Abstract
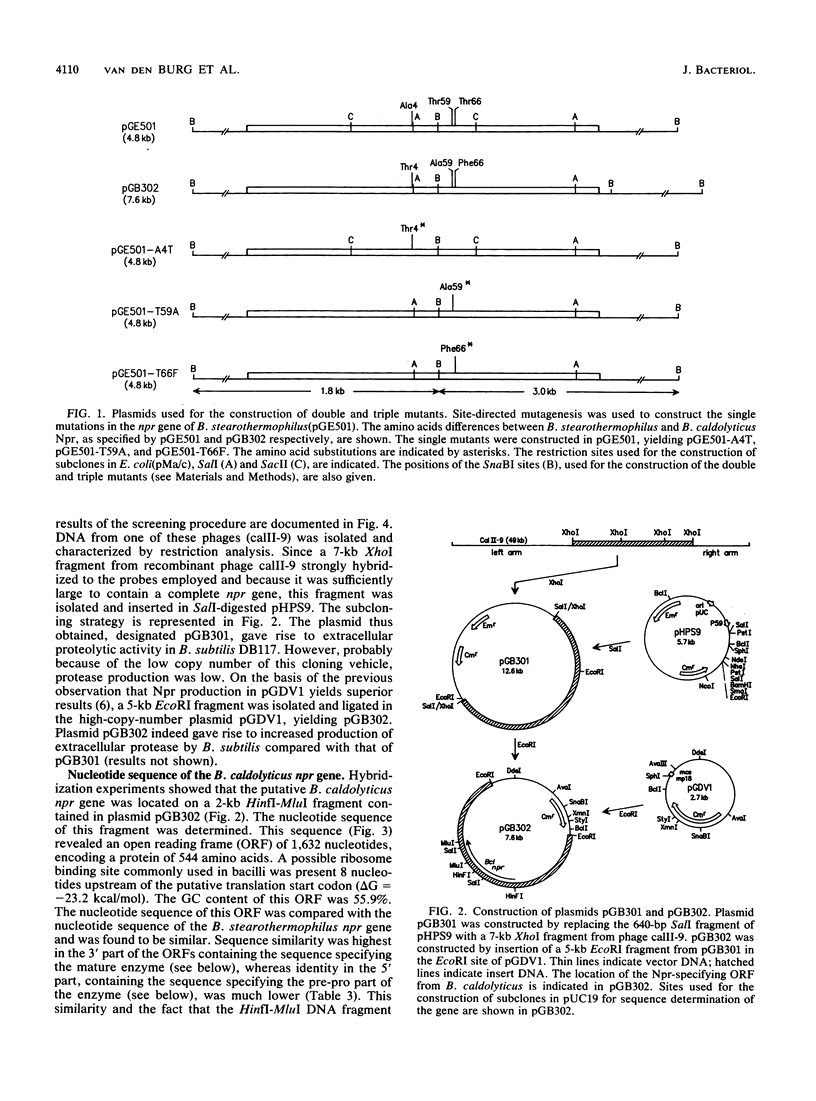
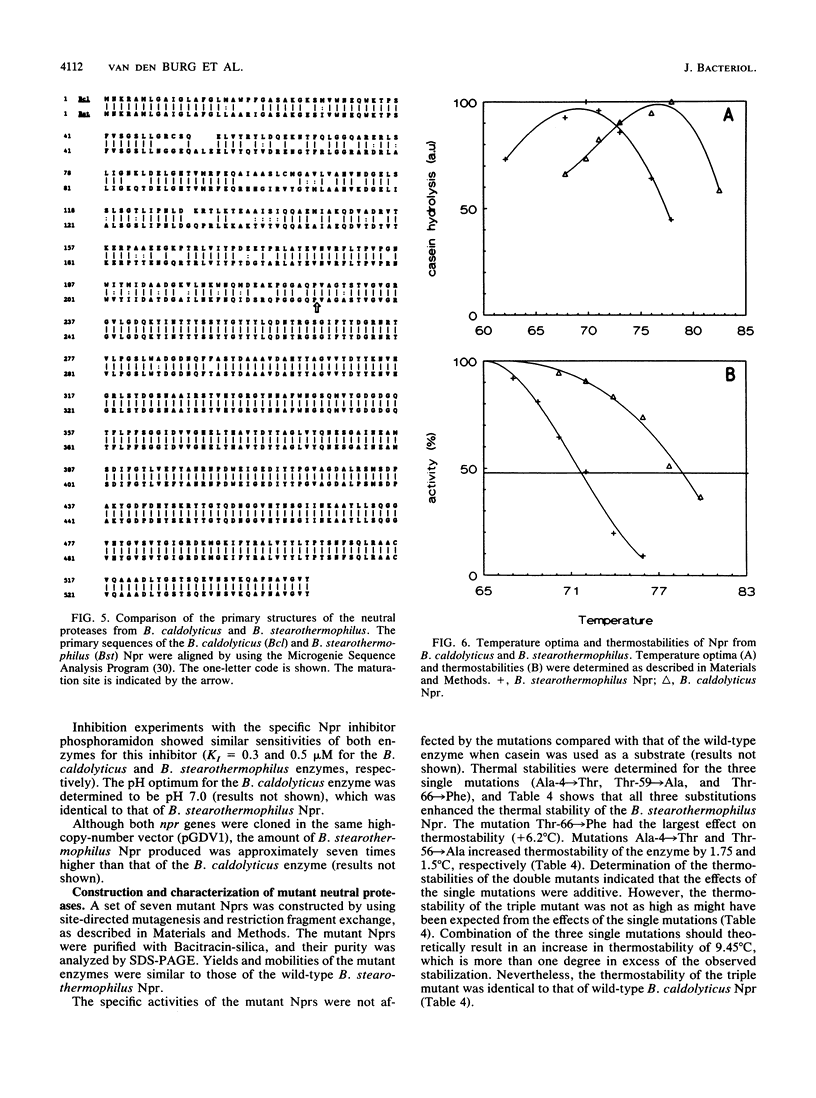
By using a gene library of Bacillus caldolyticus constructed in phage lambda EMBL12 and selecting for proteolytically active phages on plates supplemented with 0.8% skim milk, chromosomal B. caldolyticus DNA fragments that specified proteolytic activity were obtained. Subcloning of one of these fragments in a protease-deficient Bacillus subtilis strain resulted in protease proficiency of the host. The nucleotide sequence of a 2-kb HinfI-MluI fragment contained an open reading frame (ORF) that specified a protein of 544 amino acids. This ORF was denoted as the B. caldolyticus npr gene, because the nucleotide and amino acid sequences of the ORF were highly similar to that of the Bacillus stearothermophilus npr gene. Additionally, the size, pH optimum, and sensitivity to the specific Npr inhibitor phosphoramidon of the secreted enzyme indicated that the B. caldolyticus enzyme was a neutral protease. The B. sterothermophilus and B. caldolyticus enzymes differed at only three amino acid positions. Nevertheless, the thermostability and optimum temperature of the B. caldolyticus enzyme were 7 to 8 degrees C higher than those of the B. stearothermophilus enzyme. In a three-dimensional model of the B. stearothermophilus Npr the three substitutions (Ala-4 to Thr, Thr-59 to Ala, and Thr-66 to Phe) were present at solvent-exposed positions. The role of these residues in thermostability was analyzed by using site-directed mutagenesis. It was shown that all three amino acid substitutions contributed to the observed difference in thermostability between the neutral proteases from B. stearothermophilus and B. caldolyticus.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aoki T., Noguchi N., Sasatsu M., Kono M. Complete nucleotide sequence of pTZ12, a chloramphenicol-resistance plasmid of Bacillus subtilis. Gene. 1987;51(1):107–111. doi: 10.1016/0378-1119(87)90481-1. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal N., Kleinschmidt A. K., Spatz H. C., Trautner T. A. Physical properties of the DNA of bacteriophage SP50. Mol Gen Genet. 1967;100(1):39–55. doi: 10.1007/BF00425774. