Abstract
Picornavirus infectivity is dependent on the RNA poly(A) tail, which binds the poly(A) binding protein (PABP). PABP was reported to stimulate viral translation and RNA synthesis. Here, we studied encephalomyocarditis virus (EMCV) and poliovirus (PV) genome expression in Krebs-2 and HeLa cell-free extracts that were drastically depleted of PABP (96%–99%). Although PABP depletion markedly diminished EMCV and PV internal ribosome entry site (IRES)-mediated translation of a polyadenylated luciferase mRNA, it displayed either no (EMCV) or slight (PV) deleterious effect on the translation of the full-length viral RNAs. Moreover, PABP-depleted extracts were fully competent in supporting EMCV and PV RNA replication and virus assembly. In contrast, removing the poly(A) tail from EMCV RNA dramatically reduced RNA synthesis and virus yields in cell-free reactions. The advantage conferred by the poly(A) tail to EMCV synthesis was more pronounced in untreated than in nuclease-treated extract, indicating that endogenous cellular mRNAs compete with the viral RNA for a component(s) of the RNA replication machinery. These results suggest that the poly(A) tail functions in picornavirus replication largely independent of PABP.
Keywords: encephalomyocarditis virus, poliovirus, in vitro translation, RNA replication, poly(A) tail, PABP
INTRODUCTION
The positive-strand RNA genomes of picornaviruses, e.g., those of poliovirus (PV) or encephalomyocarditis virus (EMCV), function in several key processes in the infected cell. Initially, the viral RNA directs translation, using an internal ribosome entry site (IRES)-mediated initiation mechanism. After polyprotein processing, the RNA is copied and amplified by the virus-specific RNA-dependent RNA polymerase (3Dpol) with the assistance of other nonstructural proteins and cellular factors. Finally, during virus assembly, the viral RNA is packaged into a capsid protein shell (Racaniello 2001; Agol 2002).
Despite extensive studies, the mechanisms controlling picornavirus replication are not fully understood. One unresolved issue concerns initiation of negative-strand RNA synthesis at the 3′ end of the viral RNA. This process is executed by a membrane-associated replication complex and requires several conserved sequences and structures in the viral RNA. For PV and possibly other enteroviruses, these elements include the 5′ terminal cloverleaf structure, the 3′ untranslated region (UTR), and the 3′ terminal poly(A) tail (for review, see Paul 2002).
The poly(A) tail plays an important role in PV replication, as its removal dramatically reduces the infectivity of PV RNA (Spector and Baltimore 1974; Sarnow 1989). Efficient EMCV replication and 3Dpol binding to the virus RNA also requires poly(A) tail of sufficient length (Hruby and Roberts 1977; Cui and Porter 1995). HeLa S10 extracts efficiently support the translation and replication of PV RNA and the assembly of infectious virus (Molla et al. 1991). Assays conducted with this system or preinitiation-RNA replication complexes isolated thereof revealed a dramatic reduction in negative-strand RNA synthesis when poly(A) had been shortened to 12 or less nucleotides (Barton et al. 1996; Silvestri et al. 2006). The known function of the poly(A) tail in virus genome replication is to serve as the template for 3Dpol-catalyzed uridylylation of a small viral protein (VPg) that primes the initiation of RNA replication (Paul et al. 1998). Shortening the length of the poly(A) tail would be expected to interfere with this process. Interestingly, in PV RNA, a portion of the 2C coding sequence (a cis-acting replication element, CRE) can also direct the uridylylation of VPg (Goodfellow et al. 2000; Paul et al. 2000). However, VPgpU/VPgpUpU made on the CRE structure apparently primes only plus-strand RNA initiation (Goodfellow et al. 2003; Morasco et al. 2003; Murray and Barton 2003). Interestingly, although picornavirus poly(A) is genetically coded (Dorsch-Hasler et al. 1975), its length is regulated by a special mechanism, of which a portion of the 3′ UTR (oriR) is a key player (van Ooij et al. 2006).
