Abstract
The spliceosome is a large, dynamic ribonuclear protein complex, required for the removal of intron sequences from newly synthesized eukaryotic RNAs. The spliceosome contains five essential small nuclear RNAs (snRNAs): U1, U2, U4, U5, and U6. Phylogenetic comparisons of snRNAs from protists to mammals have long demonstrated remarkable conservation in both primary sequence and secondary structure. In contrast, the snRNAs of the hemiascomycetous yeast Saccharomyces cerevisiae have highly unusual features that set them apart from the snRNAs of other eukaryotes. With an emphasis on the pathogenic yeast Candida albicans, we have now identified and compared snRNAs from newly sequenced yeast genomes, providing a perspective on spliceosome evolution within the hemiascomycetes. In addition to tracing the origins of previously identified snRNA variations present in Saccharomyces cerevisiae, we have found numerous unexpected changes occurring throughout the hemiascomycetous lineages. Our observations reveal interesting examples of RNA and protein coevolution, giving rise to altered interaction domains, losses of deeply conserved snRNA-binding proteins, and unique snRNA sequence changes within the catalytic center of the spliceosome. These same yeast lineages have experienced exceptionally high rates of intron loss, such that modern hemiascomycetous genomes contain introns in only ∼5% of their genes. Also, the splice site sequences of those introns that remain adhere to an unusually strict consensus. Some of the snRNA variations we observe may thus reflect the altered intron landscape with which the hemiascomycetous spliceosome must contend.
Keywords: snRNAs, evolution, splicing, yeast, hemiascomycetes
INTRODUCTION
Eukaryotic genes are often interrupted by noncoding intron sequences, which must be spliced out of pre-mRNA transcripts before the surrounding coding sequences can be translated into protein. For accurate protein expression, intron removal must be performed with absolute precision. This process is greatly complicated by the fact that intron sequences are highly variable, with relatively little information content to signal their boundaries. There are three short (2–7 nucleotides [nt]) and often highly degenerate primary elements that define each intron—the 5′ and 3′ splice sites at either end of the intron, and an internal branch site (Lim and Burge 2001). Further complicating matters, many transcripts are alternatively spliced, giving rise to one of multiple functionally distinct mRNA products.
The nuclear machine that accomplishes the intricate task of intron removal is the spliceosome, a large, dynamic assemblage of five small nuclear RNAs (snRNAs) designated U1, U2, U4, U5, and U6, and >100 proteins (for review, see Jurica and Moore 2003). Each snRNA is associated with a set of spliceosomal proteins, together making up the small nuclear ribonucleoproteins (snRNPs). For each round of splicing, the five snRNPs and numerous associated factors assemble anew on the pre-mRNA substrate to form the catalytically active spliceosome. The highly dynamic nature of spliceosome assembly provides the precision and flexibility required to accurately identify a remarkably diverse set of substrates (Staley and Guthrie 1998), while also providing multiple opportunities for regulation in response to environmental or developmental needs (e.g., Spingola and Ares 2000; Graveley 2005; Elliott and Grellscheid 2006; Pleiss et al. 2007; Tanabe et al. 2007).
The spliceosomal snRNAs are highly structured, with multiple intramolecular RNA helices (for review, see Guthrie and Patterson 1988). The observation that U6 snRNA had sequences complementary to regions of U4 and U2 led to the discoveries that U6 can pair with either of these snRNAs to form intermolecular helices as well, and that these mutually exclusive interactions were essential for the dynamic assembly of the spliceosome (Brow and Guthrie 1988; Wu and Manley 1991; Madhani and Guthrie 1992; Sun and Manley 1995). Finally, U1, U2, U5, and U6 all have short sequences that can pair directly with elements of the pre-mRNA substrate during spliceosome assembly and splicing catalysis.
In addition to sequences involved in base-pairing interactions, the snRNAs also have single-stranded regions important for protein interactions. For example, well-conserved loop sequences provide binding sites for snRNP proteins. U1, U2, U4, and U5 also share a common single-stranded consensus sequence bound by a heptameric complex of Sm proteins (for review, see Will and Luhrmann 2001).
In studying the mechanisms of splicing and its regulation, the hemiascomycetous budding yeast Saccharomyces cerevisiae has long been one of the major model systems. Because of the ease with which genetic manipulations can be performed on S. cerevisiae, compensatory base-pair analysis of the snRNAs has been a valuable tool in establishing in vivo roles for predicted secondary structures (e.g., Madhani and Guthrie 1992). Covariation analysis, which makes use of phylogenetic sequence comparisons, is a conceptually similar approach for confirming secondary structures. With a sufficiently large set of species, covering appropriate evolutionary distances, such an approach can be quite powerful (Guthrie and Patterson 1988). The number of hemiascomycetous yeasts whose genomes have been sequenced is exceptionally high, making them particularly amenable to studies of molecular evolution (Dujon 2006).
Despite highly similar morphologies, the hemiascomycetes exhibit a remarkable diversity of environmental niches (Kurtzman and Fell 2000); for example, they can be found throughout the natural environment, from the surface of grapes (S. cerevisiae) to the refuse of insects (Pichia guilliermondii), in various processed foods, such as corn (Yarrowia lipolytica), meats (Debaryomyces hansenii), and cheeses (Kluyveromyces lactis), or growing pathogenically within human hosts (numerous Candida species). One hemiascomycete, Candida albicans, is of particular interest because of its clinical relevance. It is the most common fungal pathogen of humans, and is capable of invading virtually every human organ and tissue (Odds 1988).
In the course of studying the C. albicans snRNAs, we identified numerous unexpected differences between these snRNAs and those of S. cerevisiae. This prompted us to investigate more broadly the evolution of snRNAs and their associated proteins within the hemiascomycetes. Using established phylogenetic relationships among these yeasts and parsimonious interpretations of the variations we found, we have inferred likely evolutionary histories for the hemiascomycetous snRNAs. Where the patterns of evolutionary change hint at function, we have suggested possible explanations for and consequences of these changes. We find cases in which well-conserved interactions between the snRNAs and their associated proteins appear to have been substantially altered or lost, examples of newly arisen snRNA structural domains, and changes in the stability of the intermolecular snRNA helices within the catalytic center of the spliceosome. Overall, we believe the variation we observe draws a picture of a rapidly evolving spliceosome, adapting to the particular needs of this biologically diverse group of yeasts.
RESULTS AND DISCUSSION
Candida albicans snRNA secondary structures
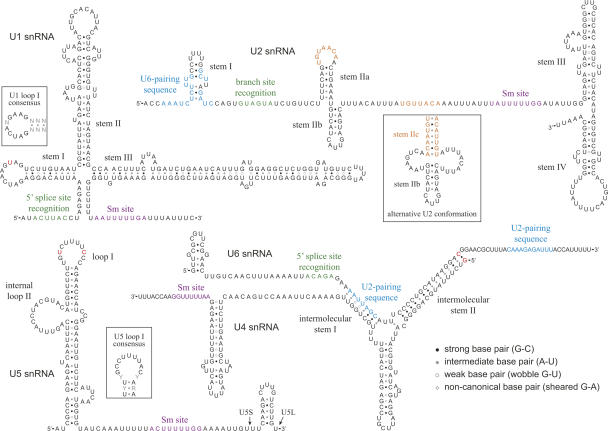
The genomic loci of C. albicans U2, U4, and U6 snRNAs have been identified previously (Bon et al. 2003). To identify the U1 and U5 loci, we searched by BLAST for sequences similar to S. cerevisiae U1 and U5 within the C. albicans genome (Jones et al. 2004). While regions of primary sequence within snRNAs are remarkably well conserved among higher eukaryotes, it is the conservation of secondary structure that is most striking (Guthrie and Patterson 1988). The secondary structures of the candidates found by BLAST support their identities as the C. albicans spliceosomal snRNAs. In Figure 1, we present our models of the C. albicans snRNA secondary structures. We determined structures primarily through manual inspection and comparison to structures conserved between human and S. cerevisiae (Guthrie and Patterson 1988). In addition, we used the m-fold algorithm to suggest possible folds within regions whose structures are less well conserved (Zuker 2003). Finally, we used comparisons with snRNAs we identified within the sequenced genomes of several close relatives of C. albicans to support or reject alternative structures within ambiguous regions (e.g., the 5′ end of U6 snRNA).
FIGURE 1.
Secondary structure predictions for the C. albicans snRNAs. Some C. albicans sequence variations discussed in the text are highlighted in red; other colored snRNA sequences are identified by labels. The two noncanonical base pairs of U4 (G•A) have been confirmed experimentally for human U4 (Vidovic et al. 2000). U5S and U5L indicate the 3′ ends of the short and long forms of U5 snRNA, respectively. (Insets) Consensus sequences for U1 loop I and U5 loop I; alternative conformation of U2 stem II (Hilliker et al. 2007). Nucleotide abbreviations for this and subsequent figures: (G) guanosine, (A) adenosine, (U) uridine, (C) cytidine; (Y) U or C; (R) G or A; (N) any nucleotide.
We expected that the structures and sequences of the C. albicans snRNAs would differ little from those of its relative S. cerevisiae. Surprisingly, we found numerous differences throughout the snRNAs, even within regions that generally show strong conservation throughout eukaryotes (discussed below). In cases where the C. albicans primary sequence deviates from consensus, it was possible the differences were due to errors in the available genomic sequence. To test this, we looked at snRNAs from the independently sequenced genome of another pathogenic Candida species, C. dubliniensis (www.sanger.ac.uk). In all cases, the unexpected deviations we found in C. albicans were conserved in C. dubliniensis (see below), demonstrating they were not simply the results of sequencing errors.
