Abstract
Many mammalian genes contain overlapping antisense RNAs, but the functions and mechanisms of action of these transcripts are mostly unknown. WT1 is a well-characterized developmental gene that is mutated in Wilms’ tumor (WT) and acute myeloid leukaemia (AML) and has an antisense transcript (WT1-AS), which we have previously found to regulate WT1 protein levels. In this study, we show that WT1-AS is present in multiple spliceoforms that are usually expressed in parallel with WT1 RNA in human and mouse tissues. We demonstrate that the expression of WT1-AS correlates with methylation of the antisense regulatory region (ARR) in WT1 intron 1, displaying imprinted monoallelic expression in normal kidney and loss of imprinting in WT. However, we find no evidence for imprinting of mouse Wt1-as. WT1-AS transcripts are exported into the cytoplasm and form heteroduplexes with WT1 mRNA in the overlapping region in WT1 exon 1. In AML, there is often abnormal splicing of WT1-AS, which may play a role in the development of this malignancy. These results show that WT1 encodes conserved antisense RNAs that may have an important regulatory role in WT1 expression via RNA:RNA interactions, and which can become deregulated by a variety of mechanisms in cancer.
Keywords: antisense, RNA, WT1, heteroduplex, epigenetics, splicing
INTRODUCTION
Antisense RNAs were first identified in prokaryotes, but are now recognized as being of great importance in the regulation of mammalian gene expression, with perhaps over 70% of mammalian transcription units containing overlapping transcripts (Lehner et al. 2002; Yelin et al. 2003; Chen et al. 2004; Katayama et al. 2005; Timmons and Good 2006). These antisense transcripts, the majority of which are noncoding, can be either concordantly or discordantly expressed temporally and spatially relative to their sense counterparts, implying possible roles as positive or negative regulators of gene expression (Chen et al. 2005; Katayama et al. 2005). In a few imprinted genes, the role of noncoding antisense RNAs is especially well established in controlling gene expression (O'Neill 2005). For example, the noncoding RNA Air, which is antisense to part of the mouse Igf2r gene, has been shown to be essential for regulating the allelic expression of the cluster of imprinted genes on proximal mouse chromosome 17 (Sleutels et al. 2002).
The WT1 gene, an important developmental locus and tumor suppressor, has been previously shown to contain noncoding antisense transcripts (WT1-AS) spanning WT1 exon 1 and continuing upstream (Campbell et al. 1994; Eccles et al. 1994). These antisense RNAs originate within WT1 intron 1 from an antisense promoter (Malik et al. 1995), which together with an adjacent differentially methylated regulatory region, are defined as the antisense regulatory region (ARR) (Malik et al. 2000). Although WT1 is not imprinted in kidney (Little et al. 1992), we have demonstrated that WT1-AS and an alternative WT1 coding RNA (AWT1) that also originates from intron 1, are both imprinted in normal kidney (Malik et al. 2000; Dallosso et al. 2004). WT1-AS and AWT1 are thought to be coregulated by the ARR, which acts as a methylation-dependent bidirectional silencer (Hancock et al. 2007). Differential allelic methylation of the ARR correlates with monoallelic expression of WT1-AS and AWT1 in normal kidney, and loss of methylation of the ARR leads to loss of imprinting of these transcripts in Wilms’ tumor (WT) (Malik et al. 2000; Dallosso et al. 2004). We have demonstrated that WT1-AS transcripts colocalize with WT1 protein and RNA in kidney development, consistent with a role for them in positively regulating WT1 expression. Furthermore, our in vitro modeling experiments in cultured renal cells have shown that expression of antisense-orientation WT1 exon 1 sequences can up-regulate WT1 protein levels (Moorwood et al. 1998).
WT1 proteins act as transcriptional and post-transcriptional regulators and have essential roles in nephrogenesis, hematopoiesis, and sex determination (Scharnhorst et al. 2001; Roberts 2005). WT1 is somatically mutated in a subset of WTs (Brown et al. 1993) and acute myeloid leukaemias (AMLs) (King-Underwood et al. 1996), as well as being overexpressed in several other malignancies (Hohenstein and Hastie 2006). Thus, it is clear that tight control of WT1 levels is essential during normal development, and that deregulated expression may contribute to malignancy.
Given the potential role for WT1-AS in the modulation of WT1 gene expression at this important disease locus, the aims of this study were to further characterize the structure of WT1-AS and its possible mechanisms of action. Our results show that there are multiple alternate spliceoforms of WT1-AS that are generally coexpressed with sense transcripts in both human and mouse. We demonstrate that DNA methylation modulates the expression of kidney WT1-AS spliceoforms, and that a previously unreported spliceoform also displays monoallelic expression in normal kidney and biallelic expression in WT, consistent with loss of imprinting. However, unlike human WT1-AS, we found no evidence for imprinting of mouse Wt1-as. The WT1-AS and WT1 RNAs colocalize intracellularly and form RNA:RNA duplexes, indicating a possible RNA stabilization role for WT1-AS transcripts. Finally, we show that AMLs often have defective splicing of WT1-AS, indicating that this RNA may be deregulated in cancer by both epigenetic and splicing defects.
RESULTS
Multiple alternate splicing of WT1-AS
A combination of cDNA library screening and EST data mining was used to characterize WT1-AS spliceoforms in human tissues (Fig. 1A). In total, 10 different transcripts were identified and expression of nine of them was confirmed by RT–PCR in the WT1-expressing cell line 7.92 (data not shown). Internal sequencing and comparison with genomic sequences (NCBI build 36) showed that in each expressed transcript, exons were flanked by canonical splice donor and acceptor sites (GT–AG rule), indicating that these mRNAs represented bona fide spliced RNAs. Of the clones that we isolated from a fetal kidney cDNA library, AS1 and AS8 contained the major splice identified by Gessler and Bruns (1993), whereas clone AS9 had a novel splicing pattern that has not been described previously. Analysis of the complete cDNA sequences showed multiple small ORFs within these clones, the largest being 92 amino acids for AS1 (as described by Campbell et al. 1994). However, this small ORF is disrupted by several of the WT1-AS splicing events that we have characterized, strongly suggesting that it is nonfunctional. None of the predicted ORFs showed homology with any known protein sequences or are conserved in the mouse sequence. Therefore, it is probable that the WT1-AS clones detailed here represent noncoding RNAs.
FIGURE 1.
Antisense transcripts at the human and mouse WT1 loci. (A) Human WT1 antisense RNAs. The top scale is numbered in kilobase pairs relative to the major WT1 transcriptional start site (Hofmann et al. 1993), which is indicated by the right-angled black arrow, along with the furthest upstream start site identified by Fraizer et al. (1994) (right-angled dashed arrow) and the WT1-AS transcriptional start site (Malik et al. 1995) (right-angled arrow below line). Below are shown the structures of previously published WT1-AS RNAs (double lines), WT1-AS transcripts identified by cDNA library screening (AS1, AS8, AS9; this study; gray lines) and human ESTs representing WT1-AS RNAs (black lines). (B) Mouse Wt1 antisense RNAs. The top scale is numbered in kilobase pairs relative to the major Wt1 transcriptional start site. Below are shown the structures of three mouse ESTs representing Wt1-as RNAs. Transcripts are annotated with GenBank accession number and tissue source. Vertical dotted lines indicate shared donor and acceptor splice sites and alignment with WT1/Wt1 transcriptional start sites.
Clones AS8 and AS9 terminated several hundred base pairs upstream of WT1 exon 1 at a common XbaI site, which is probably due to incomplete methylation of the cDNA during the fetal kidney library construction, leading to cleavage of internal XbaI sites, as described by Keirsebilck et al. (1998). Of the other clones, none spanned WT1 exon 1, most likely because of premature termination of cDNA synthesis during reverse transcription, caused by the high degree of secondary structure within the WT1 exon 1 region (Brown et al. 1992). However, it is important to note that AW194904, AI648530, the three related EST clones BI837715/BM906117/BM545678, and CR604547 all extended into exon 1 of WT1 (maximum region of overlap was 192 nucleotides from the WT1 upstream transcriptional start site identified by Fraizer et al. [1994] for BI837715) (Fig. 1A). Therefore, some of the cDNAs described here do represent antisense RNAs that overlap with the WT1 sense mRNA.