The replication complex distinguishes the viral poly(A) tail from the poly(A) tails of host mRNAs. Thus, a cis-acting element(s) must exist in the plus-strand RNA that confers specificity to virus RNA replication. Possible candidates include the 5′ cloverleaf structure (Barton et al. 2001; Herold and Andino 2001; Teterina et al. 2001) and the oriR (Pilipenko et al. 1996). The 5′ cloverleaf of PV functions in cis to promote both the uridylylation of VPg and the initiation of negative-strand synthesis (Lyons et al. 2001). Because viral negative-strand synthesis requires RNA structures from both the 3′ and 5′ UTRs, the template for viral negative-strand RNA synthesis most likely exists in a circular conformation. Consistent with this idea, studies have implicated the host poly(A) binding protein (PABP) and poly(rC) binding protein 2 (PCBP2) in the formation of the replication competent poliovirus RNA complexes (Herold and Andino 2001). It was postulated that PABP–PCBP2 interaction bridges the 3′ poly(A) and the 5′ cloverleaf of viral RNA. The circular replication complex could be further stabilized by binding of a homodimer of 3CD to both ends of the RNA (Andino et al. 1990, 1993; Harris et al. 1994; Parsley et al. 1997). The potential of EMCV double-stranded replicative form RNA to circularize has been known for a long time (Romanova and Agol 1979). EMCV single-stranded RNA has the potential to adopt a circular topology due to the presence of the 5′ poly(C) tract or another PCBP2 binding site (Chumakov and Agol 1976; Walter et al. 1999) and the 3′ poly(A) (PABP binding site). Genome circularization with the assistance of viral replication proteins and host factors should dislodge translating ribosomes, thereby engendering a switch from translation to negative-strand RNA synthesis (Gamarnik and Andino 1998; Barton et al. 1999; Herold and Andino 2001). Circularization could also serve as a checkpoint to ensure that only intact plus-strand RNAs, i.e., those possessing both the 5′ end and the 3′ end, are subjected to replication.
In eukaryotes, the poly(A) tail is also known to effect mRNA stability and translation (Mangus et al. 2003; Svitkin and Sonenberg 2006). During cap-dependent translation, the 3′ poly(A) tail cooperate with the 5′ cap structure (m7GpppN, where N is any nucleotide) to facilitate ribosome recruitment to mRNA (Jacobson 1996; Sachs 2000; Svitkin and Sonenberg 2006). This is achieved through formation of a ribonucleoprotein complex, containing PABP, the cap-binding protein eIF4E, and the scaffolding protein eIF4G. The combined cooperative interactions within this “closed loop” ribonucleoprotein among other effects enhance the affinity of eIF4E for the 5′ cap and, ultimately, facilitate the formation of ribosome initiation complexes (Kahvejian et al. 2005). Poly(A)-dependent translation is subject to regulation by the PABP-interacting proteins. The PABP-interacting protein 2 (Paip2) inhibits translation by promoting dissociation of PABP from poly(A) and by competing with eIF4G for binding to PABP (Khaleghpour et al. 2001; Karim et al. 2006).
Picornavirus' IRES-mediated translation of reporter mRNAs is also stimulated by poly(A) tail, with the effect being greater for type I (PV-human rhinovirus) than for type II (EMCV) IRESs (Bergamini et al. 2000; Michel et al. 2001; Svitkin et al. 2001). Interaction of eIF4G with the IRES is essential for internal translation initiation (Pestova et al. 2001), and it is conceivable that the PABP/poly(A) complex acts to enhance this interaction. However, although a dual function of the poly(A) tail in both viral translation and RNA replication is conceivable, for reasons that are not immediately clear, polyadenylation exerts a minor if any effect on the translation of the full-length PV mRNA in vitro (Spector et al. 1975; Barton et al. 1996; Silvestri et al. 2006).
In this study, we investigated the role of PABP/poly(A) complex in EMCV and PV replication using in vitro translation/replication systems (Molla et al. 1991; Svitkin and Sonenberg 2003). We assumed that if PABP were essential for viral functions, its depletion would result in the inhibition of these processes, leading to a decrease of virus yield. Surprisingly, the data obtained suggest that PABP is not a critical factor in virus replication. Cell-free syntheses of both EMCV and PV in translation extracts proceeded unabatedly in a virtual absence of PABP. In contrast, results of deadenylated EMCV RNA expression revealed the requirement of the poly(A) tail for EMCV RNA synthesis and virus production.