Sequence inspection alone did not allow us to determine the 3′ ends of the mature snRNAs. In S. cerevisiae, U1, U2, and U5 precursors have 3′ stem–loops that serve as sites for cleavage by RNase III (Chanfreau et al. 1997; Abou Elela and Ares 1998; Seipelt et al. 1999). These structures are important for normal 3′ end processing, but are removed from the mature snRNAs. The S. cerevisiae U4 gene also contains a potential 3′ stem–loop that is absent from the mature U4 snRNA. Thus, the presence of conserved 3′ stem–loops does not necessarily indicate they are present in mature molecules. We therefore determined the 3′ ends experimentally using a modified 3′ RACE procedure (see Materials and Methods). The structures we present in Figure 1 correspond to the predominant species identified by 3′ RACE. Interestingly, U1, U2, U4, and U5 all have potential stem–loop structures downstream of their mature 3′ ends, and these structures are conserved in other Candida species. As is the case for S. cerevisiae U1, U2, and U5, these structures may be present within snRNA precursors and play a role in proper 3′ end processing.
To gain insights into the evolution of the snRNAs in fungi, and in hemiascomycetous yeasts in particular, we also identified snRNAs from many of the sequenced fungal genomes now available. We did this primarily using BLAST to identify sequences similar to known snRNAs, and tested candidates by modeling potential secondary structures to determine whether they conformed to consensus snRNA structures. For some snRNAs whose primary sequence conservation was insufficient for identification by BLAST (e.g., U1 snRNAs from more distantly related fungi), we identified candidate sequences by searching for regular expressions that incorporated short, well-conserved primary sequence elements (Friedl 2006).
The “fungal” domains
Early phylogenetic comparisons of snRNA structures revealed several striking features unique to yeast (Ares 1986; Kretzner et al. 1987; Patterson and Guthrie 1987; Siliciano et al. 1987). S. cerevisiae has large insertions in regions of U1, U2, and U5 snRNAs that are otherwise highly constrained in both length and secondary structure. The S. cerevisiae insertions range in size from a 34-nt stem–loop structure in the internal loop II of U5 to two 400–500-nt insertions surrounding stem III of U2 (Guthrie and Patterson 1988). These insertions were termed the “fungal domains” because they were found in S. cerevisiae, and it was thought they would be common features among fungal snRNAs. It was known at the time, however, that the distantly related yeast Schizosaccharomyces pombe lacked the U2 fungal domain (Ares 1986), and subsequent examination of snRNAs from other yeasts by Northern hybridization revealed that many other species had more conventionally-sized snRNAs, suggesting the fungal domain insertions would be specific to a smaller subset of yeasts (Roiha et al. 1989).
Using data now available from the many genome sequencing projects, we have found that the U2 and U5 fungal domains are restricted to S. cerevisiae and other members of the Saccharomyces complex (Fig. 2). In C. albicans, the U2 fungal domains are entirely absent. Likewise, internal loop II of C. albicans U5 lacks the stem–loop insertion found in S. cerevisiae (Fig. 1). The function of the fungal domain insertions is unclear, as S. cerevisiae can tolerate deletion of these domains within U1 (Siliciano et al. 1991), U2 (Shuster and Guthrie 1988), and U5 (Frank et al. 1994) with little or no effect on cell growth. Conserved structures among the insertions, however, suggest there may be functional roles for these newly acquired domains. The short U5 fungal domain is in all cases capable of forming a stable stem–loop structure. There is little conservation within the primary sequence, with the possible exception of an internal AA/AA bulge found in some species. For fungal domain helices such as this that are well conserved in structure but not in sequence, it is also possible they have no functional role, but are evolutionarily constrained to helical form to avoid perturbing snRNA function.
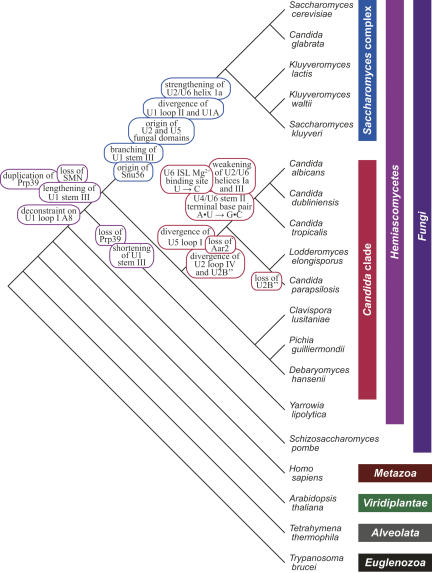
FIGURE 2.
Key events in the evolutionary history of the hemiascomycetous snRNAs. Phylogenetic tree (not to scale) represents the relationships among the eukaryotes shown, emphasizing the hemiascomycetous yeasts. The timings of evolutionary events discussed in the text, as inferred from parsimony analysis, are labeled on the tree. Branch order is based on previous analyses of others (Diezmann et al. 2004; Tsong et al. 2006; B. Tuch, pers. comm.).
The evolutionary history of the U1 fungal domain is more complex. In most species for which U1 has been characterized, stem III is highly constrained in size (22–26 nt) (e.g., Guthrie and Patterson 1988). Early in the hemiascomycete lineage, however, this size constraint appears to have been lost. In Y. lipolytica, stem III is substantially shorter than in other species (14 nt), while in an ancestor of the Candida and Saccharomyces lineages stem III appears to have lengthened dramatically (Fig. 3). In the modern descendents, there is now substantial variation in the length of this stem; in C. albicans, it is 104 nt long. While stem III has remained unbranched in the Candida clade, the main stem has acquired multiple internal stem–loop insertions in the Saccharomyces complex (Kretzner et al. 1990). These include both short, species-specific insertions within internal bulges and one large branched insertion common to all members of the Saccharomyces complex. Intriguingly, the large common insertion contains an internal stem–loop whose primary sequence is remarkably well conserved throughout the Saccharomyces complex (Fig. 3), strongly suggesting a conserved functional role. One possibility is that this structure serves as a novel binding site for a spliceosomal protein; the same may be true of the extended stem found in both the Saccharomyces complex and the Candida clade (although in this case there is no obvious conservation within the primary sequence).
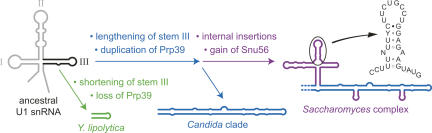
FIGURE 3.
Evolutionary model for the U1 snRNA fungal domain. U1 stem–loop III was shortened in the Y. lipolytica lineage (green) and lengthened in an ancestor of the Candida and Saccharomyces lineages (blue). There have been multiple insertions in stem III within the Saccharomyces complex (purple), including one large branched insertion that contains a well-conserved stem–loop (sequence shown). Gains and losses of U1 snRNP proteins correlating with these evolutionary events are also indicated.
Two candidates for interacting with the conserved U1 fungal domain elements are the essential U1 snRNP proteins Snu56 and Prp42, both of which are thought to be specific to budding yeast (Gottschalk et al. 1998; McLean and Rymond 1998). While Snu56 is well conserved within species of the Saccharomyces complex, we were unable to find an ortholog in any other sequenced genome. Thus, the distribution of Snu56 correlates with that of the well-conserved fungal domain stem–loop shown in Figure 3. Prp42 is similar to a more broadly distributed U1 snRNP splicing factor, Prp39 (Lockhart and Rymond 1994). Prp42 appears to have arisen as a duplication of Prp39 in a common ancestor of the Saccharomyces complex and the Candida clade, correlating with the appearance of the extended helix of U1 stem–loop III (Figs. 2, 3). Perhaps Prp39 has a more deeply conserved association with this stem, and Prp42 arose to accommodate its extension. Consistent with this idea, we could not detect an ortholog of either Prp39 or Prp42 in Y. lipolytica, the hemiascomycete in which stem III is unusually short (Fig. 3). Such a model, however, must accommodate additional observations. First, although the U1 fungal domain is not essential for viability in S. cerevisiae, both Snu56 and Prp42 are (Gottschalk et al. 1998; McLean and Rymond 1998), and so their functions could not be mediated entirely through association with the fungal domain. Second, the sequences of stem–loop III in two members of the Candida clade, P. guilliermondii and D. hansenii, have reverted to a more conventional size (23 and 29 nt, respectively), and yet both species have retained both Prp39 and Prp42, again suggesting functions that are not mediated entirely through fungal domain association.
U1 snRNA
U1 snRNA associates with the 5′ splice site prior to the first catalytic step of splicing. A single-stranded region at the 5′ end of U1 base pairs with the 5′ splice site directly, helping to define intron sequences and commit them to the splicing reaction (Guthrie and Patterson 1988).
U1A binding site
The second stem–loop of U1 snRNA has a well-studied interaction with U1A (also called Mud1 in S. cerevisiae), a protein component of the U1 snRNP. The X-ray crystal structure of this protein complexed with U1 loop II has been solved at high resolution (Oubridge et al. 1994). U1A contains two distinct RNA recognition motifs (RRMs), the most common of known RNA-binding domains (Burd and Dreyfuss 1994). An RRM is comprised of two short, well-conserved elements, RNP1 and RNP2, embedded in a larger RNA-binding domain. The amino-terminal RRM of U1A makes several direct contacts with a stretch of highly conserved nucleotides within U1 loop II (Fig. 4A; Oubridge et al. 1994). While the canonical U1A binding site is conserved in C. albicans and other Candida species, it appears to have been lost within the S. cerevisiae lineage. Instead, S. cerevisiae U1A binds with a substantially lower affinity to an internal loop of stem II, which lacks the canonical U1A binding site sequence (Tang and Rosbash 1996).
