Abstract
Decapping is a critical step in the control of gene expression and is regulated by both positive and negative trans factors. Less is known about cis elements that promote decapping. In plants, following microRNA (miRNA)-directed cleavage of an mRNA, a uridine tract can be added onto the exposed 3′ end of the resulting 5′ fragment, which can promote 5′ end decay. We now demonstrate that in mammalian cell extract, addition of five uridine residues to the 3′ end of an RNA (U5) promotes decapping relative to an RNA lacking the uridines (U0). Although the decapping stimulation observed in extract required hDcp2, recombinant hDcp2 was unable to support differential decapping of the U0 and U5 RNAs, indicating that the stimulation was likely due to an indirect recruitment of hDcp2 to the RNA. Consistent with the promotion of 5′ end decapping by the uridine tract, affinity purification with the U5 RNA revealed the presence of a decapping subcomplex at least consisting of hDcp2, Dcp1a, Edc4, LSm1, and LSm4 that were specifically bound to the U5 RNA but not the U0 RNA. In addition to promoting decapping, the U-tract stabilized the 3′ end of the RNA by preventing 3′ to 5′ exonucleolytic decay to ensure 5′ end directional degradation. These data suggest that following post-transcriptional oligo uridylation of an mRNA or mRNA fragment, the U-tract has the capacity to specifically stimulate 5′ end decapping to expedite mRNA decay.
Keywords: mRNA decapping, mRNA decay, poly(U) polymerase, polyuridylation
INTRODUCTION
Modulation of mRNA stability is an important mechanism regulating gene expression. RNAs are either actively stabilized or degraded and these processes are dictated by cis elements and trans factors. Most mRNAs share two common cis elements, monomethyl N7 guanosine cap (m7G) at the 5′ end and a polyadenosine (polyA) at the 3′ end. Both terminal cis elements serve multiple functions, including the stabilization of the mRNA from exoribonucleases (Wilusz et al. 2001b). Moreover, transcript-specific cis elements and their associated trans factors also provide critical modulatory properties onto an mRNA to control their level of accumulation.
In addition to protein trans factors that control mRNA expression, nucleic-acid-mediated control involving microRNAs (miRNA) have significantly expanded the repertoire of regulators. The post-transcriptional mode of miRNA-mediated regulation can be at the level of either translation or mRNA decay (Jackson and Standart 2007; Pillai et al. 2007). In cases where a perfect match exists between the miRNA and its target sequence, the mRNA is cleaved. Shen and Goodman (2004) reported that, in plant cells and in mammalian cells, following cleavage of the mRNA, a uridine tract can be added onto the exposed 3′ end of the resulting 5′ mRNA cleavage product. Interestingly, for two plant mRNAs tested that are targeted by miRNAs, the majority of cleaved and uridylated RNA intermediates retained the 3′ U-tract but lacked varying degrees of sequences from the 5′ end, suggesting that, at least in plants, addition of the U-tract can promote decapping and 5′ to 3′ exoribonucleolytic decay. The presence of a U-tract on miRNA targeted mammalian HoxB8 mRNA (Shen and Goodman 2004) and Epstein-Barr virus transcript (Furnari et al. 1993) as well as the recent identification of a novel class of polymerases that can add uridines to the 3′ end of mRNAs in a template-independent manner (Trippe et al. 2006; Kwak and Wickens 2007; Rissland et al. 2007) suggests a broader role for mRNA uridylation in RNA metabolism.
mRNAs are primarily degraded by exonucleolytic pathways either from the 5′ end or the 3′ end following an initial deadenylation step (for review, see Meyer et al. 2004; Parker and Song 2004; Fillman and Lykke-Andersen 2005; Liu and Kiledjian 2006; Simon et al. 2006; Garneau et al. 2007), and the directionality of decay does not necessarily have to be mutually exclusive (Murray and Schoenberg 2007). In the 5′ decay pathway, the poly(A) shortening can trigger 5′ decapping carried out by the Dcp2 catalytic subunit of an mRNA decapping complex (Dunckley and Parker 1999; Lykke-Andersen 2002; van Dijk et al. 2002; Wang et al. 2002; Steiger et al. 2003). The exposed 5′ end is subjected to digestion by the 5′ exoribonuclease, Xrn1, in the cytoplasm (Decker and Parker 1993; Hsu and Stevens 1993). The decapping step is exquisitely regulated by both positive and negative effectors. A limited number of negative regulators have thus far been identified and include the eIF4E cap binding protein (Schwartz and Parker 2000; Ramirez et al. 2002; Khanna and Kiledjian 2004), the poly(A) binding protein (Caponigro and Parker 1995; Wilusz et al. 2001a; Khanna and Kiledjian 2004), and the noncanonical cap binding protein, VCX-A (Jiao et al. 2006). In contrast, numerous proteins are known to promote decapping, including Dcp1p, Pat1p, Dhh1p (Rck/p54), Edc1p, Edc2p, Edc3p, and the Lsm1–7 protein complex (for review, see Coller and Parker 2004). In addition, the mammalian Edc4 (also known as Hedls, Ge-1, or RCD-8) protein can promote decapping both directly and by facilitating the interaction of Dcp2 with Dcp1a (Fenger-Gron et al. 2005).
In yeast, Dcp2p decapping requires Dcp1p (LaGrandeur and Parker 1998; Dunckley and Parker 1999) and together they form a Dcp2p/Dcp1p decapping-competent complex (Steiger et al. 2003). In mammals, although the Dcp2 protein is competent to hydrolyze the cap on its own, it can nevertheless be contained in distinct complexes and appears to function within various multiprotein complexes that can facilitate hDcp2 decapping activity (Fenger-Gron et al. 2005). One subcomplex could consist of the Dcp2/Dcp1a and Edc4 proteins, which could also be associated with additional facilitators of decapping, Edc3, Rck/p54 (Dhh1p), and the nonsense mediated decay factor Upf1 (Fenger-Gron et al. 2005). At present, the contribution of Dcp1a in mammalian decapping remains unclear.