RESULTS
Effects of poly(A) on cell-free synthesis of EMCV
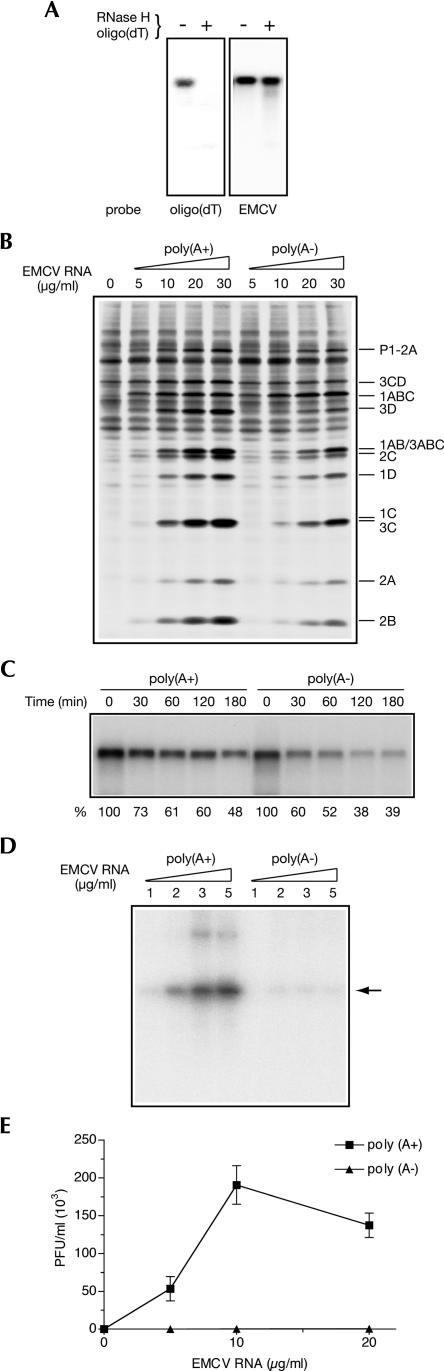
Earlier studies utilizing a PV RNA-programmed HeLa S10 extract or purified RNA replication complexes have demonstrated that negative-strand RNA synthesis is profoundly sensitive to poly(A) tail shortening (Barton et al. 1996; Silvestri et al. 2006). To extend the significance of these findings to other picornaviruses, we investigated the effect of poly(A) tail removal on EMCV synthesis using an in vitro translation–RNA replication system (Svitkin and Sonenberg 2003). The poly(A) tail in mRNAs can be specifically digested with RNase H in the presence of oligo(dT) (Spector et al. 1975). We adopted this strategy to remove poly(A) from EMCV RNA. EMCV RNA was hybridized to 25-fold molar excess of oligo(dT)16 and treated with RNase H. Removal of poly(A) was confirmed by Northern blotting using a 5′ 32P-labeled oligo(dT)16 probe. Consistent with deadenylation, oligo(dT)16 reacted with untreated, but not with RNase H/oligo(dT)-treated EMCV RNA (Fig. 1A). Importantly, ∼90% of poly(A−) EMCV RNA was intact, as revealed by hybridization to a sequence-specific EMCV probe.
FIGURE 1.
