Abstract
In bacteria, initiation of translation is kinetically controlled by factors IF1, IF2, and IF3, which work in conjunction with the 30S subunit to ensure accurate selection of the initiator tRNA (fMet-tRNAfMet) and the start codon. Here, we show that mutations G1338A and A790G of 16S rRNA decrease initiation fidelity in vivo and do so in distinct ways. Mutation G1338A increases the affinity of tRNAfMet for the 30S subunit, suggesting that G1338 normally forms a suboptimal Type II interaction with fMet-tRNAfMet. By stabilizing fMet-tRNAfMet in the preinitiation complex, G1338A may partially compensate for mismatches in the codon–anti-codon helix and thereby increase spurious initiation. Unlike G1338A, A790G decreases the affinity of IF3 for the 30S subunit. This may indirectly stabilize fMet-tRNAfMet in the preinitiation complex and/or promote premature docking of the 50S subunit, resulting in increased levels of spurious initiation.
Keywords: translation, ribosome, 16S rRNA, P site, initiation
INTRODUCTION
Initiation of translation involves recognition of the start codon by initiator tRNA in the P site of the small ribosomal subunit. This process requires both selection of the initiator tRNA from all other species of tRNA and selection of the start codon from other similar or identical triplet sequences in the mRNA. In bacteria, molecular determinants that specify the initiator tRNA (fMet-tRNAfMet) and the start codon have been identified (for review, see Gualerzi et al. 2001; Mayer et al. 2001). One unique feature of initiator tRNAfMet is a mismatch at the last base pair of the acceptor stem (C1×A72). This mismatch is important for formylation of Met-tRNAfMet (Lee et al. 1992), without which specific recognition by initiation factor 2 (IF2) is lost (Antoun et al. 2006a). The C1×A72 mismatch also prevents Met-tRNAfMet from binding EF-Tu and participating in the elongation phase of translation (Seong and RajBhandary 1987b). Another unique feature of tRNAfMet is a series of three adjacent G–C base pairs in the anti-codon stem. Two of these G–C base pairs are particularly important for efficient initiation (Mandal et al. 1996), and these (G29-C41 and G30-C40) interact with 16S rRNA nucleotides G1338 and A1339, respectively (Berk et al. 2006; Korostelev et al. 2006; Selmer et al. 2006). The importance of these G–C pairs may stem from their ability to stabilize fMet-tRNAfMet in the 30S P site (Seong and RajBhandary 1987a; Mandal et al. 1996; Lancaster and Noller 2005). Regarding start codon selection, an important determinant is a purine-rich element (e.g., AGGA) termed the Shine-Dalgarno sequence (SD) (Shine and Dalgarno 1974), which is positioned 6–12 nucleotides upstream from the start codon (Shultzaberger et al. 2001). The SD sequence base pairs with the 3′ end of the 16S rRNA (termed the anti-Shine-Dalgarno sequence or ASD) to facilitate positioning of the start codon in the 30S P site) (Steitz and Jakes 1975). The sequence of the SD, sequence of the start codon, number of nucleotides between the SD and start codon, and the mRNA secondary structure influence the efficiency of initiation (Vellanoweth and Rabinowitz 1992; Studer and Joseph 2006), indicating the importance of each of these determinants in the initiation process.
Translation initiation in bacteria is kinetically controlled by three initiation factors (IF1, IF2, and IF3), and recent studies have helped clarify the roles of these factors in the initiation process (Antoun et al. 2003, 2006a, b; Studer and Joseph 2006). The first major step in the process is assembly of the 30S preinitiation complex. In this complex, fMet-tRNAfMet is paired to the start codon of mRNA in the 30S subunit P site, IF1 occupies the 30S A site, IF2•GTP binds an adjacent position and interacts specifically with the acceptor end of fMet-tRNAfMet, and IF3 binds the platform of the 30S subunit near the P and E sites (Moazed et al. 1995; McCutcheon et al. 1999; Carter et al. 2001; Dallas and Noller 2001; Marzi et al. 2003; Allen et al. 2005). The order of assembly remains unclear, and it has been proposed that mRNA and fMet-tRNAfMet associate in random order (Gualerzi et al. 2001). During preinitiation complex formation, the initiation factors act synergistically to increase the association rate of fMet-tRNAfMet (by 400-fold), while IF1 and IF3 also increase the dissociation rate of fMet-tRNAfMet (by 300-fold) (Antoun et al. 2006a,b). Other species of tRNA exhibit considerably slower association rates and faster dissociation rates, suggesting the basis for fMet-tRNAfMet selection at this stage of initiation (Antoun et al. 2006a). The second major step of the initiation process is 50S subunit docking. The GTP-bound form of IF2 accelerates 50S docking, while IF3 inhibits the event (Antoun et al. 2003, 2006b). It has been proposed that 50S docking can occur only after release of IF3 from the complex, and thus IF3 acts to prevent premature 50S docking when fMet-tRNAfMet is absent (Antoun et al. 2006b). After 50S docking, hydrolysis of GTP facilitates release of IF2, generating a 70S initiation complex ready for the first round of elongation (Antoun et al. 2003).