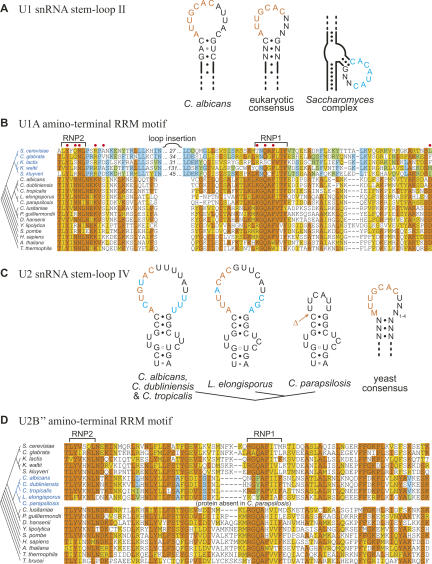
FIGURE 4.
Divergence of snRNP proteins and their snRNA binding sites within the hemiascomycetes. (A) Orange text represents the canonical U1A binding site in U1 snRNA stem–loop II. Blue text represents an alternative consensus loop sequence in the Saccharomyces complex U1 snRNAs, a candidate binding site for the altered U1A protein. (B) Alignment of the U1 snRNA-binding domain of U1A, with amino acids identical or similar to the majority consensus shaded in orange or yellow, respectively (or blue and green, for the consensus within the Saccharomyces complex subset). Phylogenetic relationships are depicted on the left, with Saccharomyces complex species in blue. Amino acids known to form direct contacts with U1 snRNA in humans are labeled with red dots (Oubridge et al. 1994). (C) Orange text represents the canonical U2B″ binding site in U2 snRNA stem–loop IV. Blue text represents deviations (nucleotide changes and insertions) from the consensus loop IV sequence. The site from which the U2B″ binding site has been deleted in C. parapsilosis is indicated (Δ). Within the yeast consensus, M represents either C or A. (D) Alignment of the U2 snRNA-binding domain of U2B″. Shading of amino acids and phylogeny are as described in B, but highlighted changes are now within a Candida subclade instead of the Saccharomyces complex. Many eukaryotic U2B″ proteins contain an additional C-terminal RRM motif; fungi and trypanosomes contain only the RRM motif shown.
This apparent loss of the U1A binding site correlates with a degeneration of the U1A RNA binding domain, suggesting that U1 and U1A may have coevolved novel interaction domains. Within S. cerevisiae (Liao et al. 1993) and other members of the Saccharomyces complex—the same yeasts that have lost the U1A consensus binding site—the U1A RNA binding domain is highly divergent, and the RNP1 and RNP2 elements are separated by a large insertion (30 amino acids in S. cerevisiae). Notably, the first two amino acids of RNP1, which are required for binding of human U1A to U1 in vitro (Boelens et al. 1991), and several of the amino acids that make direct contacts with U1 loop II in humans (Oubridge et al. 1994) are not conserved in the Saccharomyces complex (Fig. 4B). Consistent with the idea of a novel interaction domain, several of the amino acids within this region are well conserved within the Saccharomyces complex, and yet are entirely distinct from the amino acids found in other species (Fig. 4B). The variable loop inserted between RNP1 and RNP2 has also been proposed to play a role in this altered interaction (Liao et al. 1993).
It was previously noted that the internal RNA loop sequence that U1A binds in S. cerevisiae contains a stretch of CA repeats, which are also present within the same region of K. lactis U1 (Liao et al. 1993). By comparing U1 genes from sequenced genomes within the Saccharomyces complex, we identified a well-conserved consensus motif (CACAUAC) that overlaps the U1A binding site in S. cerevisiae. In some species (such as S. cerevisiae), the number of CA (or UA) repeats within this motif is expanded. The motif is generally located within a short stem–loop structure projecting from within an internal loop of stem II (Fig. 4A). It will be interesting to determine whether this motif represents the physical binding site for U1A proteins from the Saccharomyces complex. It is quite different from the U1A binding site in most species, but there are limited similarities: they both contain the sequence CAC, and they are both located within the 5′ ends of loop regions. Stem II is unusually long within the Saccharomyces complex, and this added length may have arisen as an insertion of an additional stem–loop into the end of the ancestral stem, similar to the insertions that occurred within the U1 fungal domain in this same lineage (discussed above). The Saccharomyces U1A binding site could thus be derived from the ancestral binding site, with its internalized location merely a consequence of this insertion event.
It is possible that the lower affinity of U1A for stem II in the Saccharomyces complex is augmented by additional associations, such as an association with the U1 fungal domain. Another possibility is that the association of U1A with the S. cerevisiae U1 snRNP is mediated in part through protein–protein interactions. In the basal eukaryote Trypanosoma brucei, in which the second stem of U1 is entirely absent, the apparent functional homolog of U1A (U1–24K) is unable to bind U1 directly. Instead, U1–24K associates with the conserved U1 snRNP protein U1–70K, which in turn binds to U1 directly through a conserved interaction with stem–loop I (Palfi et al. 2005). Early work with Xenopus laevis extracts demonstrated a weak association between U1A and U1 snRNAs lacking stem–loop II, again consistent with indirect interactions in addition to direct stem II binding (Hamm et al. 1987).
U1–70K binding site
The U1 snRNP protein U1–70K (called Snp1 in S. cerevisiae) contains an RRM domain that binds directly to the 5′ end of U1 loop I (Urlaub et al. 2000). As with loop II discussed above, the sequence of loop I is well conserved throughout eukaryotes. In this case, S. cerevisiae U1 loop I matches the consensus while C. albicans has a highly unusual variation (A8U; Fig. 1). This altered nucleotide is nearly invariant as deep in the eukaryotic lineage as T. brucei (Palfi et al. 2005), and is adjacent to the U1–70K binding site (Urlaub et al. 2000). By examining loop I from the various sequenced hemiascomycetes, we found the A8U variant within one species of the Saccharomyces complex (Saccharomyces castellii), the Candida clade, and the more distantly related hemiascomycete Y. lipolytica. Even among Candida species, the distribution of this variant is polyphyletic, suggesting a general relaxation in the evolutionary constraints on loop I A8 within the hemiascomycetes. We did not find this variant in nonhemiascomycetous yeasts (e.g., S. pombe, Neurospora crassa, various Aspergillus species). In human cells, introduction of an A8U mutation into U1 loop I reduces but does not eliminate association with U1–70K, while mutations at most other positions abolish this interaction completely (Surowy et al. 1989). Thus, the variability at this position within the hemiascomycetes may reflect a reduced requirement for a high-affinity association between U1 and U1–70K. Alternatively, it may reflect the evolution of an altered interaction between the two. Interestingly, variation at this position in U1 is not exclusive to the hemiascomycetes—it is also found in the vertebrates Gallus gallus (A8C; Branlant et al. 1980) and Sorex araneus (A8U; GenBank accession no. AC164871).
U2 snRNA
Like U1 snRNA, U2 associates with pre-mRNA prior to the first catalytic step. While U1 associates with the 5′ splice site, a short sequence near the 5′ end of U2 base pairs directly with the branch site (Fig. 1; Guthrie and Patterson 1988).
U2B″ binding site
In contrast to the U2 snRNAs of the Saccharomyces complex, which contain very large “fungal domain” insertions on either side of stem III, the C. albicans U2 snRNA is highly similar to consensus in both structure and sequence. One exception is the loop sequence at the end of stem IV, which is the binding site for the core U2 snRNP protein U2B″ (called Msl1 in S. cerevisiae) (for review, see Kambach et al. 1999). Like the U1 snRNP proteins U1A and U1–70K, U2B″ interacts with its binding site through an RRM domain. In fact, U2B″ and U1A are closely related proteins and share highly similar binding sites in their respective snRNAs (see Fig. 4; Scherly et al. 1990). S. cerevisiae U2B″ can associate with U2 snRNA directly in vitro (Tang et al. 1996). Unlike U1A, however, U2B″ requires association with another protein, U2A′ (called Lea1 in S. cerevisiae), before it can bind snRNA in vivo (Caspary and Seraphin 1998).
The snRNA binding site of U2B″ is slightly less evolutionarily constrained than that of U1A, but it is still easily identifiable in species as distant as T. brucei (He and Bellofatto 1995). Generally, we found that yeast vary by at most 1 nt from the consensus shown in Figure 4C. In C. albicans and its two close relatives C. dubliniensis and C. tropicalis, however, there are alternative pyrimidines at two positions within the U2B″ binding site (ACUGUAC), and the size of loop IV is 3 nt larger than the consensus size limit (Fig. 4C). The RNA-binding domain within the U2B″ protein also appears to be fairly degenerate in these three species, most notably around the beginning of the RNP1 element (Fig. 4D). (Interestingly, this same region was also notably divergent in the Saccharomyces U1A proteins, as discussed above.) The U2B″ RNP1 element is also degenerate in Lodderomyces elongisporus, a somewhat more distant relative of C. albicans (Fig. 4D). The sequence of the L. elongisporus U2B″ binding site is fairly common in yeast (AUUACAC), but the overall size of loop IV is large, comparable to that of C. albicans (Fig. 4C). Thus, if the alteration in RNP1 reflects an adaptation to a divergent binding site, it may have more to do with the size of loop IV than with the actual sequence of the binding site. As with the Saccharomyces U1A orthologs, there are several amino acids within U2B″ that are distinct and yet conserved among the four yeasts with large loop IV regions, perhaps reflecting an altered binding specificity.