Impaired EMCV replication in untreated Krebs-2 S10 extract programmed with the poly(A) tail-deficient EMCV RNA. (A) RNase H/oligo(dT) treatment of EMCV RNA. EMCV RNA that was treated with RNase H/oligo(dT) and blocked at the 3′ end with cordycepin was analyzed along with the intact RNA by denaturing agarose gel electrophoresis and Northern blotting using 32P-labeled oligo(dT) or a fragment of EMCV cDNA (EMCV) as a probe. (B) Comparison of translation efficiencies of poly(A+) and poly(A−) EMCV RNAs. Intact and deadenylated EMCV RNA, at the indicated concentrations, was translated in untreated extracts in the presence of [35S]methionine as described in Materials and Methods. The samples were analyzed by SDS-PAGE and fluorography. The positions of EMCV-specific proteins are indicated on the right. Products of translation of endogenous cellular mRNA are also shown (zero EMCV RNA concentration). (C) Stability of EMCV RNA. Poly(A+) and poly(A−) EMCV RNA was used at a concentration of 10 μg/mL to program untreated extracts. Total RNA was isolated at the indicated times from aliquots of the reaction mixtures. EMCV RNA integrity was analyzed by formaldehyde–agarose gel electrophoresis and Northern blotting as described in Materials and Methods. Relative amounts of recovered mRNAs are indicated at the bottom (values obtained for time 0 were set as 100%). (D) Poly(A+) and poly(A−) EMCV RNA dose responses of RNA synthesis. Poly(A+) and poly(A−) EMCV RNAs were added to the reaction mixtures at the indicated concentrations. The products of RNA synthesis were labeled with [α-32P]CTP and analyzed by agarose gel electrophoresis and autoradiography as described in Materials and Methods. (Arrow) Position of single-stranded EMCV RNA. (E) Poly(A+) and poly(A−) EMCV RNA dose responses of virus yield. Reaction mixtures were programmed with the indicated concentrations of poly(A+) or poly(A−) EMCV RNA for 18 h at 32°C, treated with RNase A/T1, and assayed for infectivity. The data are averages (with standard deviation from the mean) of three independent titer determinations.
The poly(A+) and poly(A−) EMCV RNAs were used at different concentrations to program translation in a Krebs-2 S10 extract. We first used the extract that contains endogenous mRNA to recreate conditions of mRNA competition characteristic to translation in virus-infected cells. In the extract that was not nuclease-treated, a high degree of [35S]methionine incorporation into endogenous cellular proteins was observed (Fig. 1B). However, most viral polypeptides in the lower portion of the gel were well discernible above background. Quantification of the 1C/3C doublet was consistent with a modest (∼50%) inhibition of EMCV protein synthesis resulting from deadenylation. This decrease in the expression of proteins, in particular proteases 3ABCPro and 3CPro, was not sufficient to affect processing of the viral precursor polypeptides, which was normal at a broad (5–30 μg/mL) range of mRNA concentrations (Fig. 1B). A faster decay of poly(A−) as compared with poly(A+) EMCV RNA could contribute to its lower template activity. To examine this possibility, the levels of input RNA in cell-free reactions were measured over time by Northern blotting. This analysis revealed that between 30 and 60 min of incubation, and coordinately with the bulk of viral protein synthesis (Svitkin and Sonenberg 2003), the cell-free reactions exhibited ∼1.2-fold less intact poly(A−) than poly(A+) EMCV RNA (Fig. 1C). Thus, destabilization of the poly(A−) EMCV RNA could be a primary cause for the decline in its template activity.
We next determined the requirement of the poly(A) tail for EMCV RNA replication. For this purpose, the reaction mixtures containing different EMCV RNA concentrations were pulse-labeled with [α-32P]CTP 4 h after the beginning of incubation. The newly synthesized RNA was extracted and analyzed by agarose gel electrophoresis and autoradiography. At all EMCV RNA concentrations, RNA synthesis was dramatically inhibited by removing poly(A) (Fig. 1D). The next step was to examine the importance of EMCV RNA polyadenylation for virus yield. We previously showed that EMCV synthesis is supported by a nuclease-untreated extract, although ∼102-fold less efficiently than by a nuclease-treated extract. While expressing the poly(A+) EMCV RNA had resulted in the generation of the infectious virus particles (up to 2 × 105 PFU/mL virus titer), there was virtually no virus synthesis in the reactions containing poly(A−) EMCV RNA (Fig. 1E). Collectively, these results suggest that EMCV RNA replication and subsequent virus production are much more poly(A)-dependent than EMCV RNA stability and translation.
