These data not only support the view that formation of RNP complexes on nascent transcripts prevents transcriptional R-loops, but also contribute to establishing a comprehensive view of the influence of cotranscriptional processes on genome stability.
We previously proposed that ASF/SF2 exerts its prophylactic function in preventing R-loops by cotranscriptionally binding to nascent mRNA precursors, which is consistent with the properties of ASF/SF2 in cotranscriptional pre-mRNA processing and/or mRNP assembly (Li and Manley 2005). Following this, an interesting question is whether additional RNA binding protein(s)/processing factor(s) also function to protect genome integrity during transcription, and here we have shown that RNPS1 functions in this way. However, it is of note that most of the RNA processing factors and/or pre-mRNP proteins we examined displayed no obvious suppressive effect on ASF/SF2 depletion-induced morphological changes and HMW DNA fragmentation. This includes UAP56, the yeast homolog of which (Sub2) is a component of the TREX complex, and has been implicated in prevention of R-looping and transcription-associated recombination in yeast (Fan et al. 2001; Jimeno et al. 2002; Strasser et al. 2002). The lack of effect we observed might reflect differences in the mechanisms in preventing transcription-induced R-looping between different species (Li and Manley 2006). For example, evidence has been presented that mammalian TREX associates with pre-mRNAs subsequent to splicing (Masuda et al. 2005), and thus would likely not be able to function in preventing R-loop formation. In contrast, splicing-related RNA binding proteins might be more suitable for this function due to the tight coupling between transcription and splicing and the near universal presence of introns in metazoan cells. In addition to ASF/SF2, genetic inactivation of SC35 in mice has recently been shown to result in DSBs and G2/M cell cycle arrest (Xiao et al. 2007). However, we did not observe significant effects on ASF/SF2 depletion-induced cell morphology and viability in DT40-ASF cells stably transfected with cDNAs encoding several different SR proteins, although a decrease in HMW fragmentation was observed in one SC35 stable transfectant. These differences might be attributable to their sequence specificity in mRNP formation, which will require further work to elucidate. Alternatively, it could be that the level of overexpression of these proteins was minimal, due to autoregulation (e.g., Wang et al. 1996).
How does RNPS1 function to alleviate the impact of ASF/SF2 depletion on genome integrity? Our previous data indicate that cotranscriptional loading of ASF/SF2 to the nascent RNA is a critical mechanism to prevent transcriptional R-looping and subsequent genome instability (Li and Manley 2005). RNPS1 is also a nuclear RNA binding protein, participating in various mRNP complexes during transcription and RNA processing (see below). Thus, an attractive explanation is that extra RNPS1 may suppress R-loop formation in ASF/SF2-depleted cells simply by binding to nascent RNAs, complementing the lack of ASF/SF2 in mRNP assembly. While ASF/SF2 is a sequence-specific RNA binding protein (Tacke and Manley 1995), ASF/SF2 does bind to nonconsensus sites in the β-actin RNA during in vitro transcription (Li and Manley 2005). This suggests that the role of ASF/SF2, and maybe other RNA binding proteins, in pre-mRNP formation is more than recognition of high-affinity RNA binding sites. In this view, it is possible that RNPS1, by cooperative interactions with other factors, is capable of being cotranscriptionally recruited to sites recognized by ASF/SF2 on nascent transcripts, substituting for ASF/SF2 to prevent mRNA precursors from hybridizing with template DNAs.
RNPS1 is also an important effecter involved in multiple steps during mRNA maturation. For example, although it can not complement ASF/SF2 function in splicing, RNPS1 was originally characterized as a pre-mRNA splicing activator that stimulates overall splicing activity in vitro, and alternative splice site selection both in vitro and in vivo (Mayeda et al. 1999). RNPS1 can influence mRNA stability when tethered to the 3′ untranslated region of β-actin mRNA, most probably due to its interaction with components of the nonsense-mediated mRNA decay pathway (Lykke-Andersen et al. 2001). Reflecting its association with the EJC, RNPS1 can also stimulate translation by enhancing mRNA polysome formation (Nott et al. 2004). On the other hand, RNPS1 is also able to interact with SAP18 and Acinus in vivo to form a protein complex, termed the apoptosis- and splicing-associated protein (ASAP) (Schwerk et al. 2003). Intriguingly, Acinus has been suggested to be involved in apoptotic DNA cleavage although the exact mechanism remains elusive (Joselin et al. 2006). Taken together, these documented properties of RNPS1 are consistent with another possibility: that excess RNPS1 influences the expression or activity of some unknown factor(s), and this in turn complements the function of ASF/SF2 in preventing transcriptional R-looping and/or R-loop-induced DNA damage. Further studies are required to decipher the precise role of RNPS1 in suppressing ASF/SF2 depletion-induced genome instability. However, whatever the mechanism, our studies have established that a nuclear RNA binding protein in addition to ASF/SF2 can function to prevent R-loop-induced genomic instability. It will be important in the future to determine how widespread this property is.