Many of the factors involved in 5′ end mRNA decay can aggregate into cytologically detectable foci termed P bodies (also referred to as GW bodies or DCP bodies) (Ingelfinger et al. 2002; Lykke-Andersen 2002; van Dijk et al. 2002; Eystathioy et al. 2003; Sheth and Parker 2003; Cougot et al. 2004). These foci are thought to be sites of mRNA decapping and/or storage (for review, see Fillman and Lykke-Andersen 2005; Eulalio et al. 2007; Parker and Sheth 2007). An interesting link exists between P bodies and miRNA-mediated regulation of gene expression. A critical component of the miRNA pathway, Argonaute, was shown to coimmunopurify with Dcp2 and colocalize along with miRNA and target mRNA to P bodies (Jakymiw et al. 2005; Liu et al. 2005; Pillai et al. 2005; Sen and Blau 2005), thus suggesting a correlation between mRNA decapping and miRNA-mediated mRNA decay.
The decapping complex can be recruited to an mRNA by distinct cis elements, as is the case for the adenosine–uracil rich element in both yeast (Vasudevan and Peltz 2001) and mammals (Gao et al. 2001; Fenger-Gron et al. 2005; Lykke-Andersen and Wagner 2005; Stoecklin et al. 2006). We now demonstrate that a uridine tract (U-tract) at the 3′ end of an RNA can recruit a Dcp2 containing decapping complex in mammalian cell extract and promote decapping.
RESULTS
A 3′ U-tract can stimulate decapping in vitro
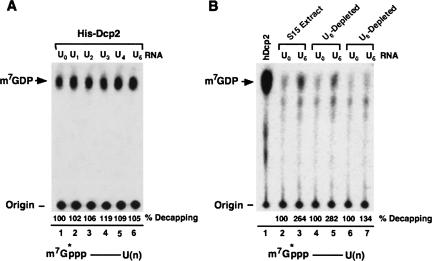
Following miRNA-mediated cleavage of a target mRNA in plants, the 3′ termini of the resulting 5′ fragment could be modified by the addition of an oligo U-tract and the presence of the U-tract correlated with the preferential loss of sequences from the 5′ end of that fragment (Shen and Goodman 2004). This observation prompted us to ask whether addition of an oligo U-tract can promote 5′ end decapping in mammalian extract. A generic RNA consisting of the polylinker region of pcDNA3 (pcP) that is routinely used for decapping studies in the laboratory (Wang and Kiledjian 2001; Wang et al. 2002) was used with or without a U-tract. The decapping assay parameters differ slightly from our standard in vitro decay assay in that they do not contain ATP, GTP, and creatine phosphate (see Materials and Methods), which are necessary for exonucleolytic decay but not for decapping activity (data not shown). Therefore, we are able to uncouple decay of the RNA from decapping to enable analysis of the decapping step with intact RNA. Decapping of the RNAs was tested in an in vitro decapping reaction using human erythroleukemia K562 cytoplasmic S15 extract. Very poor decapping is normally detected from cytoplasmic S15 extract due to the presence of decapping inhibitory trans factors in the extract (Jiao et al. 2006). Consistent with previous results, decapping was minimally detected from the pcP RNA lacking a U-tract (U0) (Fig. 1A, lane 2). In contrast, a linear increase in decapping was detected with a sequential increase in the number of uridines at the 3′ end of the RNA with a fourfold stimulation observed with five uridines (Fig. 1A, lanes 3–7). The stimulation was U-tract specific, and a stimulation of decapping was not observed with the pcP RNA containing five guanosines (Fig. 1A, lane 9). Thus the presence of a U-tract at the 3′ end of an RNA can stimulate decapping, and a direct correlation was observed between the number of uridines and an increase in decapping. However, there was a limit to the extent of stimulation, and the increase in decapping plateaued between 5 and 10 uridines and decreased upon extension of the U-tract to 15 uridines (Fig. 1B). The detected decapping was a consequence of hDcp2 since addition of anti-hDcp2-sera to the S15 extract specifically reduced decapping while addition of preimmune sera minimally affected decapping (Fig. 1C). Therefore, the presence of five uridine residues at the 3′ end of an RNA have the potential to maximally stimulate hDcp2-mediated decapping in K562 S15 extract. Considering that five uridines at the 3′ end of the pcP RNA (U5) resulted in optimal decapping stimulation, subsequent experiments utilized this RNA as the U-tract RNA substrate to minimize potential complications that could arise by the nonspecific association of general poly(U)-binding protein to a longer U-tract.
FIGURE 1.
3′ U-tract can stimulate decapping in vitro. (A) Fifty micrograms of K562 S15 extract were incubated with cap-labeled RNA containing zero to five uridines at the 3′ end at 37°C for 30 min in an in vitro decapping assay. The decapping products were resolved by PEI TLC. Position of the capped RNA substrate and m7GDP decapping product are indicated on the left and a schematic of the RNA is shown on the bottom with the line representing the RNA and the asterisks denoting the 32P-labeled phosphate. Recombinant His-hDcp2 protein was used as a positive control. The fold stimulation observed relative to the RNA lacking uridines obtained from three independent experiments is presented in the graph below with SD denoted by the error bars. (B) Maximal decapping stimulation is seen with 5 or 10 uridines. Cap-labeled RNA with 0, 5, 10, and 15 uridines or a G-tract at the 3′ end, as indicated, were used in in vitro decapping assays with 50 μg K562 S15 extract. The decapping products were separated by PEI TLC. Labeling is as described in panel A. The average fold stimulation of three independent decapping assays is shown in the bar graph below with SD. (C) The detected decapping is a function of hDcp2. In vitro decapping assays were carried out with cap-labeled U5 RNA at 37°C for 30 min in K562 S15 extract preincumbated with either preimmune or anti-hDcp2 serum. Detection of the decapping products and labeling is as described in A.