Expression of EMCV RNA in a PABP-depleted Krebs-2 S10 extract
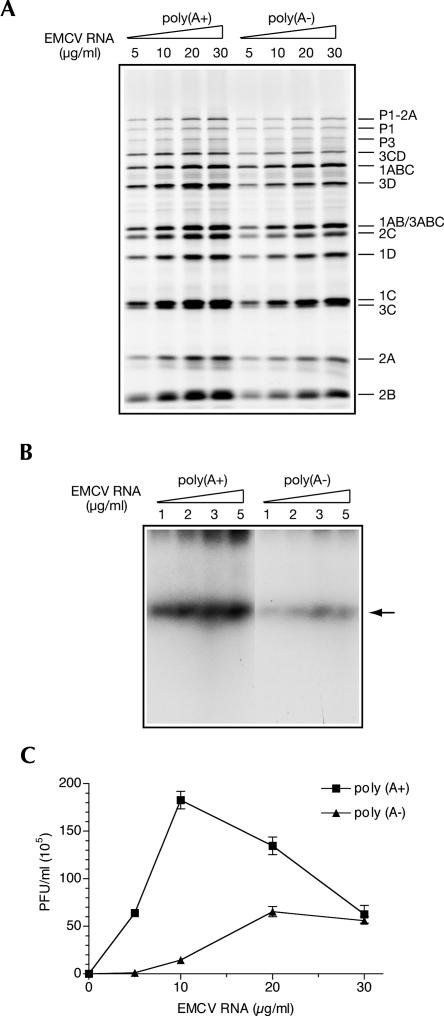
We wished to investigate whether deadenylation inhibits EMCV replication by compromising PABP's function (Herold and Andino 2001). A direct approach to determine the importance of PABP for picornavirus replication in vitro is to deplete this protein from S10 extracts. We previously showed that PABP could be efficiently and specifically removed from nuclease-treated extracts by adsorbing it to GST-Paip2 coupled to glutathione-Sepharose beads (Svitkin and Sonenberg 2004). However, the application of this technique to untreated extracts was not satisfactory, yielding a degree of depletion of <30% (data not shown). For this reason, we evaluated the role of PABP for picornavirus replication in nuclease-treated extracts. Experiments were first conducted to validate the stimulation of EMCV replication by the poly(A) tail in the nuclease-treated Krebs-2 S10 extract. The poly(A+) and poly(A−) EMCV RNAs were programmed into these reaction mixtures at different concentrations, and virus-specific processes were assessed (Fig. 2). Removing poly(A) mildly reduced EMCV RNA translation (by ∼30%), and this reduction did not cause a change in the efficiency of processing of the viral precursor polypeptides (Fig. 2A). We next determined the requirement of the poly(A) tail for EMCV RNA replication. For this, poly(A+) and poly(A−) EMCV RNAs were added to the reaction mixtures at subsaturating concentrations (≤5 μg/mL) (Svitkin and Sonenberg 2003), and [α-32P]CTP incorporation into product RNA was measured between 4 and 5 h after the beginning of incubation. Poly(A+) EMCV RNA replication was significantly (five- to 10-fold) enhanced due to the nuclease treatment of the extract, indicating that cellular mRNAs are competing with viral mRNA for a component(s) of the RNA replication complex (data not shown). Importantly, deadenylation caused a drastic (albeit less potent as compared with that observed in the untreated extract) inhibition of RNA synthesis (Fig. 2B). Virus synthesis directed by the poly(A+) EMCV RNA was similarly inhibited by removing poly(A) (Fig. 2C). Thus, at 5 μg/mL, deadenylated EMCV RNA yielded ∼80-fold fewer plaque forming units (PFU) than intact RNA (Fig. 2C). Saturating the system with input RNA diminished the differences in RNA synthesis and virus yield imparted by poly(A) (Fig. 2C; data not shown). The reduction in the difference between the virus yields from poly(A+) and poly(A−) EMCV RNA at high RNA concentrations appears to be due mainly to a decreased virus production directed by the poly(A+) EMCV RNA. As suggested previously, poly(A+) EMCV RNA excess may sequester a factor that would facilitate RNA replication (Svitkin and Sonenberg 2003). Also, a relatively efficient virus production at high concentrations of poly(A−) EMCV RNA was unexpected. This will be addressed in the Discussion.