BLASTN analysis of sequences upstream of mouse Wt1 identified three mouse newborn ovary-derived ESTs (GenBank AW552314, AK165089, and AK161318) that showed different 3′ terminal exons, but shared three common 5′ exons, the first of which originated within Wt1 exon 1 (Fig. 1B). Like human cDNAs, these Wt1-as clones do not encode extensive or phylogenetically conserved ORFs.
Expression of WT1-AS
Ribonuclease protection assay (RPA) experiments demonstrated that WT1-AS RNAs spanned from the 5′ upstream region of WT1 into exon 1 (Fig. 2A). Strand-specific RT–PCR was then used to confirm and expand this result. WT1-AS RNA was transcribed exclusively in the antisense direction upstream of WT1 (Fig. 2B, I), across the exon1–intron 1 boundary (Fig. 2B, III) and into exon 1, where WT1-AS overlapped WT1 sense RNA transcription (Fig. 2B, II). These results are consistent with WT1 antisense RNAs originating from the antisense promoter in intron 1 (Malik et al. 1995) and extending across exon 1 into the upstream region, where we have shown multiple alternate splicing (Fig. 1A).
FIGURE 2.
Expression of human WT1-AS in normal and malignant tissues. (A) Ribonuclease protection assay. Total RNA was hybridized to a probe corresponding to sequence −127 to +83 of WT1 (shaded box, left), synthesized to detect sense or antisense transcripts (shown above autoradiograph of gel of protected fragments, right). (P) Probe; (P+) probe plus RNase; (1) WT1-expressing cell line 7.92; (2) WT1 nonexpressing line 17.94. Positions of molecular weight markers are shown on the right. Full-length protected probe in antisense lane 1 indicates expression of WT1-AS from exon 1. (B) Strand–specific RT–PCR for human WT1-AS. Fetal kidney cDNA was synthesized using primers specific for WT1 antisense RNA (WT1AScsyn) or WT1 sense RNA (WT1csyn) and then amplified using the following primer pairs: (I) Primers across the AS1/8 splice (WITKBF1 and WITKBR1), (II) Primers within WT1 exon 1 (WT15 and WT8), (III) Primers spanning WT1 exon 1 and intron 1 (WT15 and WTEX1AS). (+, –) Reactions with and without reverse transcriptase, respectively. Specific signal was seen for cDNA made in the antisense (AS) direction in all cases, but only in exon 1 for sense cDNA (S). (C) Expression of WT1-AS in human tissues and Wilms’ tumor. Real-time RT–PCR of cDNA from human term placenta (P), fetal spleen (S), fetal liver (L), fetal brain (B), and fetal kidney (K), kidney adjacent to Wilms’ tumors 28 and 53 (K28 and K53) and Wilms’ tumors (T28, T53, T43, T57, and T62), with primers specific for WT1 (WTRQF and WTRQR) and WT1-AS spliceoforms AS1/AS8 (WT1-ASRQF and WT1-ASRQR), AS9 (AS9x2rnaF and AS9x3rnaR), AI648530 (AI648530rnaF and AI648530rnaR). Results were normalized relative to the housekeeping gene TBP (TBPRQF and TBPRQR). Error bars show the range of duplicate measurements; results are representative of two separate experiments. WT1-AS expression paralleled WT1 in all tissues except term placenta, where WT1 was expressed at the lowest detectable level. Primer sequences are shown in Table 1.
The expression of WT1-AS was investigated in human normal and tumor tissues by real-time RT–PCR. We used primer pairs that could selectively amplify three representative spliceoforms (AS1/8, AS9, and AI648530) out of the complex range of differentially spliced WT1-AS RNAs. WT1-AS spliceoforms were expressed in all normal tissues that expressed detectable WT1 (fetal spleen, fetal kidney, and normal kidney), except term placenta, where WT1 was expressed at the lowest detectable level (Fig. 2C). In fetal liver and fetal brain, which did not express WT1, no antisense transcription was detectable (Fig. 2C). Of the three WT1-AS spliceoforms, AI648530 showed the highest levels of expression, comprising 52%–100% of detectable WT1-AS RNA (Fig. 2C). These results reflect the previously reported expression pattern of WT1 during development (Huang et al. 1990; Pritchard Jones et al. 1990; Pelletier et al. 1991). In most Wilms’ tumors, all WT1-AS spliceoforms were overexpressed relative to normal kidney and at comparable levels to fetal kidney. The exception was tumor T53, which was a stromal-rich tumor that, as expected, expressed low levels of WT1 and correspondingly low levels of WT1-AS (Fig. 2C).
Expression of mouse Wt1-as RNA was shown to be exclusively in the antisense orientation in mouse fetal kidney, relative to Wt1 (Fig. 3A, I). Further RT–PCR analysis across the exon1–intron 1 boundary and within intron 1 (Fig. 3A, II and III), demonstrated antisense-specific transcription in those regions too. It therefore appears that as in human (Malik et al. 1995), mouse Wt1-as RNAs originate within intron 1 and probably span exon 1.
FIGURE 3.
Expression of mouse Wt1-as. (A) Mapping expression of mouse Wt1-as. Strand-specific RT–PCR for mouse Wt1-as using fetal kidney cDNA synthesized using: (I) Primers in the first 2 exons of mouse Wt1-as (552314S and 552314AS); (II) Primers spanning Wt1 exon 1 and intron 1 (WT1 and MWT1); (III) Primers within Wt1 intron 1 (MWT3A and MWT4A). In each case, specific signal was only seen for cDNA made in the antisense (AS) direction and not the sense (S). (+, –) Reactions with and without reverse transcriptase, respectively. (B) Expression of Wt1-as in mouse tissues. Real-time RT–PCR expression analysis using cDNA from mouse fetal tissues (E17.5 d); placenta (P), spleen (S), brain (B), liver (L), and kidney (K), and postnatal (P3, 7, 22, and 65 d) kidney (K), with primers specific for Wt1-as (MASPLICE1RQF and MASPLICE1RQR) and Wt1 (MWTRQF2 and MWTRQR2), normalized relative to the housekeeping gene Tbp (MTBPRQF and MTBPRQR). Error bars show the range of duplicate measurements; results are representative of two separate experiments. Wt1-as was coexpressed with Wt1 in fetal placenta, spleen, and kidney. Primer sequences are shown in Table 1.
The three known murine-spliced Wt1-as RNAs could be amplified simultaneously using a single pair of primers in a real-time RT–PCR assay, because they all shared a common first splice (Fig. 1B). Using this assay it was found that in mouse fetal tissues, Wt1-as expression paralleled that of Wt1, with highest expression in the developing kidney, similar to the results in human tissues (Fig. 3B).
These results show that there are multiple spliceoforms of WT1-AS, which are coexpressed with WT1 sense mRNA in human and mouse tissues (Pearson correlation coefficient, r = 0.99 for mouse fetal tissues and r = 0.88 for human fetal tissues).
Epigenetic regulation of WT1-AS
We have previously shown that WT1-AS spliceoform AS1/8 is imprinted in normal kidney and that expression correlates with allelic methylation of the antisense regulatory region (ARR) (Malik et al. 2000). To determine whether imprinting included other WT1-AS spliceoforms, we were able to assess the allelic expression of AS9, which contained a previously described DdeI polymorphism located in its first exon (Hancock et al. 2007). RT–PCR using AS9-specific primers showed monoallelic expression in normal kidney, but expression from both alleles in the corresponding Wilms’ tumor (Fig. 4A, left). Analysis of ARR methylation showed differential methylation in normal kidney, with complete hypomethylation in the corresponding Wilms’ tumor (Fig. 4A, right), demonstrating that DNA methylation of the ARR correlates with AS9 allelic expression, exactly as described previously for AS1/8 spliceoforms (Malik et al. 2000). Furthermore, 5-azacytidine treatment of the Wilms’ tumor cell line (17.94), which normally has undetectable levels of WT1-AS expression, was able to induce expression of AS1/8 and AS9 (Fig. 4B, left), with simultaneous reduction in methylation of the ARR (Fig. 4B, right). Taken together, these findings provide compelling evidence that methylation of the ARR plays a critical role in the epigenetic regulation of WT1-AS transcription.