The most unusual U2 loop IV sequence is that of Candida parapsilosis, a closer relative of L. elongisporus. Here, the U2B″ binding site appears to have been entirely deleted from U2 (Fig. 4C). Furthermore, we were unable to find an ortholog of either U2B″ or its associated protein U2A′ in the completed C. parapsilosis genome, suggesting that U2B″ has been lost along with its binding site. The deviations we observe in U2B″ and U2 snRNA may reflect a reduced or altered biological requirement for their interaction within this subset of the Candida lineage, culminating in complete loss of both in C. parapsilosis.
U6 snRNA and its complexes, U4/U6 and U2/U6
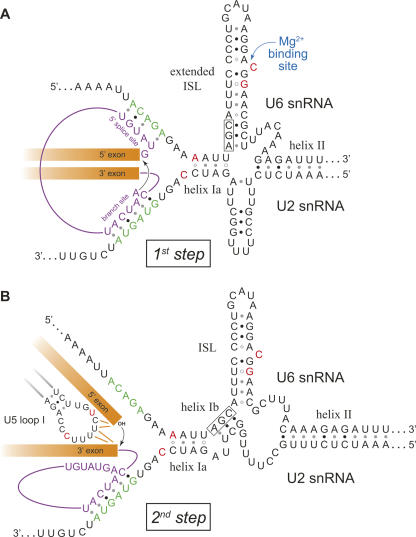
Once the 5′ splice site and branch site have been defined through associations with U1 and U2 snRNPs, U6 snRNP joins the spliceosome to facilitate the actual splicing reaction (Brow 2002). U6 enters the spliceosome in a complex that includes U4 and U5 snRNAs. Within this “tri-snRNP” complex, U4 and U6 snRNAs are directly associated with each other through extensive base-pairing (Fig. 1). This U4/U6 duplex is unwound by the U5 snRNP-associated ATPase Brr2, allowing U6 to exchange its association with U4 for an association with the U2 snRNA bound to the intron branch site (Fig. 5). U6 also base pairs directly with the 5′ splice site of the intron, displacing U1, and both U1 and U4 leave the spliceosome. Association of U6 creates the catalytic center of the spliceosome and allows the first step of splicing to occur (Fig. 5A). The 2′ hydroxyl of the branch point adenosine, which is bulged out of the helix formed by U2 and the branch site, carries out a nucleophilic attack on the 5′ exon/intron junction, resulting in a lariat-structured intermediate and a free upstream exon. In the second step of splicing (Fig. 5B), the 3′ hydroxyl of the upstream exon attacks the 3′ exon/intron junction, assisted by U5 snRNA (see below), liberating the lariat-structured intron and joining the exon sequences together, thereby completing the splicing reaction.
FIGURE 5.
Models of the two conformational states of the C. albicans U2/U6 snRNA duplex. Notable deviations from consensus sequences are highlighted in red. (A) Proposed first-step conformation of U2/U6, with an extended intramolecular stem–loop (ISL). The pre-mRNA (intron in purple, exons in orange) is shown associated with the active site (splice site recognition sequences in green). The black arrow indicates the first-step nucleophilic attack of the branch point adenosine on the 5′ exon/intron junction. (B) Proposed second-step conformation of U2/U6, with intermolecular helix Ib. U5 is shown coordinating 5′ and 3′ exon sequences. The black arrow indicates the second-step nucleophilic attack of the 5′ exon on the 3′ exon/intron junction. The invariant AGC triad that participates in both the extended ISL and helix Ib is boxed. Structures are based on models from other organisms (Madhani and Guthrie 1992; Sun and Manley 1995). (For simplification, the associations between U5 loop I and the 5′ exon during the first step and between the 5′ splice site and U6 during the second step are not shown.)
U2/U6 conformational states
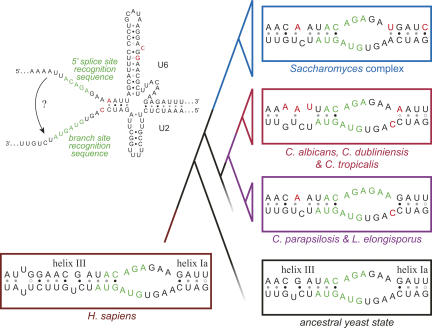
The precise secondary structure of the U2/U6 complex during the splicing reaction is unclear. Genetic experiments in yeast and humans have demonstrated the importance of intermolecular helices I and II (Fig. 5B; Datta and Weiner 1991; Wu and Manley 1991; Madhani and Guthrie 1992). Helix I is comprised of the two discontinuous helices Ia and Ib, separated by a 2-nt bulge. Helix Ia is immediately adjacent to the splice site binding sequences in the catalytic center of the spliceosome and is important for both steps of splicing catalysis (Madhani and Guthrie 1992). Distal to helix Ia and partially overlapping the splice site binding sequences is an additional intermolecular helix (helix III) required for splicing in humans (Sun and Manley 1995) but not in S. cerevisiae (Fig. 6; Yan and Ares 1996).
FIGURE 6.
Evolution of helices Ia and III in the hemiascomycetes. Regions of U2 and U6 snRNAs involved in intermolecular helix Ia and (possibly) helix III formation are shown for representative hemiascomycetes, the yeast ancestor, and humans. This region is also shown within the context of the larger C. albicans U2/U6 structure, with splice site recognition sequences highlighted in green. Sequence changes are highlighted in red. Species of the Candida clade that are not represented here all match the ancestral state, with the exception of the U6 helix III G-to-A transition, which is common to all hemiascomycetes. Although C. albicans helix Ia is shown with a terminal C•A+ wobble base pair, there is no evidence this occurs in vivo. Phylogenetic tree represents the relationships among species shown.
The sequences of U6 between helices I and II form an intramolecular stem–loop (ISL) crucial for splicing catalysis. The ISL bears a striking resemblance to domain 5 of group II self-splicing introns, which are thought to share a common ancestry with the spliceosomal snRNAs (Sashital et al. 2004). Both the ISL and domain 5 contain a bulged pyrimidine residue whose 5′ phosphate coordinates a magnesium ion essential for splicing catalysis (Seetharaman et al. 2006). Another similarity is a nearly invariant AGC triad at the base of the helix. Extension of the U6 ISL helix to incorporate this triad, however, is incompatible with formation of intermolecular helix Ib (Fig. 5). Nonetheless, data from humans support the existence of an extended ISL (Sun and Manley 1995), as does recent structural work with protein-free S. cerevisiae U2/U6 (Sashital et al. 2004). Importantly, an alternative U2/U6 structure in which the U6 ISL is extended allows coaxial stacking that could bring the ISL and the catalytic center of the spliceosome into close proximity (Sashital et al. 2004), an association supported by structural probing experiments (Rhode et al. 2006).
To reconcile these two mutually exclusive structures, it was proposed that the U2/U6 complex exists in both forms, undergoing a conformational switch between the two catalytic steps of the splicing reaction (Sashital et al. 2004). Indeed, computational modeling of U2/U6 suggests these two conformations can readily interconvert (Cao and Chen 2006). According to this model, the extended U6 ISL would associate with the catalytic center during the first step, facilitating nucleophilic attack of the branch point adenosine on the 5′ splice site (Fig. 5A). Prior to the second step, the U6 ISL would partially unwind, allowing formation of helix Ib (Fig. 5B). Consistent with this model, it has been shown that helix Ib is important for the second step of splicing in vivo (Hilliker and Staley 2004). Also, the pattern of interactions between the ISL and the catalytic center changes prior to the second step, consistent with a repositioning of the U6 ISL between the two steps (Rhode et al. 2006).
A phylogenetic comparison of yeast U2 and U6 snRNA sequences supports the existence of two conformational states. As is shown for C. albicans (Fig. 5), all sequenced hemiascomycetes have the potential to form both structures, as do the more distantly related yeasts Neurospora crassa and Aspergillus nidulans. In the yet more distant S. pombe, the extended form of the ISL would have a second bulged nucleotide, in addition to the conserved Mg2+-coordinating bulge discussed above. Curiously, this second bulge is quite similar to the first: both are bulged uridines surrounded by a C•G base pair toward the base of the stem and a potential C•A+ wobble base pair on the other side.
U6 ISL
The U6 ISL in C. albicans and its close relatives C. dubliniensis and C. tropicalis contains one highly unusual change at the essential Mg2+ binding site. Rather than the nearly invariant bulged uridine whose 5′ phosphate normally coordinates Mg2+, these three organisms contain a bulged cytidine (Fig. 5). This change correlates with another change 2 nt downstream, where a nearly invariant adenosine is replaced by a guanosine. These same two sequence changes are also present in a distantly related fungus, the parasitic microsporidian Antonospora locustae (Fast et al. 1998). The trypanosome Phytomonas sp. (though not T. brucei) has a bulged cytidine at the Mg2+ binding site, but without the A-to-G transition 2 nt downstream (Wieland and Bindereif 1995).
The consequence of the two sequence transitions in C. albicans is unclear. In S. cerevisiae, neither of these substitutions has an obvious effect on growth when introduced individually into the wild-type U6 sequence (McPheeters 1996). Interestingly, the analogous Mg2+-binding nucleotide within domain 5 of group II introns is usually a pyrimidine, but shows no marked preference for either cytidine or uridine (Michel et al. 1989).