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Protein folding. Biochem J. 1990 Aug 15;270(1):1–16. doi: 10.1042/bj2700001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijsink V. G., Vriend G., Van Den Burg B., Venema G., Stulp B. K. Contribution of the C-terminal amino acid to the stability of Bacillus subtilis neutral protease. Protein Eng. 1990 Oct;4(1):99–104. doi: 10.1093/protein/4.1.99. [DOI] [PubMed] [Google Scholar]
- Frischauf A. M., Lehrach H., Poustka A., Murray N. Lambda replacement vectors carrying polylinker sequences. J Mol Biol. 1983 Nov 15;170(4):827–842. doi: 10.1016/s0022-2836(83)80190-9. [DOI] [PubMed] [Google Scholar]
- Fujii M., Takagi M., Imanaka T., Aiba S. Molecular cloning of a thermostable neutral protease gene from Bacillus stearothermophilus in a vector plasmid and its expression in Bacillus stearothermophilus and Bacillus subtilis. J Bacteriol. 1983 May;154(2):831–837. doi: 10.1128/jb.154.2.831-837.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujishima A., Honda K. Electrochemical photolysis of water at a semiconductor electrode. Nature. 1972 Jul 7;238(5358):37–38. doi: 10.1038/238037a0. [DOI] [PubMed] [Google Scholar]
- Grosveld F. G., Dahl H. H., de Boer E., Flavell R. A. Isolation of beta-globin-related genes from a human cosmid library. Gene. 1981 Apr;13(3):227–237. doi: 10.1016/0378-1119(81)90028-7. [DOI] [PubMed] [Google Scholar]
- Haima P., van Sinderen D., Schotting H., Bron S., Venema G. Development of a beta-galactosidase alpha-complementation system for molecular cloning in Bacillus subtilis. Gene. 1990 Jan 31;86(1):63–69. doi: 10.1016/0378-1119(90)90114-7. [DOI] [PubMed] [Google Scholar]
- Heinen U. J., Heinen W. Characteristics and properties of a caldo-active bacterium producing extracellular enzymes and two related strains. Arch Mikrobiol. 1972;82(1):1–23. doi: 10.1007/BF00424925. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J Bacteriol. 1982 May;150(2):804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanaka T., Shibazaki M., Takagi M. A new way of enhancing the thermostability of proteases. Nature. 1986 Dec 18;324(6098):695–697. doi: 10.1038/324695a0. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura F., Doi R. H. Construction of a Bacillus subtilis double mutant deficient in extracellular alkaline and neutral proteases. J Bacteriol. 1984 Oct;160(1):442–444. doi: 10.1128/jb.160.1.442-444.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keay L. Neutral proteases of the genus Bacillus. Biochem Biophys Res Commun. 1969 Jul 23;36(2):257–265. doi: 10.1016/0006-291x(69)90323-4. [DOI] [PubMed] [Google Scholar]