FIGURE 4.
Epigenetic regulation of WT1-AS. (A) Allelic expression and methylation of WT1-AS in Wilms’ tumor. (Left) Allelic expression of the WT1-AS AS9 transcript (AS9-FOR and AS9-REV) in paired normal kidney (NK) and Wilms’ tumor (WT), using RT–PCR across a DdeI restriction site polymorphism. DdeI digestion gives two allelic bands (A1 and A2) and one constant band (arrowed). (+, –) Reactions with and without reverse transcriptase, respectively. DNA lane shows the alleles amplified from corresponding genomic DNA. WT1-AS AS9 was monoallelically expressed in NK and biallelically expressed in WT. (Right) Methylation of the ARR in the NK and WT assayed by methylation-sensitive Southern blotting of corresponding genomic DNAs, showing differential allelic methylation in NK (731 and 542 bp bands) and complete hypomethylation in WT (542 bp band only). (B) Azacytidine—mediated modulation of transcription at the human WT1 locus. (Left) Agarose gel (HPRT; HPRT5′ and HPRT3′) or autoradiograph (WT1-AS spliceoforms AS1/8; WITKBF1 and WITKBR1 and AS9; AS9-FOR and AS9-REV) of RT–PCR reactions using total RNA extracted from the Wilms’ tumor cell line 17.94, treated with (+AZA) or without (−AZA) 5- azacytidine for 4 d. (+, –) Reactions with and without reverse transcriptase, respectively. AZA treatment induced expression of both WT1-AS spliceoforms. (Right) Methylation-sensitive Southern blot of corresponding 17.94 cell genomic DNAs showed partial demethylation at the Antisense Regulatory Region (ARR) in WT1 intron 1 after 4 d of AZA treatment, as demonstrated by the appearance of unmethyated bands at 731 and 542 bp. (C) Wt1 (MWTRQF2 and MWTRQR2) and Wt1-as (MASPLICE1RQF and MASPLICE1RQR) expression in Pax6 Sey-H mice. RNA expression was assayed by real-time PCR of normal kidney cDNA from wild-type mice (WT) and mice heterozygous for the Pax6 Sey-H deletion (which includes Wt1), inherited from father (Pat KO) or mother (Mat KO). Expression levels are shown as fold differences compared with wild-type controls, normalized relative to the housekeeping gene Tbp (MTBPRQF and MTBPRQR). Each mouse kidney cDNA was assayed twice and the results show the mean expression values +/− SEM for five WT, five Pat KO, and two Mat KO mice. Wt1-as expression was maintained in both Pat and Mat KO mice, indicating no imprinting of Wt1-as in mice. Primer sequences are shown in Table 1.
To investigate whether antisense imprinting is conserved in mouse, we examined Wt1-as RNA expression by real-time RT–PCR in kidney tissue from small eye (Pax6 Sey-H) mice, which have a deletion that includes Wt1 (Kent et al. 1997). Human WT1-AS is only expressed from the paternal allele in normal kidney (Malik et al. 2000; Dallosso et al. 2004); thus, if Wt1-as was imprinted similarly to human WT1-AS, then it would be predicted that heterozygote mice inheriting the Pax6 Sey-H deletion from father (Pat KO) would show absent or vastly reduced Wt1-as expression compared with wild-type mice. In fact, neither set of heterozygotes (Pat KO or Mat KO) showed significantly reduced expression of Wt1 or Wt1-as relative to wild-type mice, excluding both paternal and maternal imprinting (Fig. 4C).
WT1-AS transcripts are transported into the cytoplasm and form RNA:RNA duplexes
Our previous studies have shown colocalization of sense and antisense WT1 transcripts in kidney tissue (Moorwood et al. 1998). However, that study did not determine the subcellular location of WT1-AS, which is important, because while protein-coding mRNAs are exported into the cytoplasm for protein synthesis, noncoding RNAs may be retained in the nucleus, where they can have transcriptional regulatory functions, e.g., Xist (Kelley and Kuroda 2000) and Air (Seidl et al. 2006), or associate with splicing factors, e.g., TncRNA and MALAT-1 (Hutchinson et al. 2007).
To determine the subcellular localization of the noncoding WT1-AS RNAs, the WT1/WT1-AS-expressing cell line 7.92 (Brightwell et al. 1997) was fractionated into nuclear and cytoplasmic compartments and RNA expression assayed in each fraction by real-time RT–PCR. Successful separation of nucleus and cytoplasm was demonstrated by the presence of higher molecular weight, unprocessed ribosomal RNA in only the nuclear fraction, and by the localization of unspliced WT1 pre-mRNA predominantly in the nuclear fraction (Fig. 5A). Additionally, noncoding RNAs previously shown to be localized in the nucleus (TncRNA and MALAT-1) (Hutchinson et al. 2007) were almost completely absent from the cytoplasmic fraction (<2%) (Fig. 5A). As expected, for four protein-coding mRNAs (TBP, HPRT, PCDHGA3, and WT1) (Fig. 5A), the cytoplasmic fraction contained the vast majority (91%–98%) of RT–PCR product. The noncoding RNA H19, which is predominantly cytoplasmic (Brannan et al. 1990), had a similar distribution to these mRNAs (89% cytoplasmic) (Fig. 5A). Three spliceoforms of WT1-AS RNA were also found predominantly in the cytoplasm (87%–97%) (Fig. 5A), indicating that they too are exported from the nucleus.
FIGURE 5.
Subcellular localization of WT1-AS transcripts and RNA duplex analysis. (A) Subcellular localization. The 7.92 cells were fractionated into cytoplasmic (C) and nuclear (N) extracts as described in Materials and Methods. (Left) Agarose gel of total RNA, demonstrating unprocessed pre-rRNA in the nuclear fraction. (Right) Table showing distribution of WT1 antisense RNAs, protein-coding RNAs, and other noncoding RNAs in the cytoplasmic fraction, as assessed by real-time RT–PCR. Results show that WT1-AS is transported into the cytoplasm. (B) RNA duplex analysis. Total RNA was made from 7.92 cells using nondenaturing conditions, to extract intact RNA:RNA duplexes. Panels I to V show agarose gel electrophoresis of RT–PCR products from the WT1 locus made from RNA incubated in the presence (lanes 2,3) or absence (lane 1) of RNase A, followed by first-strand cDNA synthesis in the presence (lanes 1,3) or absence (lane 2) of reverse transcriptase. I: Upstream (WITKBR1 + WITKBF2), II: Exon 1 (WT14 + WT16), III: Exon 1a (CPG-USTR + CPG-AS), IV: Exons 6–10 (WT2 + WT4), V: Exon 10 UTR (WT6 +WT7). Above is a schematic of WT1, WT1-AS, and AWT1 transcripts with the positions of the primers shown as arrowheads. Amplification products are only visible in lane 3 if duplex formation spanning the primer pair has “protected” the amplicon from RNase A digestion. The only protected product is in panel II, suggesting RNA:RNA duplex formation within the WT1 exon 1 region. Primer sequences are shown in Table 1.
To test for direct interaction between sense and antisense RNAs in the cytoplasm, RNase protection RT–PCR experiments were performed using cytoplasmic RNA extracted from 7.92 cells (Fig. 5B). In these analyses, RNA was RNase A treated prior to RT–PCR, so that only RNA:RNA duplexed regions would give rise to RT–PCR products, because they would be refractory to RNase A digestion, whereas single-stranded RNA sequences would be digested. Specific protection from RNase A digestion occurred only within WT1 exon 1 (Fig. 5B, II) and not in regions where sense and antisense transcription do not overlap (Fig. 5B, I and III–V). This suggests duplexing of sense and antisense RNAs across WT1 exon 1. Therefore, WT1 sense and antisense RNAs may interact directly in vivo as part of a physiological regulatory role for WT1-AS.