It is possible the conservation of the bulged uridine in U6 is related not to ISL function but rather U4/U6 pairing. In addition to coordinating Mg2+ when U6 is paired with U2, this nucleotide also forms the terminal A•U base pair in the longer of the two U4/U6 intermolecular helices (stem II). The U6 U-to-C transition in the Candida lineage correlates with an A-to-G transition in the U4 pairing partner, creating a terminal G•C base pair in stem II (Fig. 1). Perhaps this change influences the unwinding of U4/U6 during spliceosome activation. (The terminal base pairs of U4/U6 in the other organisms with the U6 U-to-C transition—A. locustae and Phytomonas sp.—are unknown, as U4 snRNAs from these organisms have not been reported.)
U2/U6 helices Ia and III
The essential U2/U6 helix Ia is nearly invariant throughout eukaryotes, with the exception of certain hemiascomycetes. Within the Saccharomyces complex, there are two changes in U6 that alter this helix (Fig. 6). The presence of these variants was noted almost two decades ago, before the functional significance of this region—as a pairing partner for U2—was appreciated (Roiha et al. 1989). At one end of U2/U6 helix Ia, a G•U wobble base pair is replaced with a G•C base pair, while the other end of the helix may be extended in length by the creation of an A•U base pair; both changes should increase the stability of helix Ia. The novel G•C base pair seems to be important for S. cerevisiae splicing, as a U6 mutation that reverts the base pair to the weaker ancestral state (G•C to G•U) results in a temperature-sensitive growth defect, while a compensatory mutation in U2 that creates a base pair of intermediate strength (G•U to A•U) rescues this growth defect (Madhani and Guthrie 1992).
Within the Candida lineage, there have also been two sequence changes within the helix Ia region, one within U2 and one within U6 (Fig. 6). The U2 change (A to C) is adjacent to helix Ia, and arose in an ancestor of C. albicans and C. parapsilosis. The U6 change (G to A) arose more recently in an ancestor of C. albicans, C. dubliniensis, and C. tropicalis. This latter sequence change disrupts the only C•G base pair in the ancestral helix, replacing it with a potential C•A+ wobble base pair. In contrast to the changes in Saccharomyces U6, which should strengthen helix Ia, the change in C. albicans U6 should substantially weaken the helix.
As discussed, U2/U6 helix III is not required for splicing in S. cerevisiae. Looking at other yeast species, we find that the potential for helix III formation is generally comparable to that of S. cerevisiae: the potential base-pairing is conserved, but the helices would be four base pairs shorter than helix III of humans (Fig. 6). Because the primary sequences in this region are nearly invariant, however, there is no covariation to support a conserved structural role for this region as opposed to some other conserved role. If U2/U6 helix III does exist in yeast, it appears to have been lost within a recent ancestor of C. albicans. Two sequence changes in U6 would disrupt base-pairing in the center of helix III, making it unlikely to form in either C. albicans, C. dubliniensis, or C. tropicalis (Fig. 6). One of these changes (C to A) also arose independently in the Y. lipolytica lineage, and would perhaps disrupt helix III in this hemiascomycete, as well.
Transition from U4/U6 to U2/U6
The current view of the splicing mechanism posits a series of linked transitions in the conformational state of the spliceosome (Staley and Guthrie 1998; Butcher and Brow 2005; Konarska and Query 2005). Multiple spliceosomal NTPases act at various stages of assembly, catalysis, and disassembly. Coupling NTP hydrolysis to progression through the different conformational states would allow for kinetic proofreading within the spliceosome, enhancing substrate specificity and thereby increasing the fidelity of the overall splicing reaction (Burgess et al. 1990; Mayas et al. 2006). Basically, appropriate splicing substrates would promote faster transitions within the spliceosome, while the rate of NTP hydrolysis would serve as a timer, allowing splicing to proceed if the appropriate transition has occurred, but triggering a discard pathway if it has not (Burgess and Guthrie 1993). One known example of such regulation involves the spliceosomal ATPase Prp16, which acts between the first and second steps of splicing to promote a conformational rearrangement in the spliceosome (Schwer and Guthrie 1992) and may provide a kinetic proofreading function to ensure proper splice site choice (Query and Konarska 2004; Villa and Guthrie 2005). Recent work suggests that Prp16 mediates an intramolecular conformational change in U2 stem II (Fig. 1; Hilliker et al. 2007; Perriman and Ares 2007). As discussed above, another conformational change that may accompany the transition from the first step to the second involves the base-pairing between U2 and U6 (Fig. 5).
Since controlling the transition between conformational states of the spliceosome affects splicing fidelity, evolutionary or developmental modulations in this control may be important for changing substrate specificity (Konarska and Query 2005). For example, developmental modulation can allow tissue-specific or condition-specific splicing, facilitating the use of alternative splice sites. Evolutionary modulation, on the other hand, may allow for changes in intron specificity between species, perhaps facilitating changes in splice site consensus sequences. One mechanism for altering the transition between different conformational states would be through changes in snRNA sequence, stabilizing certain structural conformations or destabilizing others.
Several of the alterations we observe in the hemiascomycetous snRNAs may impact the transition from U4/U6 to U2/U6 by affecting their relative stabilities. Within the Saccharomyces complex, the increased stability of U2/U6 helix Ia may favor formation of the active spliceosome (Fig. 6). In C. albicans and its close relatives C. dubliniensis and C. tropicalis, however, the transition from U4/U6 to U2/U6 may be slowed by several of the evolutionary changes discussed above. The terminal base pair in U4/U6 stem II is stabilized by an A•U-to-G•C covariation, perhaps decreasing accessibility to the helicase Brr2. Three additional sequence changes in U6 decrease the stability of U2/U6, by weakening helix Ia and perhaps preventing formation of helix III (Fig. 6). Notably, each of these changes is specific to the same three species, suggesting that they arose at roughly the same time. Based on the kinetic proofreading model, modifying the rate of active spliceosome formation may have allowed for differential splicing regulation or changes in how different splice sites are tolerated within different yeast lineages.
In addition to affecting catalytic activation of the spliceosome, changes in U4/U6 and U2/U6 stability could also affect spliceosome recycling—disassembly of the spliceosome following splicing (which includes disassociation of U2/U6) and reestablishment of the precatalytic snRNP complexes (which includes pairing of U4/U6). As with unwinding of U4/U6 during catalytic activation, unwinding of U2/U6 during disassembly involves the U5 snRNP proteins Brr2 and Snu114 (Small et al. 2006). The essential splicing factor Aar2, a transient component of the U5 snRNP, has also been implicated in spliceosome recycling in S. cerevisiae, although the step at which it acts is unknown (Gottschalk et al. 2001). Orthologs of Aar2 are easily identifiable in most fungi, animals, and plants, and in the basal eukaryote T. thermophila, but Aar2 appears to have been lost in the Candida lineage before the divergence of C. albicans and C. parapsilosis (Fig. 2). Thus, strengthening of the U4/U6 complex or weakening of the U2/U6 complex in the Candida lineage may have been a response to the prior loss of the putative recycling factor Aar2.
U5 snRNA
U5 snRNA is involved in the second step of splicing, recruiting multiple protein factors to the spliceosome and coordinating the exon junction sequences for proper ligation at the 5′ and 3′ splice sites. Loop I is the most well-conserved region of U5, and nucleotides within this loop make direct contacts with the exon junction sequences during splicing through non-sequence-specific base-pairing (Sontheimer and Steitz 1993; Newman et al. 1995). Deletions and insertions within loop I of S. cerevisiae U5 cause misalignment of exon junction sequences, inhibiting the second step of splicing in vitro (O'Keefe and Newman 1998). In humans, however, this highly conserved region of U5 is not required for splicing in vitro (Segault et al. 1999), suggesting a more nuanced role in promoting the efficiency or fidelity of splicing in vivo.
U5 loop I
The most striking deviation in the C. albicans U5 snRNA is within loop I. Most of the positions in this loop are absolutely conserved in all previously reported U5 sequences (e.g., Guthrie and Patterson 1988). Moreover, chemical probing experiments suggest that protein associations with this loop are also highly conserved between S. cerevisiae and humans (Mougin et al. 2002). Two of the nearly invariant loop I nucleotides are altered within the C. albicans lineage (Fig. 1). These nucleotide substitutions appear to have occurred after the divergence of the D. hansenii lineage, but before C. albicans diverged from its closer relatives, which almost all share the same deviant loop I sequence. (The exception is L. elongisporus, which has only the second substitution.) In T. brucei and other trypanosomes—the only non-Candida organisms known to vary at either of these positions—there is an adenosine at the first of these two positions, but the rest of loop I matches the consensus (Xu et al. 1997; Ambrosio et al. 2007).
The implication of the Candida substitutions is unclear. The extraordinary conservation of these nucleotides suggests a central role in splicing, but they lie outside the region known to form direct contacts with exon sequences and the splicing factor Prp8 (Urlaub et al. 2000). Interestingly, these same nucleotide substitutions in U5 loop I were identified in a S. cerevisiae suppressor screen (Bacikova and Horowitz 2005). A mutant form of S. cerevisiae U5 carrying a loop I sequence identical to that of C. albicans was able to suppress a growth defect caused by deletion of a highly conserved domain in the second-step splicing factor Prp18. Perhaps the Candida-specific U5 changes have accompanied an alteration in the structure or function of Prp18 in the Candida lineage, although sequence analysis alone does not reveal any obvious changes in Prp18 that correlate with the U5 loop I variant.
U5 terminal stem–loop
In S. cerevisiae, there are two predominant forms of U5 snRNA, differing by the lengths of their 3′ ends (Patterson and Guthrie 1987). The longer form, U5L, contains a stem–loop downstream of its Sm binding site and is similar to the only form of U5 found in humans, while the shorter form, U5S, ends just upstream of this stem–loop. Northern hybridization and cloning of U5 snRNAs from other species has suggested that the short form is also common in other yeasts (Roiha et al. 1989; Frank et al. 1994).