- Kubo M., Imanaka T. Cloning and nucleotide sequence of the highly thermostable neutral protease gene from Bacillus stearothermophilus. J Gen Microbiol. 1988 Jul;134(7):1883–1892. doi: 10.1099/00221287-134-7-1883. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Menéndez-Arias L., Argos P. Engineering protein thermal stability. Sequence statistics point to residue substitutions in alpha-helices. J Mol Biol. 1989 Mar 20;206(2):397–406. doi: 10.1016/0022-2836(89)90488-9. [DOI] [PubMed] [Google Scholar]
- Merkler D. J., Farrington G. K., Wedler F. C. Protein thermostability. Correlations between calculated macroscopic parameters and growth temperature for closely related thermophilic and mesophilic bacilli. Int J Pept Protein Res. 1981 Nov;18(5):430–442. [PubMed] [Google Scholar]
- Niaudet B., Goze A., Ehrlich S. D. Insertional mutagenesis in Bacillus subtilis: mechanism and use in gene cloning. Gene. 1982 Oct;19(3):277–284. doi: 10.1016/0378-1119(82)90017-8. [DOI] [PubMed] [Google Scholar]
- Pakula A. A., Sauer R. T. Amino acid substitutions that increase the thermal stability of the lambda Cro protein. Proteins. 1989;5(3):202–210. doi: 10.1002/prot.340050303. [DOI] [PubMed] [Google Scholar]
- Pakula A. A., Sauer R. T. Reverse hydrophobic effects relieved by amino-acid substitutions at a protein surface. Nature. 1990 Mar 22;344(6264):363–364. doi: 10.1038/344363a0. [DOI] [PubMed] [Google Scholar]
- Pauptit R. A., Karlsson R., Picot D., Jenkins J. A., Niklaus-Reimer A. S., Jansonius J. N. Crystal structure of neutral protease from Bacillus cereus refined at 3.0 A resolution and comparison with the homologous but more thermostable enzyme thermolysin. J Mol Biol. 1988 Feb 5;199(3):525–537. doi: 10.1016/0022-2836(88)90623-7. [DOI] [PubMed] [Google Scholar]