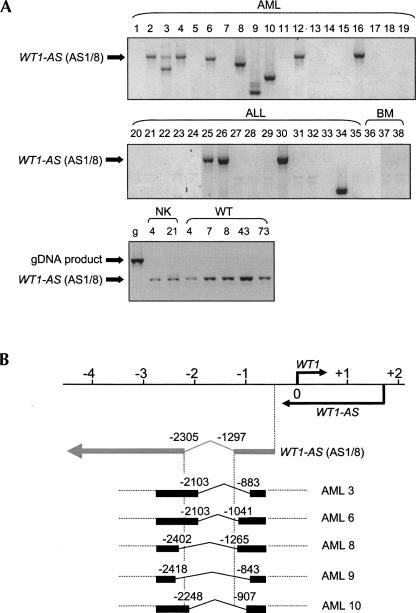
Aberrant WT1-AS splicing in acute myeloid leukaemia
Our analyses of WT1-AS function suggest that qualitative aberrations of WT1-AS could contribute to the development of malignancies, as well as the quantitative changes caused by epigenetic defects that we have previously characterized (Malik et al. 2000; Hancock et al. 2007). We therefore examined WT1-AS expression in the two main cancers previously associated with WT1 defects, namely Wilms’ tumor and acute myeloid leukaemia (AML), using RT–PCR primers flanking the AS1/8 intron. WT1-AS expression was not detectable in normal bone marrow, but was in nine of 19 (47%) AMLs and four of 16 (25%) acute lymphocytic leukaemia (ALL) samples (Fig. 6A). Interestingly, we observed aberrantly spliced WT1-AS products in six of nine (67%) AML samples that expressed WT1-AS and in one of four (25%) WT1-AS-expressing ALLs (Fig. 6A). In several cases there was more than one spliceoform identified in the leukemia patient samples (e.g., Fig. 6A, lanes 3,9). In contrast, examination of normal human kidney and Wilms’ tumors revealed no abnormal WT1-AS RT–PCR products (Fig. 6A; data not shown). Cloning and sequencing of the aberrant AML RT–PCR products demonstrated that they arose by abnormal splicing events at different positions compared with the AS1/8 splice sites (Fig. 6B), with almost all of the splice junctions lacking the normal 5′GT and 3′AG intron splice motifs (data not shown). Similar aberrant splicing was also observed in the hematopoietic cell lines CCRF–CEM (ALL), HuT78 (T-cell lymphoma), and HL-60 (promyelocytic leukemia) (data not shown). Sequencing of genomic DNA PCR products across this region did not reveal any alterations in the samples that exhibited aberrant splicing (data not shown).
FIGURE 6.
Expression of aberrant WT1-AS spliceoforms in human leukaemia. (A) Agarose gel of RT–PCR for WT1-AS expression in samples from acute myeloid leukaemia (AML; 1–19), acute lymphoblastic leukaemia (ALL; 20–35), normal bone marrow (BM; 36–38), normal kidney (NK; 4 and 21), and Wilms’ tumor (WT; 4, 7, 8, 43, 73). Nested RT–PCR was performed with primers WITKBR1 and WITKBF3A, followed by WT18 and WITKBF3B, which produced the expected 793-bp product in most tissues for AS1/8 (arrowed), but demonstrated abnormal smaller products in AMLs 3, 6, 8, 9, 10, and ALL 34. No abnormal-sized products were detected in Wilms’ tumors. The larger sized PCR product from genomic DNA (g; first lane, bottom) shows that all cDNA products derived from spliced WT1-AS RNAs. Primer sequences are shown in Table 1. (B) The structure of the normal AS1/8 transcripts are shown relative to the WT1 transcriptional start site, and underneath the structures of the new spliceoforms isolated from AMLs 3, 6, 8, 9, and 10, with the product sizes and positions of their splices indicated. None of the splice junctions coincided with those of the major WT1-AS RNAs found in normal tissues.
All of the leukemias studied here had previously been screened for WT1 mutations (King-Underwood et al. 1996, 1998). Of the WT1-AS- expressing leukemias, three of the seven (43%) with abnormal splicing had WT1 mutations, whereas zero of six with the expected splicing had WT1 mutations (P = 0.12, Fisher's exact test). Of the leukemias that demonstrated aberrant WT1-AS splicing, the ALL sample was biphenotypic and the AML samples were predominantly of the M3 subtype (one M2, four M3, and one M5). This suggests that aberrant WT1-AS splicing could be associated with specific subtypes of leukemia and possibly with WT1 mutation, although this needs to be tested in a larger series.
These results demonstrate that WT1-AS is affected by abnormal splicing in malignancy in addition to the epigenetic deregulation that we have previously identified in Wilms’ tumor.
DISCUSSION
WT1 mutations have been identified in <20% of Wilms’ tumors (Brown et al. 1993; Little and Wells 1997), which has led to the search for alternative mechanisms that could deregulate WT1 expression in WT and other malignancies. One such possible mechanism involves regulatory RNAs; thus, our detection of a coordinately expressed antisense WT1 transcript with a putative regulatory function (Malik et al. 1995, 2000; Moorwood et al. 1998) prompted this investigation into WT1-AS structure, conservation, and properties.
Structure of the WT1 antisense transcript in human and mouse
In this study we have described a range of divergently transcribed, spliced antisense transcripts, extending into WT1 exon 1 (Fig. 1). Screening of a human fetal kidney cDNA library identified two different classes of spliced cDNA, the first class of which (AS1/AS8-type) has been previously described (Gessler and Bruns 1993; Malik et al. 2000), but the cDNAs described here extend the known 5′ and 3′ termini of this spliceoform substantially. The second class of cDNA (AS9) isolated from fetal kidney represents an entirely new spliceoform lacking the 1-kb intron that is present in AS1 and AS8, being spliced further 3′ across a small (110 bp) and larger intron (7381 bp) before terminating in a 376-bp exon located 10 kbp upstream of the WT1 transcriptional start.
Previous studies have identified exon 1 and intron 1 overlapping RNAs (Campbell et al. 1994; Eccles et al. 1994; Malik et al. 1995), and in this study we present definitive evidence for WT1 antisense transcripts in the form of: (1) our identification of six novel spliced EST clones that showed overlap with the 5′ UTR of WT1 (Fig. 1A), (2) RPA and strand-specific RT–PCR showing antisense transcription in WT1 exon 1 (Fig. 2A,B), and (3) RNase protection RT–PCR experiments demonstrating RNA heteroduplexes within WT1 exon 1 (Fig. 4B). Together, these data provide the first evidence of spliced, WT1-AS RNAs expressed from the opposite strand of WT1 exon 1, and indicates that these spliceoforms could be involved in sense:antisense interactions, like those shown in Figure 4.
The potential physiological importance of WT1-AS is supported by our characterization of three mouse EST clones, which represents the first examples of a spliced mouse Wt1-as RNAs. Strand-specific RT–PCR and 5′ overlap with Wt1 exon 1 demonstrated that these ESTs are examples of true antisense mRNAs and that antisense transcription probably originates from within Wt1 intron 1 (Fig. 3). Comparison of the cDNAs in mouse and humans identified some synteny between their exon–intron architectures, but the mouse Wt1 antisense RNAs did not show sequence conservation in humans, except at their 5′ ends, which extend into Wt1 exon 1. Others have reported several regions of antisense expression located upstream and within exon 1 and intron 1 of Wt1 in mouse tissues (Campbell et al. 1994; Gong et al. 2001), which is in agreement with our results (Fig. 3), indicating the existence of multiple mouse antisense transcripts, similar to the human antisense RNAs.
The complex splicing pattern observed in WT1-AS may indicate different functional characteristics for each spliceoform. The inclusion or exclusion, by alternate splicing, of specific transcribed regions could potentially alter interactions with RNA-binding proteins or affect RNA secondary structure. These factors are thought to be important in the function of noncoding RNAs such as H19, SRA, and the RNA component of telomerase (Lanz et al. 1999; Chen et al. 2000; Juan et al. 2000). Complex alternate splicing of antisense transcripts has been observed in several other genes including IGF2-AS (Vu et al. 2003), Nespas (Williamson et al. 2002), Sphk1 (Imamura et al. 2004), and Foxl2 (Cocquet et al. 2005), along with evidence for tissue-specific expression of certain spliceoforms.