To gain a clearer understanding of the decapping stimulation by the U-tract, we asked whether uridine was required to be positioned at the 3′ termini. RNAs were generated that contained four uridines followed by any one of the 4 nt at the 3′ termini of the pcP RNA. Substitution of the last base to any of the remaining bases did not dramatically alter the ability of the RNA to stimulate decapping, and comparable stimulation was observed with an RNA of U5, U4A, U4C, and U4G (Fig. 2A). However, although the last nucleotide did not need to be a uridine, moving the U-tract 5 nucleotides (nt) upstream eliminated its stimulatory potential (Fig. 2B). Furthermore, enhancement of decapping by the U-tract is context dependent. Stimulation is detected with the pcP RNA with five U's at the 3′ termini of the pcP RNA as well as five U's added onto the same RNA containing a 10-nt 3′ terminal deletion (data not shown). However, not all RNAs with a terminal U-tract stimulated decapping, as five uridines preceded by five guanosines did not facilitate more efficient decapping (Fig. 2B, lane 6). Therefore, although the U-tract is necessary for the stimulation of decapping and required to be positioned close to the 3′ termini, it is not always sufficient to stimulate decapping.
FIGURE 2.
The U-tract stimulation of decapping is context dependent and required to be near the 3′ termini. Cap-labeled pcP RNA containing either a 5-nt tail (A) or 10-nt tail (B) with the indicated nucleotides was used in decapping assays. Labeling is as described in the legend to Figure 1. The average percent decapping of three independent experiments is listed on the bottom of each lane. The U-tract can function when placed 1 nt 5′ of the 3′ terminus but did not function when placed 5 nt upstream of the 3′ terminus.
The U-tract can recruit the decapping complex to promote decapping
The above data demonstrate that a U-tract can support hDcp2-mediated decapping of an RNA, yet it remained unknown whether this was a direct function of hDcp2 or whether it also involved additional factors. Since hDcp2 is an RNA-binding protein (Piccirillo et al. 2003), it was possible that it preferentially bound the U-tract to promote decapping of the RNA. This possibility was tested by using recombinant hDcp2 in a decapping reaction with RNAs containing 0–5 uridines at their 3′ termini in the absence of extract. Comparable decapping activities were detected with all of the RNAs in the presence of recombinant hDcp2 (Fig. 3A). The decapping reactions were carried out within the linear range of the assay and demonstrate that hDcp2, by itself, is unable to distinguish between the different U-tract-containing RNAs and is likely recruited by additional factor(s). This premise was further underscored with the use of depleted extract. K562 S15 extract depleted with biotinylated U5 RNA was unable to support stimulated decapping and was comparable to the extract depleted with biotinylated U0 RNA (Fig. 3B; cf. lanes 7 and 5). Collectively, these data demonstrate that the U-tract-directed stimulation of decapping is mediated through trans factor(s) that are capable of recruiting hDcp2. Furthermore, the ability of the biotinylated U5 RNA to deplete the essential trans factors required for the stimulated decapping indicates that it is capable of binding the trans factors and can be used as a means to identify the stimulatory factors.
FIGURE 3.
hDcp2 requires additional trans factors for the U-tract mediated stimulation of decapping. (A) Recombinant hDcp2 does not differentially stimulate decapping of RNAs lacking or containing a U-tract. In vitro decapping assays were carried out at 37°C for 30 min with 50 ng recombinant His-hDcp2 and cap-labeled pcP RNA containing zero to five uridines at the 3′ end as indicated. Labeling is as described in the legend to Figure 1. (B) A trans factor that binds the U-tract is required for decapping stimulation. K562 S15 extract depleted with biotinylated U0 (lanes 4,5) or U5 (lanes 6,7) RNA were used in in vitro decapping assays with cap-labeled U0 and U5 RNA. Resolution of the decapping product and labeling is as described in the legend to Figure 1. The average percent decapping of three independent experiments is presented in both panels.
To begin identification of the trans factor(s) that functioned through the U-tract, biotinylated U5 RNA was used to purify the protein(s) that specifically bound the U5 RNA but not the U0 RNA. The profile of proteins bound to the two RNAs detected by Sypro Ruby staining is shown in Figure 4A, where the majority of protein bands were detected to bind both RNAs except four bands that were exclusively present in the biotinylated-U5 RNA bound lane but not in the biotinylated-U0 bound lane (Fig. 4A, cf. lanes 3 and 2). Mass spectrometry analysis of these four bands revealed that they are all components of an mRNA decapping complex: the hDcp2 decapping enzyme, enhancers of decapping Edc4, Edc3, and what appears to be a proteolytic fragment of Edc4. Western analysis was used to confirm the presence of the identified proteins, and, surprisingly, we could reproducibly detect hDcp2 and Edc4 but were unable to detect Edc3 (Fig. 4B), suggesting that Edc3 is not a stable component of the decapping complex recruited by the U-tract. A search for other decapping complex components demonstrated that Dcp1a was also specifically associated with the U5 RNA while the RCK/p54 was not (Fig. 4B). The binding was not a function of recruiting generic poly(U)-binding proteins since the poly(U)-binding hnRNP C1/C2 proteins were not retained by the U5 RNA despite their retention by poly(U)-agarose beads. Therefore, we conclude that the U-tract is capable of recruiting a decapping subcomplex consisting of at least hDcp2, Dcp1a, and Edc4.
FIGURE 4.