- Priest F. G. Extracellular enzyme synthesis in the genus Bacillus. Bacteriol Rev. 1977 Sep;41(3):711–753. doi: 10.1128/br.41.3.711-753.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queen C., Korn L. J. A comprehensive sequence analysis program for the IBM personal computer. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):581–599. doi: 10.1093/nar/12.1part2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidler W., Niederer E., Suter F., Zuber H. The primary structure of Bacillus cereus neutral proteinase and comparison with thermolysin and Bacillus subtilis neutral proteinase. Biol Chem Hoppe Seyler. 1986 Jul;367(7):643–657. doi: 10.1515/bchm3.1986.367.2.643. [DOI] [PubMed] [Google Scholar]
- Stanssens P., Opsomer C., McKeown Y. M., Kramer W., Zabeau M., Fritz H. J. Efficient oligonucleotide-directed construction of mutations in expression vectors by the gapped duplex DNA method using alternating selectable markers. Nucleic Acids Res. 1989 Jun 26;17(12):4441–4454. doi: 10.1093/nar/17.12.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M., Imanaka T., Aiba S. Nucleotide sequence and promoter region for the neutral protease gene from Bacillus stearothermophilus. J Bacteriol. 1985 Sep;163(3):824–831. doi: 10.1128/jb.163.3.824-831.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VENEMA G., PRITCHARD R. H., VENEMA-SCHROEDER T. FATE OF TRANSFORMING DEOXYRIBONUCLEIC ACID IN BACILLUS SUBTILIS. J Bacteriol. 1965 May;89:1250–1255. doi: 10.1128/jb.89.5.1250-1255.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Burg B., Eijsink V. G., Stulp B. K., Venema G. One-step affinity purification of Bacillus neutral proteases using bacitracin-silica. J Biochem Biophys Methods. 1989 May;18(3):209–219. doi: 10.1016/0165-022x(89)90005-5. [DOI] [PubMed] [Google Scholar]
- Vasantha N., Thompson L. D., Rhodes C., Banner C., Nagle J., Filpula D. Genes for alkaline protease and neutral protease from Bacillus amyloliquefaciens contain a large open reading frame between the regions coding for signal sequence and mature protein. J Bacteriol. 1984 Sep;159(3):811–819. doi: 10.1128/jb.159.3.811-819.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M. Y., Ferrari E., Henner D. J. Cloning of the neutral protease gene of Bacillus subtilis and the use of the cloned gene to create an in vitro-derived deletion mutation. J Bacteriol. 1984 Oct;160(1):15–21. doi: 10.1128/jb.160.1.15-21.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]