Tissue-specific expression of WT1-AS
The expression of WT1-AS antisense spliceoforms paralleled that of WT1 mRNA levels in both human and mouse tissues (Figs. 2, 3). Previous experiments within our laboratory have identified parallel expression and colocalization of WT1 sense and antisense RNAs in human kidney (Moorwood et al. 1998). Experiments have shown an increase in WT1 mRNA and protein levels in cells overexpressing exogenous antisense sequences, suggesting a regulatory role for WT1-AS (Moorwood et al. 1998). Results from other groups have also demonstrated the existence of coordinately expressed and colocalized WT1 sense and antisense transcripts (Huang et al. 1990; Yeger et al. 1992). WT1-AS is therefore similar to the majority of sense/antisense pairs of mammalian transcripts, which show coordinate regulation, inconsistent with a simplistic negative regulatory role (Katayama et al. 2005).
Epigenetic regulation of WT1-AS expression
WT1-AS expression is thought to be epigenetically regulated by the methylation of negative regulatory elements in intron 1 of WT1 (the antisense regulatory region [ARR]) (Malik et al. 1995, 2000). Previously, we showed that WT1-AS spliceoform AS1/8 was imprinted in normal kidney (Malik et al. 2000) and we have extended this result to AS9 in this study (Fig. 4A, left). We have also shown that the monoallelic expression of AS9 correlates with differential methylation of the ARR and that loss of methylation at the ARR leads to biallelic expression (loss of imprinting) in Wilms’ tumor (Fig. 4A), as for AS1/8 (Malik et al. 2000). Thus, WT1-AS appears to belong to a class of epigenetically regulated noncoding RNAs found in imprinted genes like Air and Kcnq1ot1 (O'Neill 2005). We therefore sought direct evidence for epigenetic regulation of WT1-AS expression by modulating DNA methylation levels in cultured cells. 5-azacytidine (AZA) treatment of the WT cell line 17.94, which normally expresses undetectable levels of WT1-AS RNAs and shows hypermethylation of the ARR, resulted in the induction of WT1-AS expression and the transition from highly methylated to hypomethylated alleles at the ARR (Fig. 4B).
These results demonstrate an association between the methylation state of the ARR and WT1-AS expression, consistent with the pattern of epigenetic deregulation of WT1-AS previously found in tumors (Malik et al. 2000). Interestingly, inactivation of WT1 and WT1-AS expression, coupled with promoter hypermethylation, has recently been reported in ovarian clear-cell adenocarcinomas, further strengthening this link (Kaneuchi et al. 2005). In addition, we have demonstrated that the ARR acts as methylation-dependent silencer on the WT1-AS promoter (Hancock et al. 2007), providing a mechanistic explanation for these observations.
Interestingly, we found no evidence for imprinting of Wt1-as in mouse, implying that the epigenetic regulation of the Wt1 locus is different in mice and humans. There are quite a large number of genes that show discordant imprinting between human and mouse (Morison et al. 2005), although in most cases this involves lack of imprinting of the human gene compared with the mouse homolog, with only DLX5 reported to be imprinted in human but not in mouse (Kimura et al. 2004). Using a genome-wide statistical model, it was recently predicted that mouse Wt1 and a few nearby genes such as Rcn might be imprinted (Luedi et al. 2005). Our results did not demonstrate any conclusive evidence for imprinting of either Wt1 or Wt1-as in mice, although it should be stressed that this was limited to known spliceoforms in kidney tissue. However, our data are consistent with the phenotype of mice carrying Pax6 Sey-H deletions, where no parent-of-origin effects have been observed in heterozygous mice, suggesting an absence of imprinting at Wt1 and surrounding genes contained within the deletion (J. Peters, unpubl.).
Subcellular distribution and RNA:RNA interactions of WT1-AS
WT1-AS transcripts appear to be untranslated RNAs, suggesting that these RNA species might not be subject to the processing followed by protein-encoding mRNAs. We therefore studied the subcellular localization of these RNAs in WT1-expressing 7.92 cells to determine any variation between their localization compared with that of WT1 mRNA and other protein-coding mRNAs, which might give clues to their cellular functions (Fig. 5A). Predictably, protein-coding genes showed an extreme bias of amplified product in the cytoplasmic fraction, the location of the eukaryotic translational machinery. Real-time RT–PCR carried out using primers specific to WT1-AS showed a similar predominantly cytoplasmic distribution, which argues against a solely nuclear function such as that performed by Xist (Kelley and Kuroda 2000) and Air (Seidl et al. 2006) and proposed for the noncoding RNAs TncRNA and MALAT-1 (Hutchinson et al. 2007) that we showed to be almost entirely nuclear in localization (Fig. 5A).
Given the cellular and subcellular colocalization of WT1 sense and antisense transcripts, we used RNase protection RT–PCR experiments (Fig. 5B) to investigate possible interactions between WT1 sense and antisense RNAs. We demonstrated RNA duplex formation in the overlapping exon 1 region (Fig. 5B), suggesting a WT1 sense:antisense direct interaction that could potentially have a role in RNA stability (Podlowski et al. 2002), modulating WT1 transcript half-life in the cell and ultimately WT1 protein levels. This interaction is consistent with our previous demonstration of spatial colocalization of WT1 mRNA and WT1 protein with WT1-AS RNA in fetal kidney tissue and provides a mechanistic explanation for the up-regulation of WT1 protein levels by exogenous WT1-AS that we observed in an in vitro system (Moorwood et al. 1998).
Function of WT1-AS
The conservation of WT1-AS expression between mouse and man gives support for an important physiological function. The exact structure of the WT1 antisense transcripts is not conserved, similar to other noncoding transcripts like NESPAS and H19 (Juan et al. 2000; Williamson et al. 2002), presumably because unlike protein-coding genes, no ORFs need to be conserved. The coexpression of WT1 and WT1-AS in most tissues and parallel expression during kidney development (Figs. 2, 3), similar subcellular localization (Fig. 5A) and the potential to form RNA:RNA duplexes (Fig. 5B) suggests that direct interaction between WT1 antisense and sense transcripts may be necessary for WT1-AS function. This may regulate WT1 protein expression, as we have previously hypothesized (Moorwood et al. 1998). Additionally, it has been reported that WT1 protein can bind to WT1-AS RNA (Ye et al. 1996), suggesting that WT1 antisense transcripts may have multiple regulatory roles involving both RNA:RNA and RNA:protein interactions.
Recently we have described a coding WT1 transcript, AWT1, which originates from a novel first exon in intron 1 and appears to be under common epigenetic regulation with WT1-AS, with both transcripts being imprinted in normal kidney (Malik et al. 2000; Dallosso et al. 2004). In other imprinted genes, noncoding RNAs like Air and Kcnq1ot1 have been shown to be essential for maintaining allele-specific expression (Sleutels et al. 2002; Thakur et al. 2004). However, in the case of Air and Kcnq1ot1, the antisense RNA represses expression of the coding transcript from the same allele, which is clearly not the case in WT1, where WT1-AS and AWT1 are both expressed from the paternal allele. Possibly, WT1-AS could act by maintaining an open chromatin configuration on the expressed allele, as has been suggested to occur with the antisense RNA from the nonimprinted Sphk1 gene (Imamura et al. 2004). Definitive evidence for or against a role for WT1-AS in the control of WT1 expression awaits the results of mouse knockout experiments that are in progress.