A decapping subcomplex is recruited onto the U-tract containing RNA. (A) Biotinylated RNA with (lane 3) or without (lane 2) a U-tract was used for affinity purification to identify proteins that bound the U-tract containing RNA. Bound proteins were resolved by 12.5% SDS-polyacrylamide gel and protein bands were visualized by Sypro Ruby staining. Protein bands specifically in the U5 RNA-bound lane are designated by an arrow and their identity is listed on the right. An aliquot of total extract is shown in lane 1. (B) Western blot of proteins bound to the immobilized RNA listed at the top is shown. Identity of the protein being probed is indicated on the right.
We next focused on determining how the U-tract might recruit the decapping complex. Since hDcp2 is the only known RNA-binding protein within the identified subcomplex associated with the U5 RNA and it is not sufficient to distinguish between the U0 and U5 RNAs (Fig. 3A), we reasoned that additional protein(s) were involved that specifically bound the U-tract. One likely candidate was the LSm1–7 complex of proteins that are proposed to bind at the 3′ termini of an mRNA containing an oligo A tail and augment decapping (Boeck et al. 1998; Bouveret et al. 2000; Tharun et al. 2000). Consistent with this hypothesis, both LSm1 and LSm4 proteins can be detected to preferentially associate with the U5 RNA (Fig. 4B).
A role for the LSm1–7 complex in the U-tract stimulation of decapping
The potential role for the LSm1–7 complex in the U-tract decapping stimulation was next tested. Decapping assays were carried out with the U0 or U5 RNAs in S15 extract either depleted of LSm1 or containing antisera directed against LSm1 to determine their influence on decapping. U-tract-mediated stimulation of decapping was detected with both undepleted (Fig. 5A, lane 2) and mock-depleted extract (Fig. 5A, lane 4) but not with the LSm1-depleted extract. No difference in the level of decapping was observed between the U-tract containing or lacking RNAs in the latter case (Fig. 5A, cf. lanes 6 and 5). A similar requirement of LSm1 was also detected when LSm1-specific antisera were included in the decapping reaction. In the presence of increasing titration of antisera added to K562 S15 extract, decapping of the U0 RNA was not significantly different with either control antisera or LSm1-specific antisera (Fig. 5B). However, a reduction in decapping was observed with the U5 RNA upon addition of the LSm1 antisera relative to the control sera (Fig. 5C). Collectively, these data indicate a role for LSm1, and indirectly the LSm1–7 complex, in promoting U-tract mediated decapping.
FIGURE 5.
Decapping of the U5 RNA is compromised in the absence of LSm1. (A) In vitro decapping assays were carried out with U0 or U5 RNAs with undepleted, mock-depleted, or LSm1-depleted extract as indicated. Decapping of the U0 and U5 RNAs is comparable in the LSm1-depleted extract. The efficiency of LSm1 depletion is shown by the Western at the bottom of the panel with the LSm1 and the control DcpS protein bands indicated on the left. (B,C) In vitro decapping assays were carried out in the presence of anti-LSm1 and control serum. The percent decapping relative to the concentration of protein used is plotted. LSm1 can inhibit decapping of the U5 RNA (B) but not the U0 RNA (A) relative to their respective controls. Quantitation of three independent experiments and the corresponding standard deviation represented by the error bars are plotted in both panels.
The U-tract can inhibit 3′ to 5′ exonucleolytic decay
The above data support a model whereby a U-tract can be added to the 3′ end of an RNA to promote its demise by shunting the RNA into the 5′ to 3′ decay pathway. However, a fundamental question of whether the integrity of the RNA 3′ end would remain intact to enable the 3′ U-tract to recruit the decapping complex or whether it would be removed by 3′ to 5′ exonucleases remained. It has previously been shown that the presence of a U-tract at the 3′ end of an RNA can stabilize the RNA in an in vitro RNA decay system using nuclear extract (Ford et al. 1997). An in vitro RNA decay system consisting of cytoplasmic extract that predominantly degrades RNAs by the 3′ to 5′ exonucleolytic decay pathway (Rodgers et al. 2002) was used to test the stability of an RNA containing or lacking a 3′ terminal U-tract. Using this assay system enabled us to detect decay of the RNA from the 3′ termini. Otherwise the ability of the U-tract containing RNA to promote decapping would make this RNA more susceptible to decay due to more rapid decapping. As shown in Figure 6, the RNA containing the U-tract was considerably more stable than the corresponding RNA lacking the U-tract with a threefold greater half-life. Therefore, the U-tract appears to prevent decay of the RNA from the 3′ end and selectively promotes decay from the 5′ end.
FIGURE 6.
The U-tract can stabilize the 3′ end of an RNA. Capped and uniformly labeled U0 or U5 pcP RNA was incubated with K562 S15 extract in IVDA buffer as described in Materials and Methods for the indicated times. The isolated RNA substrates were resolved on a 7 M urea/5% polyacrylamide denaturing gel and visualized by autoradiography. 32P-labeled DNA oligonucleotide included in the stop buffer was used as an internal control to enable quantitation. Quantitations derived from three independent experiments and the standard deviations represented by the error bars are plotted to the right.
DISCUSSION
Our data demonstrate that a 3′ end oligo U-tract can facilitate decapping of the RNA. The mechanism by which it promotes decapping is through the recruitment of a decapping subcomplex consisting of the catalytic component responsible for hydrolysis of the cap, hDcp2, as well as auxiliary proteins Dcp1a and Edc4 (Fig. 4). Interestingly, the U-tract was position dependent and obligate to the 3′ terminus. The significance of the 3′ positioning appears to be the recognition of the U-tract by the LSm1–7 complex (Fig. 5) which can bind a terminal oligo A or oligo U tract (Chowdhury et al. 2007). In addition to promoting 5′ end decapping, importantly, it also impeded 3′ to 5′ decay. Therefore, at least in vitro, the oligo U tract can ensure that an RNA is shunted into the 5′ to 3′ decay pathway.