Unlike both human and S. cerevisiae U5, the predominant C. albicans U5 snRNA corresponds to U5S (lacking the 3′ terminal stem–loop), while a very small proportion of U5 is of the U5L form. Using our modified 3′ RACE procedure (discussed above), we detected sequence signal corresponding only to the short form of U5. To determine whether any long form of U5 accumulates, we visualized our 3′ RACE products by gel electrophoresis (Supplemental Fig. 1). While the vast majority of product corresponded to U5S, we did detect a small amount of U5L.
The functional significance of the two forms of U5 in S. cerevisiae is unclear. Both forms are incorporated into snRNPs (Madhani et al. 1990), but the short form is sufficient for splicing activity in vivo (Chanfreau et al. 1997). Their production depends on distinct 3′ end processing pathways, and accumulation of U5L (but not U5S) is strictly dependent on RNase III (Chanfreau et al. 1997). Thus, the reduced accumulation of U5L in C. albicans may reflect an alteration in RNase III activity or a lower affinity of RNase III for the C. albicans U5 precursor.
More generally, the differences among U5 3′ ends may reflect fundamental differences in snRNP assembly pathways. The association of Sm proteins with human snRNAs is facilitated by a multiprotein complex, at the core of which is the survival of motor neurons (SMN) protein. For human U2, U4, and U5 snRNAs, recognition by the SMN complex requires an Sm binding site and a downstream stem–loop (for review, see Yong et al. 2004). In S. cerevisiae, however, the SMN complex has not been identified, and Sm assembly may instead be mediated by binding of the La protein and consequent removal of terminal stem–loop structures (Xue et al. 2000). While we can identify an SMN ortholog in Y. lipolytica, the other hemiascomycetes appear to have lost the SMN protein (Fig. 2). The use of alternative assembly pathways in both S. cerevisiae and C. albicans may thus explain why the U5 terminal stem–loop is often removed. Consistent with this idea, the terminal stem–loops present in human U1 and U4 snRNAs are also absent from the mature snRNAs in S. cerevisiae and C. albicans (e.g., Fig. 1). It will be interesting to determine whether the absence of SMN correlates with removal of terminal stem–loops in other species.
CONCLUSIONS
Our investigation of spliceosomal snRNAs revealed an unanticipated degree of variation throughout the hemiascomycetes. This variation may reflect some of the peculiar traits found in these yeasts. They have experienced a dramatic rate of intron loss, leaving their genomes largely devoid of the introns that litter many eukaryotic genomes (Dujon 2006). Those introns that remain typically number only in the hundreds and are highly stereotyped, with splice sites adhering to unusually strict consensus sequences (Spingola et al. 1999; Bon et al. 2003; Mitrovich et al. 2007). Thus, the diversity of introns with which the spliceosome must contend is greatly reduced in the hemiascomycetes, and this may have reduced certain evolutionary constraints on the snRNA sequences. Some of the variation within the snRNAs may also be related to general changes in the intron features of different hemiascomycetes. For example, there are differences between C. albicans and S. cerevisiae in the sequence and spacing of splice sites (Mitrovich et al. 2007). An analysis of introns in other hemiascomycetes suggests substantial changes in splice site usage, although sample sizes were too small to draw general conclusions (Bon et al. 2003). Finally, some of the snRNA changes may represent adaptations to the varied environments the hemiascomycetes inhabit. Modulating the transition rates between different spliceosomal conformations, as discussed for the U4/U6-to-U2/U6 transition, could provide novel opportunities for regulating the splicing (and therefore expression) of specific transcripts, perhaps in response to different environmental demands. Indeed, we have observed examples of environmentally regulated splicing in both S. cerevisiae (Pleiss et al. 2007) and C. albicans (Mitrovich et al. 2007).
As we have discussed, evolutionary variations within the snRNAs and in some cases correlated changes within their associated proteins lead to numerous predictions about spliceosome evolution in the hemiascomycetes. It will be most interesting to test these predictions experimentally. For example, are the conserved U1 fungal domain elements associated with novel yeast U1 snRNP proteins, and what advantage does this association provide? Does the novel consensus sequence found within U1 stem II of the Saccharomyces complex provide the binding site for the established interaction with the altered U1A protein (Tang and Rosbash 1996)? Do the variations in the C. albicans U2B″ protein reflect an altered specificity for the unusual loop IV sequence in U2? Is the second intermolecular helix of C. albicans U4/U6 a less efficient substrate for Brr2 unwinding because of its terminal G•C base pair, or does this change in U6 alter its ability to coordinate Mg2+ when complexed with U2? Does the atypical C. albicans U5 sequence affect the association or function of Prp18 or other components of the U5 snRNP? Answers to these and other questions should also provide broad insights into the mechanisms of splicing in general.
S. cerevisiae has long been a leading model organism for the study of splicing. We thought that our understanding of splicing regulation in S. cerevisiae would inform the studies of its pathogenic relative C. albicans, suggesting mechanisms of splicing regulation that may have been co-opted for survival and virulence within human hosts. The surprising differences we find between the S. cerevisiae and C. albicans spliceosomes, however, demonstrate that there are also many interesting and novel aspects of splicing and its regulation to be discovered in C. albicans. Furthermore, C. albicans may provide a good complement to S. cerevisiae as a model for human splicing. The relatedness of these two yeasts means many of the techniques established for studying splicing in S. cerevisiae will likely also work for C. albicans, while the variations in their snRNAs suggest ways in which one organism or the other will more accurately model splicing of other eukaryotes.
MATERIALS AND METHODS
Modified RACE analysis of 3′ ends
To identify the 3′ ends of C. albicans snRNAs, we first ligated them to the 5′ mono-phosphate ends of 18S rRNA, reverse-transcribed the fused RNAs, and then used both 18S rRNA complementary sequences and specific internal snRNA sequences as PCR primers to amplify the intervening regions. By sequencing these PCR products directly and analyzing the sequence traces in the region of the snRNA/rRNA junctions, we were able to determine both the predominant snRNA 3′ ends as well as some minor variants (generally differing by 1–2 nt).
Total RNA was extracted from C. albicans strain SN87 (Noble and Johnson 2005) using established procedures (Mitrovich et al. 2007). Ligation products were generated en masse by treating total RNA (0.5 μg/μL) with T4 single-stranded RNA ligase (1 U/μL; NEB) and manufacturer's buffer at 37°C for 1.5 h. Purified RNA (or non-ligase-treated control RNA) was reverse transcribed (Mitrovich et al. 2007) at 100 ng/μL with a primer complementary to 18S rRNA (5 μM; see Supplemental Material for oligonucleotide sequences). Products were purified and PCR amplified at a 1:2500 dilution, then diluted further (1:20,000) for a second round of PCR using nested primers. Products were purified and sequenced directly using either of the nested PCR primers.
Genome resources
Many of the currently sequenced yeast genomes can be searched by BLAST using the NCBI Web site (http://www.ncbi.nlm.nih.gov). The Broad Institute (http://www.broad.mit.edu) provides BLAST search access to its yeast genome sequence data (e.g., C. tropicalis, C. lusitaniae, L. elongisporus, P. guilliermondii, and various nonhemiascomycetes), as does the Sanger Institute (http://www.sanger.ac.uk) for C. dubliniensis and C. parapsilosis. Extensive resources for accessing S. cerevisiae and C. albicans genome data are available at http://www.yeastgenome.org and http://www.candidagenome.org, respectively. Yeast snRNA sequence predictions are included in Supplemental Material. Accession numbers for C. albicans snRNA sequences are EU144227 (U1), EU144228 (U2), EU144229 (U4), EU144230 (U5), and EU144231 (U6).
SUPPLEMENTAL MATERIAL
Supplemental Figure 1, oligonucleotide sequences, and yeast snRNA predictions may be downloaded at http://www.candidagenome.org/download/systematic_results/Mitrovich_RNA_2007/Supplementary_Material.pdf.
ACKNOWLEDGMENTS
We thank Joan Steitz, Alexander Johnson, and members of the Guthrie laboratory for many helpful discussions and Brian Tuch for assistance with yeast phylogenies. Funding for this work was provided by NIH research grant GM21119 (C.G.) and a Sandler Postdoctoral Research Fellowship (Q.M.M.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.766607.