MATERIALS AND METHODS
Plasmid constructs
The cDNAs for the coding regions of human UAP56, TAP, Y14, SRP20, 9G8, U2AF35, SRM160, SC35, and RNPS1 were obtained by reverse transcription-PCR (RT-PCR) of HeLa cell total RNA. A 33-bp myc tag sequence was added to the 5′ end of the above cDNAs by PCR as described (Li and Manley 2005). The corresponding myc-tagged cDNAs were inserted into the expression vector pAPSV-zeo under the control of the chicken β-actin promoter (Chen and Manley 2000), generating corresponding expression constructs (pAP-UAP56/TAP/Y14/SRP20/9G8/U2AF35/SRM160/SC35/RNPS1). A 273-nucleotide (nt) cDNA, encoding residues 141–232 of the chicken RNPS1 protein, was obtained by RT-PCR of DT40 cell total RNA and cloned into pGEX-6P, giving the construct pGEX-cRNPS1-M.
Screening for suppressor(s) of ASF/SF2 depletion-induced high molecular DNA fragmentation
DT40-ASF cells were stably transfected with a cDNA pool encoding nine myc-tagged proteins as described (Li and Manley 2005). Stable transfectants were continuously subjected to tet treatment. After 3 d of tet treatment, stable transfectants were selected for further studies based on cell morphology changes including: (1) Cell shape: the selected transfectants maintained bright and intact round shape, while the parental DT40-ASF cells showed some condensed dark spots, enlarged cell volume, and shrinkage of the cytoplasmic membrane. (2) Cell debris: almost no cell debris appeared in the selected transfectants up to 3 d of tet treatment, while cell debris began to appear ∼2 d after tet addition in parental DT40-ASF cells. (3) Spindle-shaped cells: almost no spindle-shaped cells appeared in the selected transfectants up to 3 d of tet treatment, while some (5%–10%) spindle-shaped cells appeared 2 d after tet addition in DT40-ASF cells. Overexpressed exogenous proteins in those selected colonies were identified by Western blot analysis using a myc-tag-specific antibody (Invitrogen). The expected sizes of proteins encoded by the cDNAs in the cDNA pool are as follows: SRM160 (∼160 kD), TAP (∼73 kD), UAP56 (∼56 kD), RNPS1 (∼50 kD), SC35 (∼35 kD), U2AF35 (∼35 kD), 9G8 (∼35 kD), SRP20 (∼20 kD), Y14 (∼24 kD). Analysis of genomic DNA integrity, tetracycline-resistance assays, siRNA transfections, protein, and cell cycle analysis were conducted as described (Li and Manley 2005).
Protein and antibodies
GST-cRNPS1-M was purified from Escherichia coli BL21 transfected with pGEX-cRNPS1-M by glutathione-Sepharose 4B chromatography as described by the manufacturer (GE). Anti-ASF/SF2 (Invitrogen), myc-tag (Invitrogen), and actin antibodies (Sigma) are all commercially available. Polyclonal antibodies against chicken RNPS1 were raised by immunizing mice with recombinant cRNPS1-M (Antibody center of NIBS).
ACKNOWLEDGMENTS
We thank S. Bush for comments on the manuscript, I. Boluk for help in preparing the manuscript, and Z.C. Qiu and X.J. Shi for RNPS1 antibody preparation. This work was supported by NIH grant R37 GM 48259 and the National High Technology Projects 863, China.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.734407.