hDcp2 is an RNA binding protein (Piccirillo et al. 2003) and could potentially contain preferential RNA binding specificity, as does the X29 nuclear decapping protein (Ghosh et al. 2004). However, recombinant hDcp2 could not distinguish between an RNA containing or lacking a U-tract, and equally decapped both RNAs (Fig. 3A, lanes 1,6). Therefore, hDcp2 recognition of the oligo U-tract is indirect and mediated through a protein complex. The recruitment of Dcp1a, Edc4, LSm1, and LSm4 specifically to the U5 RNA but not the U0 RNA (Fig. 4) and its promotion of decapping supports a model whereby the LSm1–7 complex binds to the U-tract and in turn recruits a subset of the decapping complex (hDcp2, Dcp1a, and Edc4) to subject the 5′ terminus of the RNA to decapping (Fig. 7). Moreover, in conjunction with promoting 5′ decapping, the U-tract also contributed to stabilization of the 3′ end from exonucleolytic decay. Analogous to the contribution of Lsm1–7 complex in protecting the 3′ end of an mRNA from trimming in yeast (He and Parker 2001), we propose that the LSm1–7 can similarly function in mammalian cells. Importantly, the U-tract only functioned when positioned at the 3′ termini and did not support efficient decapping when it was moved 5 nt upstream from the termini (Fig. 2B) although the last nucleotide was variable and could be a different nucleobase. However, as also noted in Figure 2B, although the U-tract is essential, it is not necessarily sufficient in all cases to promote decapping, suggesting a further complexity of context dependence that has not yet been addressed. One possibility is that accessibility of the U-tract to the LSm1–7 might vary depending on the sequence context around the U-tract.
FIGURE 7.
Model of U-tract stimulation of decapping. An RNA targeted as substrate for miRNA mediated cleavage and/or an mRNA that is polyuridylated at the 3′ end to generate an oligo U-tract-containing RNA. We propose that the U-tract can bind the LSm1–7 complex that in turn recruits the decapping complex to hydrolyze the cap. Similarly, the U-tract, perhaps through the binding of LSm1–7 protein complex, can stabilize the 3′ end. Collectively, the oligo U-tract containing RNA is shunted into the 5′ to 3′ decay pathway to minimize translation of aberrant protein.
A family of polymerases previously thought to be poly(A) polymerases were recently shown to contain potent poly(U) polymerase activity (Kwak and Wickens 2007; Rissland et al. 2007). A variety of substrate RNAs are known to be uridylated including RNA editing in Trypanosoma brucei (Aphasizhev et al. 2002; Ernst et al. 2003), miRNAs and siRNAs in Arabidopsis (Li et al. 2005), as well as their target mRNA substrate following mRNA cleavage in plants and mammals (Shen and Goodman 2004), human U6 snRNA (Trippe et al. 2006), and a single uridine to the actin mRNA in Schizosaccharomyces pombe upon S-phase arrest (Rissland et al. 2007). The S. pombe Cid1 is proposed to be responsible for the uridylation of the actin mRNA, and its role in checkpoint control during S-phase arrest (Wang et al. 2000) suggests uridylation could be a predominant mechanism used in a cell-cycle specific manner to tag a subset of RNAs for rapid decay. Our data showing an incremental increase in the stimulation of decapping from one uridine up to five uridines demonstrate that even short oligo U-tracts can promote decapping. Interestingly, the stimulation of decapping plateaued between 5 uridines and 10 uridines and decreased with greater number of uridines. The significance of this could be that the LSm1–7 complex can access the shorter U-tracts, but the larger U-tracts are susceptible to competition with cellular poly(U)-binding proteins (Swanson and Dreyfuss 1988).
A 3′ terminal oligo U-tract has been shown to both stabilize RNA (Ford and Wilusz 1999; Chen et al. 2000) as well as promote decay of an RNA (Shen and Goodman 2004). An RNA containing a 3′ U-tract was resistant to 3′ end decay in nuclear extract (Ford and Wilusz 1999). However, since cap-labeled RNA was not used in this study, in conjunction with the low level of decapping observed in in vitro assays, 5′ end decay would have been difficult to detect. Alternatively, since not all RNAs with a U-tract necessarily promote decapping, it is possible that the particular RNA used in the study does not support recruitment of the decapping complex. Addition of a U-tract to the 3′ end of the U6 snRNA has also been proposed to promote stabilization of this RNA by virtue of regenerating the U6 snRNA 3′ end following 3′ end trimming (Chen et al. 2000; Trippe et al. 2006). However, the lack of a cap on the U6 snRNA would preclude detection of stimulated decapping. Conversely, a U-tract was shown to promote 5′ end decay in plants (Shen and Goodman 2004). Our data are consistent with both the 3′ end stabilization and 5′ end decay. By uncoupling decapping from exonucleolytic decay, we can demonstrate that the U-tract can simultaneously prevent 3′ to 5′ decay, most likely from the exosome, and stimulate decapping at the 5′ end (Figs. 1, 6). One question arises: What would the significance of this activity be? One hypothesis is that, at least in the case of miRNA-directed cleavage of an mRNA, facilitation of decapping while impeding 3′ end decay could prevent ribosomal loading onto an mRNA targeted for decay to minimize the generation of aberrantly truncated proteins that could otherwise be generated from 3′ end decay intermediates.
Similar to the post-transcriptional 3′ terminal oligo uridylation (Shen and Goodman 2004) an oligo A-tract can be added to the 3′ termini of a 5′ fragment resulting from miRNA-directed mRNA cleavage in Chlamydomonas reinhardtii (Ibrahim et al. 2006). Moreover, rather than promoting decapping, the A-tract promoted 3′ to 5′ decay by the exosome (Ibrahim et al. 2006). Therefore, it is likely that, upon addition of a U-tract, the mRNA is primarily degraded from the 5′ end while addition of an A-tract facilitates 3′ end decay. However, whether the A-tract only promotes 3′ end decay is still not clear since the status of the 5′ end was not tested in the C. reinhardtii study (Ibrahim et al. 2006). As shown in Figure 2B, although the greatest stimulation of decapping was observed with the oligo U-tract, an intermediate stimulation of decapping was observed with the terminal oligo A-tract. As the LSm1–7 complex can bind both a terminal U-tract and a terminal A-tract (Chowdhury et al. 2007), can promote decapping (Fig. 2B; Bouveret et al. 2000; Tharun et al. 2000), and can promote 3′ end decay (Ibrahim et al. 2006), it will be interesting to determine whether the A-tract could promote bidirectional decay.