REFERENCES
- Abou Elela, S., Ares M., Jr Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J. 1998;17:3738–3746. doi: 10.1093/emboj/17.13.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosio, D.L., Silva, M.T., Cicarelli, R.M. Cloning and molecular characterization of Trypanosoma cruzi U2, U4, U5, and U6 small nuclear RNAs. Mem. Inst. Oswaldo Cruz. 2007;102:97–105. doi: 10.1590/s0074-02762007000100017. [DOI] [PubMed] [Google Scholar]
- Ares M., Jr U2 RNA from yeast is unexpectedly large and contains homology to vertebrate U4, U5, and U6 small nuclear RNAs. Cell. 1986;47:49–59. doi: 10.1016/0092-8674(86)90365-x. [DOI] [PubMed] [Google Scholar]
- Bacikova, D., Horowitz, D.S. Genetic and functional interaction of evolutionarily conserved regions of the Prp18 protein and the U5 snRNA. Mol. Cell. Biol. 2005;25:2107–2116. doi: 10.1128/MCB.25.6.2107-2116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boelens, W., Scherly, D., Jansen, E.J., Kolen, K., Mattaj, I.W., van Venrooij, W.J. Analysis of in vitro binding of U1-A protein mutants to U1 snRNA. Nucleic Acids Res. 1991;19:4611–4618. doi: 10.1093/nar/19.17.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bon, E., Casaregola, S., Blandin, G., Llorente, B., Neuveglise, C., Munsterkotter, M., Guldener, U., Mewes, H.W., Van Helden, J., Dujon, B., et al. Molecular evolution of eukaryotic genomes: Hemiascomycetous yeast spliceosomal introns. Nucleic Acids Res. 2003;31:1121–1135. doi: 10.1093/nar/gkg213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branlant, C., Krol, A., Ebel, J.P., Lazar, E., Gallinaro, H., Jacob, M., Sri-Widada, J., Jeanteur, P. Nucleotide sequences of nuclear U1A RNAs from chicken, rat, and man. Nucleic Acids Res. 1980;8:4143–4154. doi: 10.1093/nar/8.18.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow, D.A. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- Brow, D.A., Guthrie, C. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature. 1988;334:213–218. doi: 10.1038/334213a0. [DOI] [PubMed] [Google Scholar]
- Burd, C.G., Dreyfuss, G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Burgess, S.M., Guthrie, C. Beat the clock: Paradigms for NTPases in the maintenance of biological fidelity. Trends Biochem. Sci. 1993;18:381–384. doi: 10.1016/0968-0004(93)90094-4. [DOI] [PubMed] [Google Scholar]
- Burgess, S., Couto, J.R., Guthrie, C. A putative ATP binding protein influences the fidelity of branchpoint recognition in yeast splicing. Cell. 1990;60:705–717. doi: 10.1016/0092-8674(90)90086-t. [DOI] [PubMed] [Google Scholar]
- Butcher, S.E., Brow, D.A. Toward understanding the catalytic core structure of the spliceosome. Biochem. Soc. Trans. 2005;33:447–449. doi: 10.1042/BST0330447. [DOI] [PubMed] [Google Scholar]
- Cao, S., Chen, S.J. Free energy landscapes of RNA/RNA complexes: With applications to snRNA complexes in spliceosomes. J. Mol. Biol. 2006;357:292–312. doi: 10.1016/j.jmb.2005.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary, F., Seraphin, B. The yeast U2A′/U2B complex is required for pre-spliceosome formation. EMBO J. 1998;17:6348–6358. doi: 10.1093/emboj/17.21.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau, G., Elela, S.A., Ares M., Jr, Guthrie, C. Alternative 3′-end processing of U5 snRNA by RNase III. Genes & Dev. 1997;11:2741–2751. doi: 10.1101/gad.11.20.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, B., Weiner, A.M. Genetic evidence for base pairing between U2 and U6 snRNA in mammalian mRNA splicing. Nature. 1991;352:821–824. doi: 10.1038/352821a0. [DOI] [PubMed] [Google Scholar]
- Diezmann, S., Cox, C.J., Schonian, G., Vilgalys, R.J., Mitchell, T.G. Phylogeny and evolution of medical species of Candida and related taxa: A multigenic analysis. J. Clin. Microbiol. 2004;42:5624–5635. doi: 10.1128/JCM.42.12.5624-5635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon, B. Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet. 2006;22:375–387. doi: 10.1016/j.tig.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Elliott, D.J., Grellscheid, S.N. Alternative RNA splicing regulation in the testis. Reproduction. 2006;132:811–819. doi: 10.1530/REP-06-0147. [DOI] [PubMed] [Google Scholar]
- Fast, N.M., Roger, A.J., Richardson, C.A., Doolittle, W.F. U2 and U6 snRNA genes in the microsporidian Nosema locustae: Evidence for a functional spliceosome. Nucleic Acids Res. 1998;26:3202–3207. doi: 10.1093/nar/26.13.3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, D.N., Roiha, H., Guthrie, C. Architecture of the U5 small nuclear RNA. Mol. Cell. Biol. 1994;14:2180–2190. doi: 10.1128/mcb.14.3.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl, J. Mastering regular expressions. O'Reilly Media, Inc; Sebastopol, CA: 2006. [Google Scholar]
- Gottschalk, A., Tang, J., Puig, O., Salgado, J., Neubauer, G., Colot, H.V., Mann, M., Seraphin, B., Rosbash, M., Luhrmann, R., et al. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA. 1998;4:374–393. [PMC free article] [PubMed] [Google Scholar]
- Gottschalk, A., Kastner, B., Luhrmann, R., Fabrizio, P. The yeast U5 snRNP coisolated with the U1 snRNP has an unexpected protein composition and includes the splicing factor Aar2p. RNA. 2001;7:1554–1565. [PMC free article] [PubMed] [Google Scholar]
- Graveley, B.R. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell. 2005;123:65–73. doi: 10.1016/j.cell.2005.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., Patterson, B. Spliceosomal snRNAs. Annu. Rev. Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- Hamm, J., Kazmaier, M., Mattaj, I.W. In vitro assembly of U1 snRNPs. EMBO J. 1987;6:3479–3485. doi: 10.1002/j.1460-2075.1987.tb02672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, P., Bellofatto, V. Structure of the Leptomonas seymouri trans-spliceosomal U2 snRNA-encoding gene; potential U2-U6 snRNA interactions conform to the cis-splicing counterpart. Gene. 1995;165:131–135. doi: 10.1016/0378-1119(95)00413-z. [DOI] [PubMed] [Google Scholar]
- Hilliker, A.K., Staley, J.P. Multiple functions for the invariant AGC triad of U6 snRNA. RNA. 2004;10:921–928. doi: 10.1261/rna.7310704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker, A.K., Mefford, M.A., Staley, J.P. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes & Dev. 2007;21:821–834. doi: 10.1101/gad.1536107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T., Federspiel, N.A., Chibana, H., Dungan, J., Kalman, S., Magee, B.B., Newport, G., Thorstenson, Y.R., Agabian, N., Magee, P.T., et al. The diploid genome sequence of Candida albicans . Proc. Natl. Acad. Sci. 2004;101:7329–7334. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica, M.S., Moore, M.J. Pre-mRNA splicing: Awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- Kambach, C., Walke, S., Nagai, K. Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles. Curr. Opin. Struct. Biol. 1999;9:222–230. doi: 10.1016/s0959-440x(99)80032-3. [DOI] [PubMed] [Google Scholar]
- Konarska, M.M., Query, C.C. Insights into the mechanisms of splicing: More lessons from the ribosome. Genes & Dev. 2005;19:2255–2260. doi: 10.1101/gad.1363105. [DOI] [PubMed] [Google Scholar]
- Kretzner, L., Rymond, B.C., Rosbash, M. S. cerevisiae U1 RNA is large and has limited primary sequence homology to metazoan U1 snRNA. Cell. 1987;50:593–602. doi: 10.1016/0092-8674(87)90032-8. [DOI] [PubMed] [Google Scholar]
- Kretzner, L., Krol, A., Rosbash, M. Saccharomyces cerevisiae U1 small nuclear RNA secondary structure contains both universal and yeast-specific domains. Proc. Natl. Acad. Sci. 1990;87:851–855. doi: 10.1073/pnas.87.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman, C.P., Fell, J.W. The yeasts: A taxonomic study. Elsevier Science B.V; Amsterdam, The Netherlands: 2000. [Google Scholar]
- Liao, X.C., Tang, J., Rosbash, M. An enhancer screen identifies a gene that encodes the yeast U1 snRNP A protein: Implications for snRNP protein function in pre-mRNA splicing. Genes & Dev. 1993;7:419–428. doi: 10.1101/gad.7.3.419. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Burge, C.B. A computational analysis of sequence features involved in recognition of short introns. Proc. Natl. Acad. Sci. 2001;98:11193–11198. doi: 10.1073/pnas.201407298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart, S.R., Rymond, B.C. Commitment of yeast pre-mRNA to the splicing pathway requires a novel U1 small nuclear ribonucleoprotein polypeptide, Prp39p. Mol. Cell. Biol. 1994;14:3623–3633. doi: 10.1128/mcb.14.6.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani, H.D., Guthrie, C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71:803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- Madhani, H.D., Bordonne, R., Guthrie, C. Multiple roles for U6 snRNA in the splicing pathway. Genes & Dev. 1990;4:2264–2277. doi: 10.1101/gad.4.12b.2264. [DOI] [PubMed] [Google Scholar]
- Mayas, R.M., Maita, H., Staley, J.P. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat. Struct. Mol. Biol. 2006;13:482–490. doi: 10.1038/nsmb1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean, M.R., Rymond, B.C. Yeast pre-mRNA splicing requires a pair of U1 snRNP-associated tetratricopeptide repeat proteins. Mol. Cell. Biol. 1998;18:353–360. doi: 10.1128/mcb.18.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPheeters, D.S. Interactions of the yeast U6 RNA with the pre-mRNA branch site. RNA. 1996;2:1110–1123. [PMC free article] [PubMed] [Google Scholar]