A number of mutations have been isolated in Escherichia coli that increase translation from noncanonical start codons (i.e., codons other than AUG, UUG, and GUG) (Sacerdot et al. 1996; Sussman et al. 1996; Haggerty and Lovett 1997). These mutations mapped to infC, the gene encoding IF3, strongly implicating the factor in the fidelity of initiation. Reducing the concentration of wild-type IF3 in the cell also increased spurious initiation (Olsson et al. 1996), suggesting that the isolated mutations may reduce either the activity or level of IF3. Overexpression of IF3 conferred the opposite phenotype: repression of translation from noncanonical start codons relative to control strains (Butler et al. 1986; Sacerdot et al. 1996). The ability of IF3 to influence start codon selection is well exemplified by its ability to negatively regulate its own gene, infC. Translation of infC mRNA depends on initiation from the noncanonical start codon AUU (Butler et al. 1986, 1987).
In theory, spurious initiation could involve either an elongator tRNA paired to a cognate codon (i.e., an error in tRNA selection) or fMet-tRNAfMet paired to a non- or near-cognate codon (i.e., an error in codon selection). N-terminal sequencing of polypeptide products of spurious initiation events indicated that, in infC and wild-type strains, translation began with the initiator tRNA (O'Connor et al. 1997, 2001). Moreover, spurious initiation was observed specifically from codons termed Class IIA (i.e., AUA, AUC, AUU, ACG, and CUG) (Sacerdot et al. 1996; Sussman et al. 1996), which differ from cognate start codons (AUG, GUG, or UUG) at only one position. This sequence dependence would not be expected if elongator tRNAs played a substantial role in spurious initiation. These observations suggest that IF3 increases fidelity in vivo primarily by preventing errors in start codon selection.
Mutations in the 16S rRNA genes were not recovered in the aforementioned genetic studies, even though IF3 makes extensive contact with the platform domain of the 30S subunit (Dallas and Noller 2001). There are seven copies of the 16S rRNA gene in E. coli, which raises the possibility that these mutations were missed because their effects were masked by the remaining wild-type copies. Here, using a specialized ribosome system, we show that mutations G1338A and A790G of 16S rRNA increase translation from noncanonical start codons and do so in distinct ways. Mutation G1338A stabilizes tRNAfMet in the 30S P site, whereas A790G decreases the affinity of IF3 for the 30S subunit.
RESULTS AND DISCUSSION
G1338A and A790G increase translation from noncanonical start codons
To investigate the role of 16S rRNA in start codon selection, we screened a collection of P-site mutations for those that increase initiation from noncanonical start codons using a specialized ribosome system (Abdi and Fredrick 2005). In this system, the plasmid-encoded 16S rRNA contains the altered (specialized) ASD sequence 5′-GGGGU-3′ to allow specific translation of the lacZ reporter mRNA, which contains the complementary SD sequence 5′-AUCCC-3′. Because the specialized ribosomes recognize the lacZ mRNA but not the endogenous mRNA, the effects of 16S rRNA mutations on translation can be quantified in vivo without secondary effects on cell growth. Prior to the screen, sensitivity was optimized in this system by strengthening the promoter of the chromosomal lacZ reporter gene and by assaying β-galactosidase activity with the substrate CPRG (see Materials and Methods). We found that two mutations, G1338A and A790G, enhanced translation from ACG, AUC, and CUG but not from AUG (Table 1). Both of these mutations increased spurious initiation modestly (by 30%–110%) but significantly (P<0.05). This phenotype was not attributable to a number of other P-site mutations (e.g., A790C, A790U, G926A, G926C, G926U, m2G966A, m2G966C, m2G966U, G1338C, G1338U, A1339C, A1339G, A1339U, C1400A, C1400G, and C1400U).
TABLE 1.
Effects of 16S rRNA mutations G1338A and A790G on translation of mRNA containing noncanonical start codons
Next, we measured the effects of G1338A and A790G in the presence of infC362, a mutant allele of the IF3 gene. In strains expressing the control 16S rRNA, infC362 increased translation specifically from noncanonical start codons by five- to ninefold (Table 1), consistent with previous studies (Sussman et al. 1996). These data indicate that the alternative SD-ASD helix has no appreciable effect on the ability of IF3 to increase the stringency of start codon selection. When 16S rRNA containing G1338A was expressed in the infC362 background, a further increase of 60%–80% was observed. In each case (ACG, AUC, and CUG), the increase attributed to G1338A was significant (P<0.05). Thus, the effects of G1338A and infC362 are additive, suggesting that they act independently to increase spurious initiation. By contrast, A790G did not confer an increase in spurious initiation in the presence of infC362. Instead, A790G decreased translation from ACG, AUC, and CUG by approximately twofold. Similar results were obtained when these experiments were repeated with infC561 (data not shown), arguing against allele-specific effects. These data provide evidence that G1338A and A790G decrease the fidelity of initiation in distinct ways.
Interestingly, in the infC362 background, G1338A increased translation from AUG modestly but significantly (P<0.05) (Table 1). It has been shown that IF3 acts in concert with the other initiation factors to stimulate initiation complex formation at AUG start codons. Additionally, IF3 negatively regulates the process to enhance accuracy (Antoun et al. 2006a,b). It is possible that in the infC362 background, the efficiency of initiation complex formation at the canonical AUG start codon is a bit compromised and G1338A compensates to restore high-level translation (Table 1).