The identification of an evolutionarily conserved family of PUPs that can add 3′ terminal U-tracts (Trippe et al. 2006; Kwak and Wickens 2007; Rissland et al. 2007) and the capacity of the U-tract to stimulate decapping indicates a broad role for these proteins in mRNA decay. Future efforts to identify mRNA substrates that are uridylated in cells and the cellular conditions that promote uridylation will begin to address the generality of polyuridylation and its impact on gene expression.
MATERIALS AND METHODS
Extract preparation
Frozen K562 cells were obtained from the National Cell Culture Center. For preparation of S15 cytoplasmic protein extract, K562 cells were resuspended in hypotonic buffer (10 mM Tris-HCl at pH 7.5, 10 mM KCl, 1.5 mM MgOAc, protease inhibitor cocktail; Roche Diagnostics) on ice for 20 min, and passed through a tuberculin syringe 20 times. The KCl concentration of the extract was adjusted to 100 mM followed by an initial 2000g centrifugation for 10 min at 4°C to remove nuclei and debris and a second centrifugation at 15,000g for 30 min at 4°C to remove organelles. The resulting supernatant was supplemented with 10% glycerol (v/v), and stored at −80°C.
RNA production
The pcDNA3 poly linker (pcP) RNA was transcribed by SP6 polymerase using PCR generated DNA template with SP6 and T7 primers. For RNAs with different uridine moieties at the 3′ end, a primer containing the corresponding number of adenosine nucleotides at the 5′ end of the T7 primer sequence was used for the PCR reactions to generate in vitro transcription templates that could generate RNAs with a varying length U-tract. The in vitro transcribed RNAs were cap labeled using the vaccinia virus capping enzyme (Shuman 1990) in the presence of [α-32P]GTP and S-adenosyl-dismethionine (SAM), as described (Wang and Kiledjian 2001). The uniform labeled and capped U0 and U5 RNA were transcribed using SP6 polymerase in the presence of [α-32P]UTP and m7GpppG cap analog. For the biotinylated and capped U0 and U5 RNAs, the in vitro transcription was carried out in the presence of biotin-14-CTP (Invitrogen) and m7GpppG cap analog.
In vitro decapping and decay assays
In vitro decapping assays were carried out as previously described (Wang et al. 2002). Briefly, 50 μg of K562 S15 cell extract and cap labeled RNA were used in a reaction containing decapping buffer (10 nM Tris-HCl at pH 7.5, 100 mM KOAc, 1.5 mM MgCl, 2 mM DTT, 0.5 mM MnCl2, 40 U/mL recombinant RNasin inhibitor) with 200 mM m7GpppG cap analog to prevent endogenous DcpS activity and minimize m7GMP production (Liu et al. 2002; Wang et al. 2002). Decapping reactions were carried out at 37°C for 30 min and stopped by extracting once with phenol:chloroform (1:1). Decapping products were resolved by polyethyleneimine-cellulose TLC plates (Sigma-Aldrich) developed in 0.45 M (NH4)2SO4 in a TLC chamber at room temperature (Wang and Kiledjian 2001) and exposed to a PhosphorImager. Quantifications were carried out using a Molecular Dynamics PhosphorImager using ImageQuante-5 software.
In vitro RNA decay assays were carried out with 50 μg K562 S15 cell extract and uniform labeled and capped U0 and U5 RNA in 20-μL reactions containing IVDA buffer (10 mM Tris-HCl at pH 7.5, 100 mM potassium acetate, 2 mM magnesium acetate, 2 mM DTT, 10 mM creatine phosphate, 1 mM rATP, 0.4 mM rGTP, 1000 U/mL creatine phosphokinase) at 37°C for the indicated times. Reactions were stopped with 200 μL ULB (10 mM Tris-HCl at pH 7.5, 350 mM NaCl, 7 M urea, 10 mM EDTA, 2% SDS) containing 32P end labeled DNA oligonucleotide as an internal control for quantitations, extracted once with phenol:chloroform (1:1) and twice with chloroform, and then precipitated with ethanol in the presence of 20 μg glycogen. The samples were resolved by 7 M urea/5% polyacrylamide denaturing gel and visualized and quantitated with a Molecular Dynamics PhosphorImager.
Biotin labeled RNA affinity purification
Five milligrams of S15 extract precleared on streptavidin beads were incubated with capped and biotinylated U0 and U5 RNA immobilized on streptavidin beads in the presence of 0.25% tritonX-100 at 4°C for 1 h. Following washes in washing buffer (10 mM Tris-HCl at pH 7.5, 300 mM KCl, 1.5 mM MgCl2, 0.25% TritonX-100), associated proteins were eluted with protein loading buffer by boiling 3 min and separated by 12.5% SDS-polyacrylamide gel. The bands were visualized by staining with Sypro Ruby (Molecular Probes). Bands specifically bound to biotin-U5 RNA were excised and identified by MALDI-TOF Mass Spectrometry analysis (UMDNJ).