Aberrant splicing in AML
Epigenetic deregulation of WT1-AS expression appears to be a common event in Wilms’ tumor (Fig. 4A) (Malik et al. 2000; K.W. Brown, F. Power, B. Moore, and K.T.A. Malik, in prep.) and methylation changes in WT1 intron 1 have also been reported in other cancers (Huang et al. 1997; Kleymenova et al. 1998; Plass et al. 1999; Costello et al. 2000; Kaneuchi et al. 2005). However, we have now found evidence for a possible nonepigenetic deregulation of WT1-AS in malignancy by our discovery of a number of leukemia-specific alternative antisense transcripts, almost exclusively in AML patients (Fig. 6). Few of these new splices followed canonical splicing rules, with most exhibiting a minimal trimer repeat at either end of the intron. Thus, it is assumed that they represent abnormal cancer-associated spliceoforms, possibly caused by defects in the splicing machinery, although, in this case, there was no aberrant WT1 mRNA splicing (data not shown), arguing for a specific WT1-AS defect. Aberrations in alternative splicing have been suggested as contributing factors in the development of various diseases including cancer (Caceres and Kornblihtt 2002; Roy et al. 2005). Such disease-associated alternatively spliced transcripts may be extremely useful as cancer markers, because often there is a greater difference in the usage of alternatively spliced variants between normal and tumor tissue than there is in the overall level of expression of a gene (Caballero et al. 2001). This could be the case for the alternative antisense WT1-AS transcripts in AML patients, which show some evidence of an association with specific leukemia subtypes.
Summary
The experiments described in this study demonstrate a complex range of RNA transcripts at the 5′ end of the WT1 gene in both mice and humans. These RNAs appear to share common regulatory machinery, because antisense RNA levels and their allelic expression are coregulated between spliceoforms in both normal and tumor tissues, and their expression correlates with the methylation status of the ARR. Our results argue for a primarily cytoplasmic function of WT1-AS, possibly via direct interaction with sense transcripts, indicating how a putative regulatory role could be mediated. The functional importance of WT1-AS is further suggested by deregulation in cancer by both epigenetic defects (in Wilms’ tumor) and by abnormal splicing (in AML). Despite the large number of mammalian noncoding RNAs recently identified, few have been ascribed biological functions. This work defines a novel interaction of a noncoding RNA with its sense counterpart, which may have a physiologically important regulatory function in a developmental gene, and demonstrates both qualitative and quantitative defects of antisense RNA expression in cancer.
MATERIALS AND METHODS
Bioinformatics
BLASTN searching of public databases (http://www.ncbi.nlm.nih.gov) was used to detect expressed sequence tags (ESTs) representing cDNAs of interest at the WT1 locus. The source clones for these ESTs were obtained from the IMAGE Consortium via MRC Geneservices (http://www.geneservice.co.uk/home/).
Cell lines and tissues
The 7.92 (Brightwell et al. 1997) and 17.94 human cell lines (K.W. Brown, unpubl.) were derived from a rhabdoid tumor of the kidney and an anaplastic Wilms’ tumor, respectively, using standard methods. Frozen human fetal tissue (15–31 wk gestation), human placenta (term), kidney taken adjacent to Wilms’ tumor, Wilms’ tumor, normal bone marrow, and leukemic marrow were obtained from local hospitals with appropriate ethical approval. Mouse fetal tissue samples were pooled from several E17.5 d fetuses, whereas postnatal kidney samples (P3, 7, 22, and 65 d) came from individual mice. Mouse kidney tissue for the imprinting experiments (Fig. 4C) was taken from 7-d-old heterozygote Pax6 Sey-H/+ or +/Pax6 Sey-H mice and wild-type sibs. All mouse studies were done under the guidance issued by the Medical Research Council in “Responsibility in the Use of Animals for Medical Research” (July 1993) and under the authority of Home Office Project Licence Number 30/1517.
Library screening
A human fetal kidney λ cDNA library (Clontech) was screened using a radiolabeled probe corresponding to the WIT-1 cDNA (Gessler and Bruns 1993), according to the manufacturers’ protocols. Positive clones were identified on duplicate plates and the complete DNA sequence was obtained by automated sequencing (University of Durham). The accession numbers of the sequences of AS1, AS8, and AS9 are DQ289490, DQ289489, and DQ289488, respectively.
Ribonuclease protection assay
RPA was carried out exactly as described in Dallosso et al. (2004).
PCR
Total RNA was purified using TRI-reagent (Sigma) and between 0.5 and 1 μg total RNA was used to synthesize cDNA with MMLV RT (RNase H-) Reverse Transcriptase (Promega) or Thermoscript RT (Invitrogen) in the presence of RNasin Ribonuclease Inhibitor (Promega). First-strand cDNA was first synthesized using the reverse or forward primer for each transcript to confirm the orientation, then subsequent experiments used cDNA synthesized with oligo d(T)15 primer (Promega). One-twentieth of this cDNA was used per PCR reaction. PCR primer sequences are listed in Table 1.
TABLE 1.
PCR primers
Subcellular fractionation
The 7.92 tissue culture cells were lysed in ice-cold cell lysis buffer (14 mM NaCl, 1.5 mM MgCl2, 10 mM Tris-HCl at pH8.6, 0.5% Nonidet P40) and centrifuged through a 24% (w/v) sucrose cushion (in cell lysis buffer) to separate nuclear and cytoplasmic compartments. Total RNA was purified from each fraction by phenol-chloroform extraction and ethanol precipitation before DNase I treatment and synthesis of cDNA for RT–PCR amplification. Approximately 10 times more total RNA was isolated from the cytoplasmic compared with the nuclear fraction, but cDNA synthesis was carried out using equivalent amounts of total RNA so as not to vary the efficiency of cDNA synthesis in either reaction.
RNase protection RT–PCR
These experiments were adapted from a previously described method (Krystal et al. 1990; Podlowski et al. 2002). Briefly, cytoplasmic RNA was isolated from the 7.92 rhabdoid tumor cell line under nondenaturing conditions, subjected to RNase A digestion (to digest single-stranded RNA), and then cDNA was synthesized using random hexamers for subsequent PCR.
Demethylation of cultured cells
Cells were grown in DMEM complete medium supplemented with 10% fetal bovine serum, with the addition of 1 μM 5-azacytidine (AZA) for 4 d. Stock AZA was made in PBS and mock-treated cells were treated with an equivalent volume of PBS.
Methylation-sensitive Southern blotting
Southern analysis of genomic DNAs was carried out as previously described (Malik et al. 2000).
Real-time RT–PCR
Comparative quantitation of RNA levels real-time PCR was performed using SYBR green technology on an Mx3000P detection system (Stratagene), using primers listed in Table 1. The 20 μL reactions were performed using Platinum SYBR Green qPCR supermix-UDG (InVitrogen), each containing cDNA from 25 ng RNA, 200 nM forward and reverse primers and 50 nM ROX reference dye. The cycle program consisted of an initial 50°C step for 2 min, followed by 95°C for 2 min, then 45 cycles of 95°C for 15 sec, 60°C for 30 sec, and 72°C for 30 sec.
ACKNOWLEDGMENTS
This work was supported by the CLIC Sargent Charity, the Wellcome Trust, Cancer Research UK, and an MRC studentship to A.S.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.562907.