G1338A increases the affinity of tRNAfMet for the 30S subunit P site
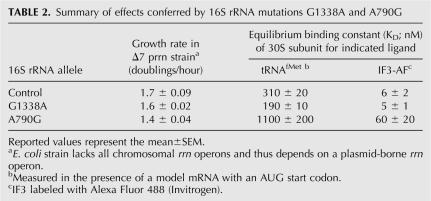
Previous studies showed that G1338A can suppress phenotypes conferred by other P-site mutations, suggesting that G1338A might stabilize tRNA in the 30S P site (Abdi and Fredrick 2005). To investigate this possibility, we first needed to purify mutant 30S subunits. Plasmids encoding rrnB with either G1338A or A790G in the 16S gene (containing a wild-type ASD) were moved into an E. coli strain lacking all chromosomal rrn operons (Δ7 prrn), replacing the resident plasmid containing rrnC (see Materials and Methods). Although G1338 and A790 are universally conserved, ribosomes carrying either G1338A or A790G supported robust cell growth (Table 2). These mutations decreased the growth rate of the Δ7 prrn strain by only 6% and 18%, respectively. From these strains, mutant and control 30S subunits were purified, and their affinity for tRNAfMet was compared (Fig. 1; Table 2). Subunits containing G1338A exhibited higher affinity for tRNAfMet than control subunits. Because this difference in affinity was modest (approximately twofold), mutant and control 30S subunits were repurified two more times, and the binding experiment was repeated. In each independent experiment, subunits with G1338A bound tRNAfMet with approximately twofold higher affinity than control subunits. By contrast, A790G decreased the affinity of tRNAfMet by approximately fourfold (Fig. 1; Table 2).
TABLE 2.
Summary of effects conferred by 16S rRNA mutations G1338A and A790G
FIGURE 1.
Mutation G1338A increases the affinity of tRNAfMet for the 30S subunit P site. Wild-type (○) or mutant 30S subunits harboring G1338A (△) or A790G (□) were incubated at various concentrations with a model mRNA (containing an AUG codon; 2 μM) and 3′-[32P]-tRNAfMet. Then, the fraction of 3′-[32P]-tRNAfMet bound was determined by filtration through a bilayer of nitrocellulose and nylon membranes (see Materials and Methods). Equilibrium binding constants (KD) listed in Table 2 come from at least three independent binding experiments like the one shown here.
Structural studies have shown that A1339 and G1338 dock into the minor groove of the anti-codon stem of P-site tRNA (Berk et al. 2006; Korostelev et al. 2006; Selmer et al. 2006). A1339 and G1338 are positioned to form Type I and II interactions with tRNA base pairs 30–40 and 29–41, respectively. Our data suggest that G1338 forms a suboptimal interaction with tRNAfMet in the 30S P site, since tRNAfMet binding is enhanced by mutation G1338A (Fig. 1). Study of an analogous Type II interaction showed that replacement of the docking A for G caused a modest decrease in stability (Doherty et al. 2001), in line with our results. We propose that the ability of G1338A to stabilize fMet-tRNAfMet in the 30S P site partially compensates for mismatches in the codon–anti-codon helix and thereby allows increased levels of spurious initiation.
Although our data show that G1338 helps increase the stringency of start codon selection, the contribution of G1338 to fidelity appears minor compared with that of IF3. Mutations in IF3 generally derepress spurious initiation by fivefold to 40-fold (Table 1; (Sacerdot et al. 1996; Sussman et al. 1996; O'Connor et al. 2001), while the effects of G1338A are twofold or less (Table 1). Furthermore, the ability of IF3 to discriminate the start codon does not depend on the Type II “G-minor” interaction, because the degree of repression by IF3 (infC+ versus infC362) is comparable for ribosomes harboring either G or A at position 1338 (Table 1).
A790G decreases the affinity of IF3 for the 30S subunit
The genetic data suggested that A790G and G1338A differentially perturb the initiation process (Table 1). Although both contribute to the 30S P site, A790 and G1338 lie in different domains of the subunit and contact opposite sides of the P-site tRNA (Korostelev et al. 2006; Selmer et al. 2006). IF3 interacts predominantly with the platform domain of 16S rRNA, protecting A790 and nearby 16S rRNA nucleotides from chemical probes (Moazed et al. 1995; Dallas and Noller 2001). A mutation in this region (G791A) has been shown to reduce the affinity of IF3 for the 30S subunit by ninefold (Tapprich et al. 1989). With these observations in mind, we considered the possibility that decreased fidelity conferred by A790G was due to a defect in IF3 binding.