- Michel, F., Umesono, K., Ozeki, H. Comparative and functional anatomy of group II catalytic introns—A review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- Mitrovich, Q.M., Tuch, B.B., Guthrie, C., Johnson, A.D. Computational and experimental approaches double the number of known introns in the pathogenic yeast Candida albicans . Genome Res. 2007;17:492–502. doi: 10.1101/gr.6111907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougin, A., Gottschalk, A., Fabrizio, P., Luhrmann, R., Branlant, C. Direct probing of RNA structure and RNA–protein interactions in purified HeLa cell's and yeast spliceosomal U4/U6.U5 tri-snRNP particles. J. Mol. Biol. 2002;317:631–649. doi: 10.1006/jmbi.2002.5451. [DOI] [PubMed] [Google Scholar]
- Newman, A.J., Teigelkamp, S., Beggs, J.D. snRNA interactions at 5′ and 3′ splice sites monitored by photoactivated crosslinking in yeast spliceosomes. RNA. 1995;1:968–980. [PMC free article] [PubMed] [Google Scholar]
- Noble, S.M., Johnson, A.D. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans . Eukaryot. Cell. 2005;4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds, F.C. Candida and Candidosis. Baillière Tindall; London, UK: 1988. [Google Scholar]
- O'Keefe, R.T., Newman, A.J. Functional analysis of the U5 snRNA loop 1 in the second catalytic step of yeast pre-mRNA splicing. EMBO J. 1998;17:565–574. doi: 10.1093/emboj/17.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oubridge, C., Ito, N., Evans, P.R., Teo, C.H., Nagai, K. Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- Palfi, Z., Schimanski, B., Gunzl, A., Lucke, S., Bindereif, A. U1 small nuclear RNP from Trypanosoma brucei: A minimal U1 snRNA with unusual protein components. Nucleic Acids Res. 2005;33:2493–2503. doi: 10.1093/nar/gki548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, B., Guthrie, C. An essential yeast snRNA with a U5-like domain is required for splicing in vivo. Cell. 1987;49:613–624. doi: 10.1016/0092-8674(87)90537-x. [DOI] [PubMed] [Google Scholar]
- Perriman, R.J., Ares M., Jr Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes & Dev. 2007;21:811–820. doi: 10.1101/gad.1524307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiss, J.A., Whitworth, G.B., Bergkessel, M., Guthrie, C. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol. Cell. 2007;27:928–937. doi: 10.1016/j.molcel.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Query, C.C., Konarska, M.M. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol. Cell. 2004;14:343–354. doi: 10.1016/s1097-2765(04)00217-5. [DOI] [PubMed] [Google Scholar]
- Rhode, B.M., Hartmuth, K., Westhof, E., Luhrmann, R. Proximity of conserved U6 and U2 snRNA elements to the 5′ splice site region in activated spliceosomes. EMBO J. 2006;25:2475–2486. doi: 10.1038/sj.emboj.7601134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roiha, H., Shuster, E.O., Brow, D.A., Guthrie, C. Small nuclear RNAs from budding yeasts: Phylogenetic comparisons reveal extensive size variation. Gene. 1989;82:137–144. doi: 10.1016/0378-1119(89)90038-3. [DOI] [PubMed] [Google Scholar]
- Sashital, D.G., Cornilescu, G., McManus, C.J., Brow, D.A., Butcher, S.E. U2-U6 RNA folding reveals a group II intron-like domain and a four-helix junction. Nat. Struct. Mol. Biol. 2004;11:1237–1242. doi: 10.1038/nsmb863. [DOI] [PubMed] [Google Scholar]
- Scherly, D., Boelens, W., Dathan, N.A., van Venrooij, W.J., Mattaj, I.W. Major determinants of the specificity of interaction between small nuclear ribonucleoproteins U1A and U2B″ and their cognate RNAs. Nature. 1990;345:502–506. doi: 10.1038/345502a0. [DOI] [PubMed] [Google Scholar]
- Schwer, B., Guthrie, C. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J. 1992;11:5033–5039. doi: 10.1002/j.1460-2075.1992.tb05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seetharaman, M., Eldho, N.V., Padgett, R.A., Dayie, K.T. Structure of a self-splicing group II intron catalytic effector domain 5: Parallels with spliceosomal U6 RNA. RNA. 2006;12:235–247. doi: 10.1261/rna.2237806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segault, V., Will, C.L., Polycarpou-Schwarz, M., Mattaj, I.W., Branlant, C., Luhrmann, R. Conserved loop I of U5 small nuclear RNA is dispensable for both catalytic steps of pre-mRNA splicing in HeLa nuclear extracts. Mol. Cell. Biol. 1999;19:2782–2790. doi: 10.1128/mcb.19.4.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seipelt, R.L., Zheng, B., Asuru, A., Rymond, B.C. U1 snRNA is cleaved by RNase III and processed through an Sm site-dependent pathway. Nucleic Acids Res. 1999;27:587–595. doi: 10.1093/nar/27.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster, E.O., Guthrie, C. Two conserved domains of yeast U2 snRNA are separated by 945 nonessential nucleotides. Cell. 1988;55:41–48. doi: 10.1016/0092-8674(88)90007-4. [DOI] [PubMed] [Google Scholar]
- Siliciano, P.G., Jones, M.H., Guthrie, C. Saccharomyces cerevisiae has a U1-like small nuclear RNA with unexpected properties. Science. 1987;237:1484–1487. doi: 10.1126/science.3306922. [DOI] [PubMed] [Google Scholar]
- Siliciano, P.G., Kivens, W.J., Guthrie, C. More than half of yeast U1 snRNA is dispensable for growth. Nucleic Acids Res. 1991;19:6367–6372. doi: 10.1093/nar/19.23.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, E.C., Leggett, S.R., Winans, A.A., Staley, J.P. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol. Cell. 2006;23:389–399. doi: 10.1016/j.molcel.2006.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer, E.J., Steitz, J.A. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science. 1993;262:1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- Spingola, M., Ares M., Jr A yeast intronic splicing enhancer and Nam8p are required for Mer1p-activated splicing. Mol. Cell. 2000;6:329–338. doi: 10.1016/s1097-2765(00)00033-2. [DOI] [PubMed] [Google Scholar]
- Spingola, M., Grate, L., Haussler, D., Ares M., Jr Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae . RNA. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley, J.P., Guthrie, C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Sun, J.S., Manley, J.L. A novel U2-U6 snRNA structure is necessary for mammalian mRNA splicing. Genes & Dev. 1995;9:843–854. doi: 10.1101/gad.9.7.843. [DOI] [PubMed] [Google Scholar]
- Surowy, C.S., van Santen, V.L., Scheib-Wixted, S.M., Spritz, R.A. Direct, sequence-specific binding of the human U1-70K ribonucleoprotein antigen protein to loop I of U1 small nuclear RNA. Mol. Cell. Biol. 1989;9:4179–4186. doi: 10.1128/mcb.9.10.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe, N., Yoshimura, K., Kimura, A., Yabuta, Y., Shigeoka, S. Differential expression of alternatively spliced mRNAs of Arabidopsis SR protein homologues, atSR30 and atSR45a, in response to environmental stress. Plant Cell Physiol. 2007;48:1036–1049. doi: 10.1093/pcp/pcm069. [DOI] [PubMed] [Google Scholar]
- Tang, J., Rosbash, M. Characterization of yeast U1 snRNP A protein: Identification of the N-terminal RNA binding domain (RBD) binding site and evidence that the C-terminal RBD functions in splicing. RNA. 1996;2:1058–1070. [PMC free article] [PubMed] [Google Scholar]
- Tang, J., Abovich, N., Rosbash, M. Identification and characterization of a yeast gene encoding the U2 small nuclear ribonucleoprotein particle B″ protein. Mol. Cell. Biol. 1996;16:2787–2795. doi: 10.1128/mcb.16.6.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsong, A.E., Tuch, B.B., Li, H., Johnson, A.D. Evolution of alternative transcriptional circuits with identical logic. Nature. 2006;443:415–420. doi: 10.1038/nature05099. [DOI] [PubMed] [Google Scholar]
- Urlaub, H., Hartmuth, K., Kostka, S., Grelle, G., Luhrmann, R. A general approach for identification of RNA–protein cross-linking sites within native human spliceosomal small nuclear ribonucleoproteins (snRNPs). Analysis of RNA–protein contacts in native U1 and U4/U6.U5 snRNPs. J. Biol. Chem. 2000;275:41458–41468. doi: 10.1074/jbc.M007434200. [DOI] [PubMed] [Google Scholar]
- Vidovic, I., Nottrott, S., Hartmuth, K., Luhrmann, R., Ficner, R. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol. Cell. 2000;6:1331–1342. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- Villa, T., Guthrie, C. The Isy1p component of the NineTeen complex interacts with the ATPase Prp16p to regulate the fidelity of pre-mRNA splicing. Genes & Dev. 2005;19:1894–1904. doi: 10.1101/gad.1336305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland, B., Bindereif, A. Unexpected diversity in U6 snRNA sequences from trypanosomatids. Gene. 1995;161:129–133. doi: 10.1016/0378-1119(95)00247-4. [DOI] [PubMed] [Google Scholar]
- Will, C.L., Luhrmann, R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- Wu, J.A., Manley, J.L. Base-pairing between U2 and U6 snRNAs is necessary for splicing of a mammalian pre-mRNA. Nature. 1991;352:818–821. doi: 10.1038/352818a0. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Ben-Shlomo, H., Michaeli, S. The U5 RNA of trypanosomes deviates from the canonical U5 RNA: The Leptomonas collosoma U5 RNA and its coding gene. Proc. Natl. Acad. Sci. 1997;94:8473–8478. doi: 10.1073/pnas.94.16.8473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, D., Rubinson, D.A., Pannone, B.K., Yoo, C.J., Wolin, S.L. U snRNP assembly in yeast involves the La protein. EMBO J. 2000;19:1650–1660. doi: 10.1093/emboj/19.7.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, D., Ares M., Jr Invariant U2 RNA sequences bordering the branchpoint recognition region are essential for interaction with yeast SF3a and SF3b subunits. Mol. Cell. Biol. 1996;16:818–828. doi: 10.1128/mcb.16.3.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong, J., Wan, L., Dreyfuss, G. Why do cells need an assembly machine for RNA–protein complexes? Trends Cell Biol. 2004;14:226–232. doi: 10.1016/j.tcb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]