REFERENCES
- Brannan, C.I., Dees, E.C., Ingram, R.S., Tilghman, S.M. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brightwell, G., Poirier, V., Cole, E., Ivins, S., Brown, K.W. Serum-dependent and cell cycle-dependent expression from a cytomegalovirus-based mammalian expression vector. Gene. 1997;194:115–123. doi: 10.1016/s0378-1119(97)00178-9. [DOI] [PubMed] [Google Scholar]
- Brown, K.W., Watson, J.E., Poirier, V., Mott, M.G., Berry, P.J., Maitland, N.J. Inactivation of the remaining allele of the WT1 gene in a Wilms’ tumour from a WAGR patient. Oncogene. 1992;7:763–768. [PubMed] [Google Scholar]
- Brown, K.W., Wilmore, H.P., Watson, J.E., Mott, M.G., Berry, P.J., Maitland, N.J. Low frequency of mutations in the WT1 coding region in Wilms’ tumor. Genes Chromosomes Cancer. 1993;8:74–79. doi: 10.1002/gcc.2870080203. [DOI] [PubMed] [Google Scholar]
- Caballero, O.L., Souza, S.J., Brentani, R.R., Simpson, A.J. Alternative spliced transcripts as cancer markers. Dis. Markers. 2001;17:67–75. doi: 10.1155/2001/184856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres, J.F., Kornblihtt, A.R. Alternative splicing: Multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18:186–193. doi: 10.1016/s0168-9525(01)02626-9. [DOI] [PubMed] [Google Scholar]
- Campbell, C.E., Huang, A., Gurney, A.L., Kessler, P.M., Hewitt, J.A., Williams, B.R.G. Antisense transcripts and protein binding motifs within the Wilms tumour (WT1) locus. Oncogene. 1994;9:583–595. [PubMed] [Google Scholar]
- Chen, J., Sun, M., Kent, W.J., Huang, X., Xie, H., Wang, W., Zhou, G., Shi, R.Z., Rowley, J.D. Over 20% of human transcripts might form sense–antisense pairs. Nucleic Acids Res. 2004;32:4812–4820. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Sun, M., Hurst, L.D., Carmichael, G.G., Rowley, J.D. Genome-wide analysis of coordinate expression and evolution of human cis-encoded sense–antisense transcripts. Trends Genet. 2005;21:326–329. doi: 10.1016/j.tig.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Chen, J.L., Blasco, M.A., Greider, C.W. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- Cocquet, J., Pannetier, M., Fellous, M., Veitia, R.A. Sense and antisense Foxl2 transcripts in mouse. Genomics. 2005;85:531–541. doi: 10.1016/j.ygeno.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Costello, J.F., Fruhwald, M.C., Smiraglia, D.J., Rush, L.J., Robertson, G.P., Gao, X., Wright, F.A., Feramisco, J.D., Peltomaki, P., Lang, J.C., et al. Aberrant CpG-island methylation has nonrandom and tumour-type-specific patterns. Nat. Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- Dallosso, A.R., Hancock, A.L., Brown, K.W., Williams, A.C., Jackson, S., Malik, K. Genomic imprinting at the WT1 gene involves a novel coding transcript (AWT1) that shows deregulation in Wilms’ tumours. Hum. Mol. Genet. 2004;13:405–415. doi: 10.1093/hmg/ddh038. [DOI] [PubMed] [Google Scholar]
- Eccles, M.R., Grubb, G., Ogawa, O., Szeto, J., Reeve, A.E. Cloning of novel Wilms’ tumor gene (WT1) cDNAs: Evidence for antisense transcription of WT1. Oncogene. 1994;9:2059–2063. [PubMed] [Google Scholar]
- Fraizer, G.C., Wu, Y.J., Hewitt, S.M., Maity, T., Ton, C.C.T., Huff, V., Saunders, G.F. Transcriptional regulation of the human Wilms’ tumor gene (WT1). Cell type-specific enhancer and promiscuous promoter. J. Biol. Chem. 1994;269:8892–8900. [PubMed] [Google Scholar]
- Gessler, M., Bruns, G.A.P. Sequence of the WT1 upstream region including the Wit-1 gene. Genomics. 1993;17:499–501. doi: 10.1006/geno.1993.1355. [DOI] [PubMed] [Google Scholar]
- Gong, Y., Eggert, H., Englert, C. The murine Wilms’ tumor suppressor gene (wt1) locus. Gene. 2001;279:119–126. doi: 10.1016/s0378-1119(01)00757-0. [DOI] [PubMed] [Google Scholar]
- Hancock, A.L., Brown, K.W., Moorwood, K., Moon, H., Holmgren, C., Mardikar, S.H., Dallosso, A.R., Klenova, E., Loukinov, D., Ohlsson, R., et al. A CTCF-binding silencer regulates the imprinted genes AWT1 and WT1-AS, and exhibits sequential epigenetic defects during Wilms’ tumourigenesis. Hum. Mol. Genet. 2007;16:344–354. doi: 10.1093/hmg/ddl478. [DOI] [PubMed] [Google Scholar]
- Hofmann, W., Royer, H.D., Drechsler, M., Schneider, S., Royerpokora, B. Characterization of the transcriptional regulatory region of the human WT1 gene. Oncogene. 1993;8:3123–3132. [PubMed] [Google Scholar]
- Hohenstein, P., Hastie, N.D. The many facets of the Wilms’ tumour gene, WT1. Hum. Mol. Genet. 2006;15:R196–R201. doi: 10.1093/hmg/ddi196. [DOI] [PubMed] [Google Scholar]
- Huang, A., Campbell, C.E., Bonetta, L., McAndrews Hill, M.S., Chiltonm MacNeill, S., Coppes, M.J., Law, D.J., Feinberg, A.P., Yeger, H., Williams, B.R. Tissue, developmental, and tumor-specific expression of divergent transcripts in Wilms’ tumor. Science. 1990;250:991–994. doi: 10.1126/science.2173145. [DOI] [PubMed] [Google Scholar]
- Huang, T.H.-M., Laux, D.E., Hamlin, B.C., Tran, P., Tran, H., Lubahn, D.B. Identification of DNA methylation markers for human breast carcinomas using the methylation-sensitive restriction fingerprinting technique. Cancer Res. 1997;57:1030–1034. [PubMed] [Google Scholar]
- Hutchinson, J.N., Ensminger, A.W., Clemson, C.M., Lynch, C.R., Lawrence, J.B., Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura, T., Yamamoto, S., Ohgane, J., Hattori, N., Tanaka, S., Shiota, K. Non-coding RNA directed DNA demethylation of Sphk1 CpG island. Biochem. Biophys. Res. Commun. 2004;322:593–600. doi: 10.1016/j.bbrc.2004.07.159. [DOI] [PubMed] [Google Scholar]
- Juan, V., Crain, C., Wilson, C. Evidence for evolutionarily conserved secondary structure in the H19 tumor suppressor RNA. Nucleic Acids Res. 2000;28:1221–1227. doi: 10.1093/nar/28.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneuchi, M., Sasaki, M., Tanaka, Y., Shiina, H., Yamada, H., Yamamoto, R., Sakuragi, N., Enokida, H., Verma, M., Dahiya, R. WT1 and WT1-AS genes are inactivated by promoter methylation in ovarian clear cell adenocarcinoma. Cancer. 2005;104:1924–1930. doi: 10.1002/cncr.21397. [DOI] [PubMed] [Google Scholar]
- Katayama, S., Tomaru, Y., Kasukawa, T., Waki, K., Nakanishi, M., Nakamura, M., Nishida, H., Yap, C.C., Suzuki, M., Kawai, J., et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Keirsebilck, A., Bonne, S., Staes, K., van Hengel, J., Nollet, F., Reynolds, A., van Roy, F. Molecular cloning of the human p120ctn catenin gene (CTNND1): Expression of multiple alternatively spliced isoforms. Genomics. 1998;50:129–146. doi: 10.1006/geno.1998.5325. [DOI] [PubMed] [Google Scholar]
- Kelley, R.L., Kuroda, M.I. Noncoding RNA genes in dosage compensation and imprinting. Cell. 2000;103:9–12. doi: 10.1016/s0092-8674(00)00099-4. [DOI] [PubMed] [Google Scholar]
- Kent, J., Lee, M., Schedl, A., Boyle, S., Fantes, J., Powell, M., Rushmere, N., Abbott, C., Van, H.V., Bickmore, W.A. The reticulocalbin gene maps to the WAGR region in human and to the Small eye Harwell deletion in mouse. Genomics. 1997;42:260–267. doi: 10.1006/geno.1997.4706. [DOI] [PubMed] [Google Scholar]