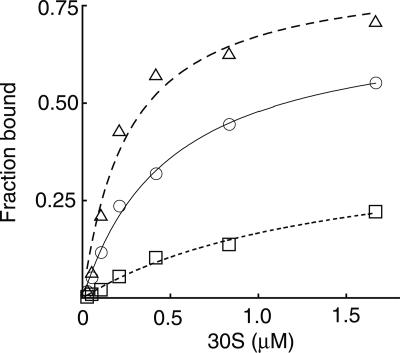
To study the effects of these 16S rRNA mutations on IF3 binding, we labeled IF3 using an amino-reactive fluorophore (Alexa Fluor 488; see Materials and Methods). The degree of labeling was estimated to be 0.6 dyes per IF3. In the first binding experiment, fluorescently labeled IF3 (IF3-AF; 1 μM) was incubated with each preparation of 30S subunits (1 μM), reactions were subjected to sucrose gradient sedimentation, and the relative amount of IF3-AF migrating at 30S was assessed (Fig. 2A). With wild-type and G1338A subunits, an obvious peak of fluorescence was detected in fractions corresponding to the 30S subunit peak, deduced by absorbance at 260 nm. In contrast, IF3-AF did not comigrate with 30S subunits containing A790G. Instead, increased fluorescence signal was observed at the top of the gradient, where free IF3-AF was expected to remain. Consistent with this interpretation, control reactions containing 50S subunits or lacking subunits gave identical distributions, with IF3-AF detected only at the top of the gradient.
FIGURE 2.
Mutation A790G decreases the affinity of IF3 for the 30S subunit. (A) Fluorescently labeled IF3 (IF3-AF) was incubated with mutant or wild-type 30S subunits, with 50S subunits, or in absence of subunits (as indicated), and the reactions were subjected to sucrose gradient sedimentation analysis. To determine the distribution of IF3-AF in each gradient, 0.5 mL fractions were collected and the fluorescence intensity (excitation 495 nm; emission 520 nm) of each fraction was quantified. To determine the distribution of ribosomal subunits in each gradient, A260 was monitored during the fractionation process using an in-line UV detector. (B) Fluorescence anisotropy was used to estimate the affinity of IF3-AF for 30S subunits without (○) or with mutation G1338A (△) or A790G (□). Subunits at various concentrations were incubated with 10 nM IF3-AF, and fluorescence anisotropy was measured. Equilibrium binding constants (KD) listed in Table 2 come from at least three independent binding experiments like those shown here.
Next, we estimated the affinity of IF3-AF for each of the 30S subunits using fluorescence anisotropy (Fig. 2B; Table 2). In each case, an increase in anisotropy attributable to binary complex formation was observed when 30S subunits were added to IF3-AF. Mutation G1338A did not affect the affinity of IF3-AF for 30S subunits, while A790G decreased the affinity for IF3-AF by 10-fold (Fig. 2B; Table 2). These data corroborate results from the sucrose gradient sedimentation analysis and suggest that decreased fidelity conferred by A790G is due to decreased affinity of IF3 for the 30S subunit.
How IF3 increases the accuracy of start codon selection remains unclear, but recent studies provide clues about the mechanism (Antoun et al. 2006a,b; Studer and Joseph 2006). IF3 negatively regulates formation of the 30S preinitiation complex by destabilizing tRNA (including fMet-tRNAfMet) in the 30S P site. IF3 also negatively regulates the subsequent step, 50S subunit docking. Based on these observations, reduced IF3 activity in the cell is predicted to both (1) stabilize fMet-tRNAfMet in the preinitiation complex and (2) increase the rate of 50S subunit docking. These effects may allow initiation despite mismatches in the codon–anti-codon helix, thus decreasing the stringency of start codon selection. Mutation A790G, which inhibits IF3 binding to the 30S subunit (Fig. 2), may stimulate spurious initiation in an analogous way. Even though A790G decreases the affinity of tRNAfMet by fourfold in the absence of factors (Fig. 1), this mutation may effectively stabilize fMet-tRNAfMet in the preinitiation complex because it inhibits IF3 binding. Alternatively or additionally, by decreasing the affinity of IF3 for the preinitiation complex, A790G may promote premature 50S docking, thereby increasing spurious initiation. Further characterization of these mutant ribosomes will be necessary to distinguish among these possibilities.
Conclusions
In this study, we show that mutations G1338A and A790G of 16S rRNA increase translation from noncanonical start codons in vivo. This phenotype is conferred directly, because the endogenous wild-type ribosomes fail to recognize the reporter mRNA in our strains. Mutation G1338A increases the affinity of the initiator tRNA for the 30S P site, suggesting that G1338 normally forms a suboptimal Type II interaction with fMet-tRNAfMet. We propose that an enhanced Type II interaction conferred by G1338A partially compensates for mismatches in the codon–anti-codon helix and thereby increases spurious initiation. Mutation A790G, on the other hand, decreases the affinity of IF3 for the 30S subunit. Reduced IF3 binding affinity may stabilize fMet-tRNAfMet in the preinitiation complex and/or promote premature 50S docking, thereby increasing spurious initiation. These mutations should prove useful in further elucidating the kinetic pathway of initiation complex formation and the molecular basis for fidelity during the process.