Western blot and immunoprecipitation
Fifty micrograms of K562 S15 extract were resolved on a 10% SDS polyacrylamide gel and transferred to nitrocellulose membrane (Schleicher & Schuell BioScience, Inc.) using a semi-dry apparatus. Immunodetection was performed with 1:1000 dilution of the primary antibodies and visualized by ECL (GE Healthcare) using horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse secondary antibody (1:10,000 dilution; Jackson Immunoresearch). The antibodies used were anti-p-P70 S6 kinase (A-6) antibody (Santa Cruz Biotechnology), which can cross-react with EDC4, anti-Dcp1a antibody (Abnova), anti-EDC3 antibody (Abnova), and anti-RCK/p54 antibody (Bethyl Laboratories). The LSm1 and LSm4 antisera (Ingelfinger et al. 2002) were a gift from Reinhardt Luhrmann (Max Planck Institute, Göttingen, Germany). The anti-hnRNP C1/C2 antibody, 4F4, was a gift from Gideon Dreyfuss (University of Pennsylvania, Philadelphia).
For the generation of LSm1 depleted extract, chicken anti-LSm1 antibody (ProSci Inc.) was first immobilized on anti-chicken IgY agrose beads (Gallus Immunotech Inc.). After washing three times with PBS, K562 S15 extract was added to the beads and incubated for 2 h at 4°C. After two rounds of depletion, the extract was used directly in the decapping assays.
ACKNOWLEDGMENTS
We thank members of the Kiledjian laboratory for helpful discussions and critical reading of the manuscript. This work was supported by NIH grant GM67005 to M.K.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.765807.
REFERENCES
- Aphasizhev, R., Sbicego, S., Peris, M., Jang, S.H., Aphasizheva, I., Simpson, A.M., Rivlin, A., Simpson, L. Trypanosome mitochondrial 3′ terminal uridylyl transferase (TUTase): The key enzyme in U-insertion/deletion RNA editing. Cell. 2002;108:637–648. doi: 10.1016/s0092-8674(02)00647-5. [DOI] [PubMed] [Google Scholar]
- Boeck, R., Lapeyre, B., Brown, C.E., Sachs, A.B. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol. Cell. Biol. 1998;18:5062–5072. doi: 10.1128/mcb.18.9.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret, E., Rigaut, G., Shevchenko, A., Wilm, M., Seraphin, B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro, G., Parker, R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes & Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- Chen, Y., Sinha, K., Perumal, K., Reddy, R. Effect of 3′ terminal adenylic acid residue on the uridylation of human small RNAs in vitro and in frog oocytes. RNA. 2000;6:1277–1288. doi: 10.1017/s1355838200000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury, A., Mukhopadhyay, J., Tharun, S. The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA. 2007;13:998–1016. doi: 10.1261/rna.502507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller, J., Parker, R. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- Cougot, N., Babajko, S., Seraphin, B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker, C.J., Parker, R. A turnover pathway for both stable and unstable mRNAs in yeast: Evidence for a requirement for deadenylation. Genes & Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Dunckley, T., Parker, R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst, N.L., Panicucci, B., Igo R.P., Jr, Panigrahi, A.K., Salavati, R., Stuart, K. TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol. Cell. 2003;11:1525–1536. doi: 10.1016/s1097-2765(03)00185-0. [DOI] [PubMed] [Google Scholar]
- Eulalio, A., Behm-Ansmant, I., Izaurralde, E. P bodies: At the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Eystathioy, T., Jakymiw, A., Chan, E.K., Seraphin, B., Cougot, N., Fritzler, M.J. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Gron, M., Fillman, C., Norrild, B., Lykke-Andersen, J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Fillman, C., Lykke-Andersen, J. RNA decapping inside and outside of processing bodies. Curr. Opin. Cell Biol. 2005;17:326–331. doi: 10.1016/j.ceb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Ford, L.P., Wilusz, J. 3′-Terminal RNA structures and poly(U) tracts inhibit initiation by a 3′→5′ exonuclease in vitro. Nucleic Acids Res. 1999;27:1159–1167. doi: 10.1093/nar/27.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford, L.P., Bagga, P.S., Wilusz, J. The poly(A) tail inhibits the assembly of a 3′-to-5′ exonuclease in an in vitro RNA stability system. Mol. Cell. Biol. 1997;17:398–406. doi: 10.1128/mcb.17.1.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari, F.B., Adams, M.D., Pagano, J.S. Unconventional processing of the 3′ termini of the Epstein-Barr virus DNA polymerase mRNA. Proc. Natl. Acad. Sci. 1993;90:378–382. doi: 10.1073/pnas.90.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M., Wilusz, C.J., Peltz, S.W., Wilusz, J. A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J. 2001;20:1134–1143. doi: 10.1093/emboj/20.5.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau, N.L., Wilusz, J., Wilusz, C.J. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Ghosh, T., Peterson, B., Tomasevic, N., Peculis, B.A. Xenopus U8 snoRNA binding protein is a conserved nuclear decapping enzyme. Mol. Cell. 2004;13:817–828. doi: 10.1016/s1097-2765(04)00127-3. [DOI] [PubMed] [Google Scholar]
- He, W., Parker, R. The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3′ termini from partial degradation. Genetics. 2001;158:1445–1455. doi: 10.1093/genetics/158.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, C.L., Stevens, A. Yeast cells lacking 5→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim, F., Rohr, J., Jeong, W.J., Hesson, J., Cerutti, H. Untemplated oligoadenylation promotes degradation of RISC-cleaved transcripts. Science. 2006;314:1893. doi: 10.1126/science.1135268. [DOI] [PubMed] [Google Scholar]
- Ingelfinger, D., Arndt-Jovin, D.J., Luhrmann, R., Achsel, T. The human LSm1–7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Jackson, R.J., Standart, N. How do microRNAs regulate gene expression? Sci. STKE. 2007;367 doi: 10.1126/stke.3672007re1. re1. [DOI] [PubMed] [Google Scholar]