- Kimura, M.I., Kazuki, Y., Kashiwagi, A., Kai, Y., Abe, S., Barbieri, O., Levi, G., Oshimura, M. Dlx5, the mouse homologue of the human-imprinted DLX5 gene, is biallelically expressed in the mouse brain. J. Hum. Genet. 2004;49:273–277. doi: 10.1007/s10038-004-0139-2. [DOI] [PubMed] [Google Scholar]
- King-Underwood, L., Renshaw, J., Pritchardjones, K. Mutations in the Wilms’ tumor gene WT1 in leukemias. Blood. 1996;87:2171–2179. [PubMed] [Google Scholar]
- King-Underwood, L., Pritchard-Jones, K. Wilms’ tumor (WT1) gene mutations occur mainly in acute myeloid leukemia and may confer drug resistance. Blood. 1998;91:2961–2968. [PubMed] [Google Scholar]
- Kleymenova, E.V., Yuan, X., LaBate, M.E., Walker, C.L. Identification of a tumor-specific methylation site in the Wilms’ tumor suppressor gene. Oncogene. 1998;16:713–720. doi: 10.1038/sj.onc.1201583. [DOI] [PubMed] [Google Scholar]
- Krystal, G.W., Armstrong, B.C., Battey, J.F. N-myc mRNA forms an RNA–RNA duplex with endogenous antisense transcripts. Mol. Cell. Biol. 1990;10:4180–4191. doi: 10.1128/mcb.10.8.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz, R.B., McKenna, N.J., Onate, S.A., Albrecht, U., Wong, J., Tsai, S.Y., Tsai, M.J., O'Malley, B.W. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- Lehner, B., Williams, G., Campbell, R.D., Sanderson, C.M. Antisense transcripts in the human genome. Trends Genet. 2002;18:63–65. doi: 10.1016/s0168-9525(02)02598-2. [DOI] [PubMed] [Google Scholar]
- Little, M., Wells, C. A clinical overview of WT1 gene mutations. Hum. Mutat. 1997;9:209–225. doi: 10.1002/(SICI)1098-1004(1997)9:3<209::AID-HUMU2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Little, M.H., Dunn, R., Byrne, J.A., Seawright, A., Smith, P.J., Pritchard-Jones, K., Vanheyningen, V., Hastie, N.D. Equivalent expression of paternally and maternally inherited WT1 alleles in normal fetal tissue and Wilms’ tumours. Oncogene. 1992;7:635–641. [PubMed] [Google Scholar]
- Luedi, P.P., Hartemink, A.J., Jirtle, R.L. Genome-wide prediction of imprinted murine genes. Genome Res. 2005;15:875–884. doi: 10.1101/gr.3303505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, K., Salpekar, A., Hancock, A., Moorwood, K., Jackson, S., Charles, A., Brown, K.W. Identification of differential methylation of the WT1 antisense regulatory region and relaxation of imprinting in Wilms’ tumor. Cancer Res. 2000;60:2356–2360. [PubMed] [Google Scholar]
- Malik, K.T., Wallace, J.I., Ivins, S.M., Brown, K.W. Identification of an antisense WT1 promoter in intron 1: Implications for WT1 gene regulation. Oncogene. 1995;11:1589–1595. [PubMed] [Google Scholar]
- Moorwood, K., Charles, A.K., Salpekar, A., Wallace, J.I., Brown, K.W., Malik, K. Antisense WT1 transcription parallels sense mRNA and protein expression in fetal kidney and can elevate protein levels in vitro. J. Pathol. 1998;185:352–359. doi: 10.1002/(SICI)1096-9896(199808)185:4<352::AID-PATH119>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Morison, I.M., Ramsay, J.P., Spencer, H.G. A census of mammalian imprinting. Trends Genet. 2005;21:457–465. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- O'Neill, M.J. The influence of noncoding RNAs on allele-specific gene expression in mammals. Hum. Mol. Genet. 2005;14(Suppl. 1):R113–R120. doi: 10.1093/hmg/ddi108. [DOI] [PubMed] [Google Scholar]
- Pelletier, J., Schalling, M., Buckler, A.J., Rogers, A., Haber, D.A., Housman, D. Expression of the Wilms’ tumor gene WT1 in the murine urogenital system. Genes & Dev. 1991;5:1345–1356. doi: 10.1101/gad.5.8.1345. [DOI] [PubMed] [Google Scholar]
- Plass, C., Yu, F., Yu, L., Strout, M.P., El Rifai, W., Elonen, E., Knuutila, S., Marcucci, G., Young, D.C., Held, W.A., et al. Restriction landmark genome scanning for aberrant methylation in primary refractory and relapsed acute myeloid leukemia; involvement of the WIT-1 gene. Oncogene. 1999;18:3159–3165. doi: 10.1038/sj.onc.1202651. [DOI] [PubMed] [Google Scholar]
- Podlowski, S., Bramlage, P., Baumann, G., Morano, I., Luther, H.P. Cardiac troponin I sense-antisense RNA duplexes in the myocardium. J. Cell. Biochem. 2002;85:198–207. [PubMed] [Google Scholar]
- Pritchard Jones, K., Fleming, S., Davidson, D., Bickmore, W., Porteous, D., Gosden, C., Bard, J., Buckler, A., Pelletier, J., Housman, D. The candidate Wilms’ tumour gene is involved in genitourinary development. Nature. 1990;346:194–197. doi: 10.1038/346194a0. [DOI] [PubMed] [Google Scholar]
- Roberts, S.G. Transcriptional regulation by WT1 in development. Curr. Opin. Genet. Dev. 2005;15:542–547. doi: 10.1016/j.gde.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Roy, M., Xu, Q., Lee, C. Evidence that public database records for many cancer-associated genes reflect a splice form found in tumors and lack normal splice forms. Nucleic Acids Res. 2005;33:5026–5033. doi: 10.1093/nar/gki792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharnhorst, V., Van der Eb, A.J., Jochemsen, A.G. WT1 proteins: Functions in growth and differentiation. Gene. 2001;273:141–161. doi: 10.1016/s0378-1119(01)00593-5. [DOI] [PubMed] [Google Scholar]
- Seidl, C.I., Stricker, S.H., Barlow, D.P. The imprinted Air ncRNA is an atypical RNAPII transcript that evades splicing and escapes nuclear export. EMBO J. 2006;25:3565–3575. doi: 10.1038/sj.emboj.7601245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels, F., Zwart, R., Barlow, D.P. The noncoding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- Thakur, N., Tiwari, V.K., Thomassin, H., Pandey, R.R., Kanduri, M., Gondor, A., Grange, T., Ohlsson, R., Kanduri, C. An antisense RNA regulates the bidirectional silencing property of the Kcnq1 imprinting control region. Mol. Cell. Biol. 2004;24:7855–7862. doi: 10.1128/MCB.24.18.7855-7862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons, J.A., Good, L. Does everything now make (anti)sense? Biochem. Soc. Trans. 2006;34:1148–1150. doi: 10.1042/BST0341148. [DOI] [PubMed] [Google Scholar]
- Vu, T.H., Chuyen, N.V., Li, T., Hoffman, A.R. Loss of imprinting of IGF2 sense and antisense transcripts in Wilms’ tumor. Cancer Res. 2003;63:1900–1905. [PubMed] [Google Scholar]
- Williamson, C.M., Skinner, J.A., Kelsey, G., Peters, J. Alternative non-coding splice variants of Nespas, an imprinted gene antisense to Nesp in the Gnas imprinting cluster. Mamm. Genome. 2002;13:74–79. doi: 10.1007/s00335-001-2102-2. [DOI] [PubMed] [Google Scholar]
- Ye, Y., Raychaudhuri, B., Gurney, A., Campbell, C.E., Williams, B.R.G. Regulation of WT1 by phosphorylation: Inhibition of DNA binding, alteration of transcriptional activity and cellular translocation. EMBO J. 1996;15:5606–5615. [PMC free article] [PubMed] [Google Scholar]
- Yeger, H., Cullinane, C., Flenniken, A., Chilton-MacNeil, S., Campbell, C., Huang, A., Bonetta, L., Coppes, M.J., Thorner, P., Williams, B.R. Coordinate expression of Wilms’ tumor genes correlates with Wilms’ tumor phenotypes. Cell Growth Differ. 1992;3:855–864. [PubMed] [Google Scholar]
- Yelin, R., Dahary, D., Sorek, R., Levanon, E.Y., Goldstein, O., Shoshan, A., Diber, A., Biton, S., Tamir, Y., Khosravi, R., et al. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]