MATERIALS AND METHODS
Specialized ribosome strains
Translation activity of specialized ribosomes was measured in indicator strains similar to those described previously (Abdi and Fredrick 2005). Each strain contained the lacZ gene (with the alternative SD sequence 5′-ATCCC-3′) in single copy on the chromosome. To construct these strains, DNA fragments containing a consensus variant of the Pant promoter (Moyle et al. 1991), the alternative SD sequence 5′-ATCCC-3′, and a start codon (ATG, ACG, ATC, or CTG) were generated as described (Abdi and Fredrick 2005). These fragments were digested with EcoRI and BamHI and cloned upstream of lacZ in pRS552, and the resulting fusions were transferred to λRS45 by homologous recombination in vivo (Simons et al. 1987). The recombinant λ phage were then used to lysogenize strain JK382 [F- ara-600 Δ(gpt-lac)5 relA1 spoT1 thi-1 zdi-925∷Tn10], JK378 [F- ara-600 Δ(gpt-lac)5 relA1 spoT1 thi-1 zdi-925∷Tn10 infC362] and JK530 [F- ara-600 Δ(gpt-lac)5 relA1 spoT1 thi-1 zdi-925∷Tn10 infC561] (Sussman et al. 1996), which were obtained from the Coli Genetic Stock Center (Yale University, New Haven, CT). Each of the resulting lysogens was confirmed by PCR to contain a single prophage (Powell et al. 1994).
Plasmid pKF207 contains the 16S rRNA gene with the altered ASD 5′-GGGGT-3′ (Lee et al. 1996) under transcriptional control of the PBAD promoter. The construction of pKF207 and its derivatives was described previously (Abdi and Fredrick 2005). Expression of these plasmid-borne 16S rRNA alleles in the indicator strains described above allowed translation activity of the corresponding mutant ribosomes to be measured.
Activity of mutant ribosomes in vivo
Cells from a saturated culture were diluted 500-fold into fresh Luria broth (LB) containing ampicillin (Amp; 100 μg/mL), kanamycin (Kan; 30 μg/mL), and L-arabinose (5 mM) and grown for 6 h at 37°C. The cells were washed once in Z buffer (100 mM sodium phosphate at pH 7.0, 10 mM KCl, 10 mM MgSO4), and β-galactosidase activity was measured as described (Abdi and Fredrick 2005), except that chlorophenol red-β-d-galactopyranoside (CPRG; Sigma) was used as the substrate. Reported units were defined by the following equation: 1 unit=1000×(A573)/(OD600×v×t), where A573 is absorbance at 573 nm (characteristic of the product), OD600 is optical density of the cell suspension used, v is the volume of cell suspension used (in microliters), and t is time of incubation (in minutes) at room temperature. Differences were deemed statistically significant based on Student's t-test with the Bonferroni correction.
Δ7 prrn strains
Mutations G1338A and A790G were introduced into plasmid p278MS2 (Youngman et al. 2004), which contains (a tagged version of) the rrnB operon, to generate plasmids pNA56 and pNA58, respectively. Plasmids p278MS2, pNA56, and pNA58 were then transformed into the Δ7 prrn strain SQZ10 (kindly provided by C. Squires, Tufts University, Boston, MA), selecting for resistance to Amp. Resulting transformants were grown in liquid media and spread onto LB plates containing Amp and sucrose (5%) to select against pHKrrnC-sacB (KanR), the resident plasmid of SQZ10. Isolates resistant to sucrose and Amp but sensitive to Kan were identified in which the p278MS2 derivative had replaced pHKrrnC-sacB. Plasmid replacement was confirmed in strains expressing mutant ribosomes by (1) purifying the plasmid and sequencing the region containing the mutation and (2) purifying 16S rRNA and confirming the presence of the mutation using primer extension (data not shown).
Purification of 30S subunits
Each Δ7 prrn strain was grown to mid-logarithmic phase (OD600≈0.4) at 37°C in 1 L of LB, the culture was chilled on ice, and all subsequent steps were performed at 4°C. The cells were collected by centrifugation, washed in 30 mL of buffer A (20 mM Tris-HCl at pH 7.5, 10.5 mM MgCl2, 100 mM NH4Cl, 0.5 mM EDTA, 6 mM β-ME), resuspended in 30 mL of the same buffer, and lysed by passage through a French press. The cell lysate was clarified by centrifugation at 15,000 rpm for 15 min. The ribosomes were pelleted through a 10 mL sucrose cushion (1.1 M) in buffer B (20 mM Tris-HCl at pH 7.5, 10.5 mM MgCl2, 500 mM NH4Cl, 0.5 mM EDTA, 6 mM β-ME) by centrifugation at 41,000 rpm for 18 h in a Beckman Ti60 rotor. The ribosome pellet was dissolved in buffer C (50 mM Tris-HCl at pH 7.5, 1 mM MgCl2, 100 mM NH4Cl, 6 mM β-ME) and dialyzed against the same buffer to dissociate the ribosomes into subunits. Subunits were separated by centrifugation through 34 mL sucrose gradients (10%–30% sucrose in buffer C) at 20,000 rpm for 13 h in a Beckman SW32 rotor. 30S subunits were collected, the magnesium concentration was raised to 10 mM, and the subunits were pelleted by centrifugation in a Beckman Ti60 rotor at 38,000 rpm for 17 h. Finally, the subunits were dissolved in buffer D (50 mM Tris-HCl at pH 7.5, 10 mM MgCl2, 100 mM NH4Cl, 6 mM β-ME), separated into small aliquots, flash frozen, and stored at −70°C.