REFERENCES
- Badolato, J., Gardiner, E., Morrison, N., Eisman, J. Identification and characterisation of a novel human RNA-binding protein. Gene. 1995;166:323–327. doi: 10.1016/0378-1119(95)00571-4. [DOI] [PubMed] [Google Scholar]
- Chen, Z., Manley, J.L. Robust mRNA transcription in chicken DT40 cells depleted of TAF(II)31 suggests both functional degeneracy and evolutionary divergence. Mol. Cell. Biol. 2000;20:5064–5076. doi: 10.1128/mcb.20.14.5064-5076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss, G., Kim, V.N., Kataoka, N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Drolet, M. Growth inhibition mediated by excess negative supercoiling: The interplay between transcription elongation, R-loop formation and DNA topology. Mol. Microbiol. 2006;59:723–730. doi: 10.1111/j.1365-2958.2005.05006.x. [DOI] [PubMed] [Google Scholar]
- Drolet, M., Broccoli, S., Rallu, F., Hraiky, C., Fortin, C., Masse, E., Baaklini, I. The problem of hypernegative supercoiling and R-loop formation in transcription. Front. Biosci. 2003;8:d210–d221. doi: 10.2741/970. [DOI] [PubMed] [Google Scholar]
- Fan, H.Y., Merker, R.J., Klein, H.L. High-copy-number expression of Sub2p, a member of the RNA helicase superfamily, suppresses hpr1-mediated genomic instability. Mol. Cell. Biol. 2001;21:5459–5470. doi: 10.1128/MCB.21.16.5459-5470.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, H., Manley, J.L. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- Ge, H., Zuo, P., Manley, J.L. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell. 1991;66:373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- Huang, Y., Gattoni, R., Stevenin, J., Steitz, J.A. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Huang, Y., Yario, T.A., Steitz, J.A. A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl. Acad. Sci. 2004;101:9666–9670. doi: 10.1073/pnas.0403533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas, P., Aguilera, A. Co-transcriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Jimeno, S., Rondon, A.G., Luna, R., Aguilera, A. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 2002;21:3526–3535. doi: 10.1093/emboj/cdf335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joselin, A.P., Schulze-Osthoff, K., Schwerk, C. Loss of Acinus inhibits oligonucleosomal DNA fragmentation but not chromatin condensation during apoptosis. J. Biol. Chem. 2006;281:12475–12484. doi: 10.1074/jbc.M509859200. [DOI] [PubMed] [Google Scholar]
- Krainer, A.R., Conway, G.C., Kozak, D. The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell. 1990a;62:35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- Krainer, A.R., Conway, G.C., Kozak, D. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes & Dev. 1990b;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- Le Hir, H., Izaurralde, E., Maquat, L.E., Moore, M.J. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire, R., Prasad, J., Kashima, T., Gustafson, J., Manley, J.L., Lafyatis, R. Stability of a PKCI-1-related mRNA is controlled by the splicing factor ASF/SF2: A novel function for SR proteins. Genes & Dev. 2002;16:594–607. doi: 10.1101/gad.939502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Manley, J.L. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Li, X., Manley, J.L. Cotranscriptional processes and their influence on genome stability. Genes & Dev. 2006;20:1838–1847. doi: 10.1101/gad.1438306. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen, J., Shu, M.D., Steitz, J.A. Communication of the position of exon–exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293:1836–1839. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- Masuda, S., Das, R., Cheng, H., Hurt, E., Dorman, N., Reed, R. Recruitment of the human TREX complex to mRNA during splicing. Genes & Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda, A., Badolato, J., Kobayashi, R., Zhang, M.Q., Gardiner, E.M., Krainer, A.R. Purification and characterization of human RNPS1: A general activator of pre-mRNA splicing. EMBO J. 1999;18:4560–4570. doi: 10.1093/emboj/18.16.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott, A., Le Hir, H., Moore, M.J. Splicing enhances translation in mammalian cells: An additional function of the exon junction complex. Genes & Dev. 2004;18:210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert, V.L., Le Hir, H., Jurica, M.S., Moore, M.J. 5′ exon interactions within the human spliceosome establish a framework for exon junction complex structure and assembly. Genes & Dev. 2002;16:2778–2791. doi: 10.1101/gad.1030602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford, J.R., Gray, N.K., Beckmann, K., Caceres, J.F. A novel role for shuttling SR proteins in mRNA translation. Genes & Dev. 2004;18:755–768. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerk, C., Prasad, J., Degenhardt, K., Erdjument-Bromage, H., White, E., Tempst, P., Kidd, V.J., Manley, J.L., Lahti, J.M., Reinberg, D. ASAP, a novel protein complex involved in RNA processing and apoptosis. Mol. Cell. Biol. 2003;23:2981–2990. doi: 10.1128/MCB.23.8.2981-2990.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser, K., Masuda, S., Mason, P., Pfannstiel, J., Oppizzi, M., Rodriguez-Navarro, S., Rondon, A.G., Aguilera, A., Struhl, K., Reed, R., et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- Tacke, R., Manley, J.L. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange, T.O., Nott, A., Moore, M.J. The ever-increasing complexities of the exon junction complex. Curr. Opin. Cell Biol. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Wang, J., Takagaki, Y., Manley, J.L. Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes & Dev. 1996;10:2588–2599. doi: 10.1101/gad.10.20.2588. [DOI] [PubMed] [Google Scholar]
- Xiao, R., Sun, Y., Ding, J.H., Lin, S., Rose, D.W., Rosenfeld, M.G., Fu, X.D., Li, X. Splicing regulator SC35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Mol. Cell. Biol. 2007;27:5393–5402. doi: 10.1128/MCB.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X., Yang, D., Ding, J.H., Wang, W., Chu, P.H., Dalton, N.D., Wang, H.Y., Bermingham J.R., Jr, Ye, Z., Liu, F., et al. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation–contraction coupling in cardiac muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]