- Jakymiw, A., Lian, S., Eystathioy, T., Li, S., Satoh, M., Hamel, J.C., Fritzler, M.J., Chan, E.K. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- Jiao, X., Wang, Z., Kiledjian, M. Identification of an mRNA-decapping regulator implicated in X-linked mental retardation. Mol. Cell. 2006;24:713–722. doi: 10.1016/j.molcel.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna, R., Kiledjian, M. Poly(A)-binding-protein-mediated regulation of hDcp2 decapping in vitro. EMBO J. 2004;23:1968–1976. doi: 10.1038/sj.emboj.7600213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak, J.E., Wickens, M. A family of poly(U) polymerases. RNA. 2007;13:860–867. doi: 10.1261/rna.514007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaGrandeur, T.E., Parker, R. Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J. 1998;17:1487–1496. doi: 10.1093/emboj/17.5.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Yang, Z., Yu, B., Liu, J., Chen, X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis . Curr. Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Kiledjian, M. Decapping the message: A beginning or an end. Biochem. Soc. Trans. 2006;34:35–38. doi: 10.1042/BST20060035. [DOI] [PubMed] [Google Scholar]
- Liu, H., Rodgers, N.D., Jiao, X., Kiledjian, M. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J. 2002;21:4699–4708. doi: 10.1093/emboj/cdf448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Valencia-Sanchez, M.A., Hannon, G.J., Parker, R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen, J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen, J., Wagner, E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes & Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, S., Temme, C., Wahle, E. Messenger RNA turnover in eukaryotes: Pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- Murray, E.L., Schoenberg, D.R. A+U-rich instability elements differentially activate 5′–3′ and 3′–5′ mRNA decay. Mol. Cell. Biol. 2007;27:2791–2799. doi: 10.1128/MCB.01445-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, R., Sheth, U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Parker, R., Song, H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Piccirillo, C., Khanna, R., Kiledjian, M. Functional characterization of the mammalian mRNA decapping enzyme hDcp2. RNA. 2003;9:1138–1147. doi: 10.1261/rna.5690503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai, R.S., Bhattacharyya, S.N., Artus, C.G., Zoller, T., Cougot, N., Basyuk, E., Bertrand, E., Filipowicz, W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Pillai, R.S., Bhattacharyya, S.N., Filipowicz, W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Ramirez, C.V., Vilela, C., Berthelot, K., McCarthy, J.E. Modulation of eukaryotic mRNA stability via the cap-binding translation complex eIF4F. J. Mol. Biol. 2002;318:951–962. doi: 10.1016/S0022-2836(02)00162-6. [DOI] [PubMed] [Google Scholar]
- Rissland, O.S., Mikulasova, A., Norbury, C.J. Efficient RNA polyuridylation by noncanonical poly(a) polymerases. Mol. Cell. Biol. 2007;27:3612–3624. doi: 10.1128/MCB.02209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers, N.D., Wang, Z., Kiledjian, M. Regulated α-globin mRNA decay is a cytoplasmic event proceeding through 3′-to-5′ exosome-dependent decapping. RNA. 2002;8:1526–1537. [PMC free article] [PubMed] [Google Scholar]
- Schwartz, D.C., Parker, R. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 2000;20:7933–7942. doi: 10.1128/mcb.20.21.7933-7942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen, G.L., Blau, H.M. Argonaute2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- Shen, B., Goodman, H.M. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- Sheth, U., Parker, R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman, S. Catalytic activity of vaccinia mRNA capping enzyme subunits coexpressed in Escherichia coli . J. Biol. Chem. 1990;265:11960–11966. [PubMed] [Google Scholar]
- Simon, E., Camier, S., Seraphin, B. New insights into the control of mRNA decapping. Trends Biochem. Sci. 2006;31:241–243. doi: 10.1016/j.tibs.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Steiger, M., Carr-Schmid, A., Schwartz, D.C., Kiledjian, M., Parker, R. Analysis of recombinant yeast decapping enzyme. RNA. 2003;9:231–238. doi: 10.1261/rna.2151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin, G., Mayo, T., Anderson, P. ARE-mRNA degradation requires the 5′–3′ decay pathway. EMBO Rep. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, M.S., Dreyfuss, G. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol. Cell. Biol. 1988;8:2237–2241. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun, S., He, W., Mayes, A.E., Lennertz, P., Beggs, J.D., Parker, R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- Trippe, R., Guschina, E., Hossbach, M., Urlaub, H., Luhrmann, R., Benecke, B.J. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA. 2006;12:1494–1504. doi: 10.1261/rna.87706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk, E., Cougot, N., Meyer, S., Babajko, S., Wahle, E., Seraphin, B. Human Dcp2: A catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan, S., Peltz, S.W. Regulated ARE-mediated mRNA decay in Saccharomyces cerevisiae . Mol. Cell. 2001;7:1191–1200. doi: 10.1016/s1097-2765(01)00279-9. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Kiledjian, M. Functional link between the mammalian exosome and mRNA decapping. Cell. 2001;107:751–762. doi: 10.1016/s0092-8674(01)00592-x. [DOI] [PubMed] [Google Scholar]
- Wang, S.W., Toda, T., MacCallum, R., Harris, A.L., Norbury, C. Cid1, a fission yeast protein required for S-M checkpoint control when DNA polymerase δ or ε is inactivated. Mol. Cell. Biol. 2000;20:3234–3244. doi: 10.1128/mcb.20.9.3234-3244.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Jiao, X., Carr-Schmid, A., Kiledjian, M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl. Acad. Sci. 2002;99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz, C.J., Gao, M., Jones, C.L., Wilusz, J., Peltz, S.W. Poly(A)-binding proteins regulate both mRNA deadenylation and decapping in yeast cytoplasmic extracts. RNA. 2001a;7:1416–1424. [PMC free article] [PubMed] [Google Scholar]
- Wilusz, C.J., Wormington, M., Peltz, S.W. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2001b;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]