P-site tRNA binding experiments
The affinity of tRNAfMet for the 30S subunit P site was measured using a double-filter method described previously (Fahlman and Uhlenbeck 2004). 3′-[32P]-tRNAfMet (10 nM), mRNA (5′-UAAACGAGGAAACAAAUGGCUCGCACAACA-3′; 2 μM), and heat-activated 30S subunits at various concentrations were incubated for 10 min at 37°C in 20 μL TNM buffer (50 mM Tris-HCl at pH 7.5, 20 mM MgCl2, 100 mM NH4Cl, and 6 mM β-ME). Binding reactions were diluted with 100 μL of cold TNM and immediately filtered through bilayer of nitrocellulose (GE Corporation) and Hybond-N+ (Amersham Biosciences) membranes, using a 96-well dot-blot apparatus (Schleicher and Schuell). The membranes were separated, dried, and exposed to a phosphorimager screen for quantification. The fraction of counts trapped on the nitrocellulose was taken as the fraction of tRNAfMet bound and plotted as a function of 30S subunit concentration using Kaleidagraph. Dissociation constants (K D) were determined using the following equation:
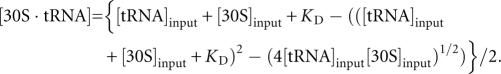 |
IF3 binding experiments
IF3 containing a C-terminal hexahistidine tag was purified as described previously (Dallas and Noller 2001) and labeled with an amino-reactive fluorophore (Alexa Fluor 488) as recommended by the supplier (Invitrogen). In a 0.1 mL reaction, IF3 (1 mg) was stirred with Alexa Fluor 488 carboxylic acid, 2,3,5,6-tetrafluorophenyl ester (50 μg; Invitrogen) in 0.1 M sodium bicarbonate buffer (pH 9.0) for 1 h at room temperature. The reaction was stopped by adding 10 μL of freshly prepared 1.5 M hydroxylamine (pH 8.5). The labeled protein (IF3-AF) was separated from unreacted labeling reagent by extensive dialysis in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4) buffer (pH 7.4), separated into small aliquots, flash frozen, and stored at −70°C. The degree of labeling, determined by measuring the absorbance of the protein at 280 nm and the absorbance of the dye at 495 nm, was estimated to be 0.6 dyes per IF3.
IF-AF (1 μM) was incubated with control or mutant 30S subunits (1 μM) in 10 mM Tris-HCl (pH 7.5), 60 mM NH4Cl, 10 mM Mg(OAc)2, 0.03% Nikkol, and 6 mM β-mercaptoethanol for 10 min at room temperature. Samples were then loaded onto 11 mL sucrose gradients (10%–30% sucrose in the same buffer) and centrifuged at 26,000 rpm for 16 h in a Beckmann SW41 rotor. Gradients were fractionated while A260 was monitored using an in-line UV detector (ISCO/Brandel system). Fractions (0.5 mL) were collected and the fluorescence intensity (excitation and emission at 495 nm 520 nm, respectively) of each was measured.
Steady-state fluorescence measurements were made using a Fluorolog-3 spectrofluorometer (Horiba Jobin Yvon, Inc.) and Type 16F Sub-Micro cuvettes (160 μL volume; Starna Cells, Inc.). The excitation and emission wavelengths were 495 nm and 520 nm, respectively, and the slit widths were set at 5 nm. To estimate equilibrium dissociation constants, fluorescence anisotropy was employed as described (An and Musier-Forsyth 2005). IF3-AF (10 nM) was incubated with 30S subunits (at various concentrations) in 10 mM Tris-HCl (pH 7.5), 60 mM NH4Cl, 10 mM Mg(OAc)2, and 6 mM β-mercaptoethanol for 10 min at 25°C prior to measuring anisotropy in a thermostated chamber. Anisotropy values were plotted as a function of 30S subunit concentration, and dissociation constants (K D) were obtained from fitting the data to the following equation: A=A min+(A max−A min) {([IF3-AF]input+[30S]input+K D) −(([IF3-AF]input+[30S]input+K D)2−(4[IF3-AF]input [30S]input)) 1/2}/2[IF3-AF]input, where A is the measured anisotropy, A min is the minimum anisotropy, and A max is the maximum anisotropy.
ACKNOWLEDGMENTS
We thank C. Squires for providing the Δ7 prrn strain and M. O'Connor for helpful discussions. This work was supported by NIH grant GM072528.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.715307.
REFERENCES
- Abdi, N.M., Fredrick, K. Contribution of 16S rRNA nucleotides forming the 30S subunit A and P sites to translation in Escherichia coli . RNA. 2005;11:1624–1632. doi: 10.1261/rna.2118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, G.S., Zavialov, A., Gursky, R., Ehrenberg, M., Frank, J. The cryo-EM structure of a translation initiation complex from Escherichia coli . Cell. 2005;121:703–712. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- An, S., Musier-Forsyth, K. Cys-tRNAPro editing by Haemophilus influenzae YbaK via a novel synthetase-YbaK-tRNA ternary complex. J. Biol. Chem. 2005;280:34465–34472. doi: 10.1074/jbc.M507550200. [DOI] [PubMed] [Google Scholar]
- Antoun, A., Pavlov, M.Y., Andersson, K., Tenson, T., Ehrenberg, M. The roles of initiation factor 2 and guanosine triphosphate in initiation of protein synthesis. EMBO J. 2003;22:5593–5601. doi: 10.1093/emboj/cdg525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoun, A., Pavlov, M.Y., Lovmar, M., Ehrenberg, M. How initiation factors maximize the accuracy of tRNA selection in initiation of bacterial protein synthesis. Mol. Cell. 2006a;23:183–193. doi: 10.1016/j.molcel.2006.05.030. [DOI] [PubMed] [Google Scholar]
- Antoun, A., Pavlov, M.Y., Lovmar, M., Ehrenberg, M. How initiation factors tune the rate of initiation of protein synthesis in bacteria. EMBO J. 2006b;25:2539–2550. doi: 10.1038/sj.emboj.7601140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk, V., Zhang, W., Pai, R.D., Cate, J.H.D. Structural basis for mRNA and tRNA positioning on the ribosome. Proc. Natl. Acad. Sci. 2006;103:15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, J.S., Springer, M., Dondon, J., Graffe, M., Grunberg-Manago, M. Escherichia coli protein synthesis initiation factor IF3 controls its own gene expression at the translational level in vivo. J. Mol. Biol. 1986;192:767–780. doi: 10.1016/0022-2836(86)90027-6. [DOI] [PubMed] [Google Scholar]
- Butler, J.S., Springer, M., Grunberg-Manago, M. AUU-to-AUG mutation in the initiator codon of the translation initiation factor IF3 abolishes translational autocontrol of its own gene (infC) in vivo. Proc. Natl. Acad. Sci. 1987;84:4022–4025. doi: 10.1073/pnas.84.12.4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, A.P., Clemons W.M., Jr, Brodersen, D.E., Morgan-Warren, R.J., Hartsch, T., Wimberly, B.T., Ramakrishnan, V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- Dallas, A., Noller, H.F. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol. Cell. 2001;8:855–864. doi: 10.1016/s1097-2765(01)00356-2. [DOI] [PubMed] [Google Scholar]
- Doherty, E.A., Batey, R.T., Masquida, B., Doudna, J.A. A universal mode of helix packing in RNA. Nat. Struct. Biol. 2001;8:339–343. doi: 10.1038/86221. [DOI] [PubMed] [Google Scholar]
- Fahlman, R.P., Uhlenbeck, O.C. Contribution of the esterified amino acid to the binding of aminoacylated tRNAs to the ribosomal P- and A-sites. Biochemistry. 2004;43:7575–7583. doi: 10.1021/bi0495836. [DOI] [PubMed] [Google Scholar]
- Gualerzi, C.O., Brandi, L., Caserta, E., Garofalo, C., Lammi, M., La Teana, A., Petrelli, D., Spurio, R., Tomsic, J., Pon, C.L. The Ribosome. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. Initiation factors in the early events of mRNA translation in bacteria; pp. 363–376. [DOI] [PubMed] [Google Scholar]
- Haggerty, T.J., Lovett, S.T. IF3-mediated suppression of a GUA initiation codon mutation in the recJ gene of Escherichia coli . J. Bacteriol. 1997;179:6705–6713. doi: 10.1128/jb.179.21.6705-6713.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev, A., Trakhanov, S., Laurberg, M., Noller, H.F. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1–13. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Lancaster, L., Noller, H.F. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol. Cell. 2005;20:623–632. doi: 10.1016/j.molcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Lee, C., Dyson, M.R., Mandal, N., Varshney, U., Bahramian, B., Rajbhandary, U.L. Striking effects of coupling mutations in the acceptor stem on recognition of tRNAs by Escherichia coli Met-tRNA synthetase and Met-tRNA transformylase. Proc. Natl. Acad. Sci. 1992;89:9262–9266. doi: 10.1073/pnas.89.19.9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K., Holland-Staley, C.A., Cunningham, P.R. Genetic analysis of the Shine–Dalgarno interaction: Selection of alternative functional mRNA-rRNA combinations. RNA. 1996;2:1270–1285. [PMC free article] [PubMed] [Google Scholar]
- Mandal, N., Mangroo, D., Dalluge, J.J., McCloskey, J.A., Raj-Bhandary, U.L. Role of the three consecutive G:C base pairs conserved in the anticodon stem of initiator tRNAs in initiation of protein synthesis in Escherichia coli . RNA. 1996;2:473–482. [PMC free article] [PubMed] [Google Scholar]
- Marzi, S., Knight, W., Brandi, L., Caserta, E., Soboleva, N., Hill, W.E., Gualerzi, C.O., Lodmell, J.S. Ribosomal localization of translation initiation factor IF2. RNA. 2003;9:958–969. doi: 10.1261/rna.2116303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, C., Stortchevoi, A., Kohrer, C., Varshney, U., RajBhandary, U.L. Cold Spring Harbor Symposia on Quantitative Biology. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. Initiator tRNA and its role in initiation of protein synthesis; pp. 195–206. [DOI] [PubMed] [Google Scholar]
- McCutcheon, J.P., Agrawal, R.K., Philips, S.M., Grassucci, R.A., Gerchman, S.E., Clemons W.M., Jr, Ramakrishnan, V., Frank, J. Location of translational initiation factor IF3 on the small ribosomal subunit. Proc. Natl. Acad. Sci. 1999;96:4301–4306. doi: 10.1073/pnas.96.8.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed, D., Samaha, R.R., Gualerzi, C., Noller, H.F. Specific protection of 16 S rRNA by translational initiation factors. J. Mol. Biol. 1995;248:207–210. doi: 10.1016/s0022-2836(95)80042-5. [DOI] [PubMed] [Google Scholar]
- Moyle, H., Waldburger, C., Susskind, M.M. Hierarchies of base-pair preferences in the P22 ant promoter. J. Bacteriol. 1991;173:1944–1950. doi: 10.1128/jb.173.6.1944-1950.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor, M., Thomas, C.L., Zimmermann, R.A., Dahlberg, A.E. Decoding fidelity at the ribosomal A and P sites: Influence of mutations in three different regions of the decoding domain in 16S rRNA. Nucleic Acids Res. 1997;25:1185–1193. doi: 10.1093/nar/25.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor, M., Gregory, S.T., Raj-Bhandary, U.L., Dahlberg, A.E. Altered discrimination of start codons and initiator tRNAs by mutant initiation factor 3. RNA. 2001;7:969–978. doi: 10.1017/s1355838201010184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson, C.L., Graffe, M., Springer, M., Hershey, J.W.B. Physological effects of translation initiation factor IF3 and ribosomal protein L20 in Escherichia coli . Mol. Gen. Genet. 1996;250:705–714. doi: 10.1007/BF02172982. [DOI] [PubMed] [Google Scholar]
- Powell, B.S., Court, D.L., Nakamura, Y., Rivas, M.P., Turnbough, C.L. Rapid confirmation of single copy λ prophage integration by PCR. Nucleic Acids Res. 1994;22:5765–5766. doi: 10.1093/nar/22.25.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacerdot, C., Chiaruttini, C., Engst, K., Graffe, M., Milet, M., Mathy, N., Dondon, J., Springer, M. The role of the AUU initiation codon in the negative feedback regulation of the gene for translation initiation factor IF3 in Escherichia coli . Mol. Microbiol. 1996;21:331–346. doi: 10.1046/j.1365-2958.1996.6361359.x. [DOI] [PubMed] [Google Scholar]
- Selmer, M., Dunham, C.M., Murphy, F.V., Weixlbaumer, A., Petry, S., Kelley, A.C., Weir, J.R., Ramakrishnan, V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Seong, B.L., Raj-Bhandary, U.L. Escherichia coli formylmethionine tRNA: Mutations in the GGG/CCC sequence conserved in anticodon stem of initiator tRNAs affect initiation of protein synthesis and conformation of anticodon loop. Proc. Natl. Acad. Sci. 1987a;84:334–338. doi: 10.1073/pnas.84.2.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong, B.L., Raj-Bhandary, U.L. Mutants of Escherichia coli formylmethionine tRNA: A single base change enables initiator tRNA to act as an elongator in vitro . Proc. Natl. Acad. Sci. 1987b;84:8859–8863. doi: 10.1073/pnas.84.24.8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine, J., Dalgarno, L. The 3′-terminal sequence of E. coli 16S ribosomal RNA complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultzaberger, R.K., Bucheimer, R.E., Rudd, K.E., Schneider, T.D. Anatomy of Escherichia coli ribosome binding sites. J. Mol. Biol. 2001;313:215–228. doi: 10.1006/jmbi.2001.5040. [DOI] [PubMed] [Google Scholar]
- Simons, R.W., Houman, F., Kleckner, N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Steitz, J.A., Jakes, K. How ribosomes select initiator regions in mRNA: Base pair formation between the 3′ terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli . Proc. Natl. Acad. Sci. 1975;72:4734–4738. doi: 10.1073/pnas.72.12.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer, S.M., Joseph, S. Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Mol. Cell. 2006;22:105–115. doi: 10.1016/j.molcel.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Sussman, J.K., Simons, E.L., Simons, R.W. Escherichia coli translation initiation factor 3 discriminates the initiation codon in vivo. Mol. Microbiol. 1996;21:347–360. doi: 10.1046/j.1365-2958.1996.6371354.x. [DOI] [PubMed] [Google Scholar]
- Tapprich, W.E., Goss, D.J., Dahlberg, A.E. Mutation at position 791 in Escherichia coli 16S ribosomal RNA affects processes involved in the initiation of protein synthesis. Proc. Natl. Acad. Sci. 1989;86:4927–4931. doi: 10.1073/pnas.86.13.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellanoweth, R.L., Rabinowitz, J.C. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol. Microbiol. 1992;6:1105–1114. doi: 10.1111/j.1365-2958.1992.tb01548.x. [DOI] [PubMed] [Google Scholar]
- Youngman, E.M., Brunelle, J.L., Kochaniak, A.B., Green, R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]