FIGURE 2.
Inhibition of EMCV replication by poly(A) tail deficiency in nuclease-treated Krebs-2 S10 extract. (A) Translation efficiencies of poly(A+) and poly(A−) EMCV RNA. The translation of the indicated concentrations of the intact and deadenylated EMCV RNA was carried out as described for Fig. 1B with the exception that nuclease-treated extract was used. Samples were analyzed by SDS-PAGE and fluorography. The positions of EMCV-specific proteins are indicated on the right. Products of translation of endogenous cellular mRNA were undetectable (data not shown). (B) Poly(A+) and poly(A−) EMCV RNA dose responses of RNA synthesis. The indicated concentrations of poly(A+) and poly(A−) EMCV RNA were added to the reaction mixtures containing nuclease-treated extract. The products of RNA synthesis were labeled with [α-32P]CTP and analyzed as described for Fig. 1D. (Arrow) Position of single-stranded EMCV RNA. (C) Poly(A+) and poly(A−) EMCV RNA dose responses of virus yield. Reaction mixtures containing nuclease-treated extract were programmed with the indicated concentrations of poly(A+) or poly(A−) EMCV RNAs for 18 h at 32°C, treated with RNase A/T1, and assayed for infectivity. The data are averages (with standard deviation from the mean) of three independent titer determinations.
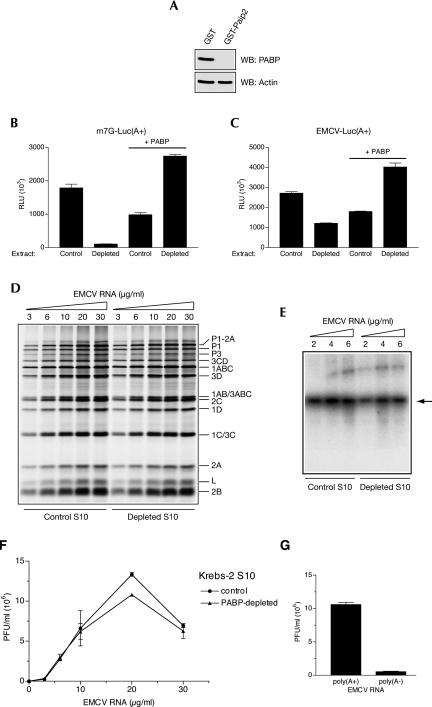
Having validated the poly(A) dependence of EMCV replication in nuclease-treated Krebs-2 extracts, we addressed the role of PABP by its depletion (Svitkin and Sonenberg 2004). Western blotting analysis of PABP performed on aliquots of control and PABP-depleted extract showed a near-complete (∼99%) removal of PABP (Fig. 3A). PABP depletion severely (∼16-fold) inhibited the translation of capped and polyadenylated luciferase mRNA (Fig. 3B). EMCV IRES-driven translation of a poly(A+) reporter mRNA was also inhibited by PABP depletion, albeit to a significantly lesser extent (∼2.3-fold inhibition) (Fig. 3C). Reductions in both cap-dependent and EMCV IRES-mediated translation were specifically due to PABP-depletion, as translation could be restored by the addition of recombinant PABP (Fig. 3B,C).
FIGURE 3.
Effects of PABP depletion on EMCV genome expression in nuclease-treated Krebs-2 S10 extract. (A) Extracts were treated with glutathione-Sepharose-bound GST or GST-Paip2. Equal aliquots of these extracts (4 μL, 50 μg of protein) were analyzed by Western blotting for PABP or actin, as indicated. Signals were detected by ECL and quantified. The degree of PABP depletion was 99.5%. (B) Cap-dependent translation of a reporter mRNA is strongly reduced by PABP depletion. Capped poly(A+) luciferase mRNA (1 μg/mL) was translated in extracts that were either depleted of PABP or mock-depleted (Control). The reaction mixtures were supplemented with recombinant human PABP where indicated. (C) EMCV IRES-mediated translation of a reporter mRNA is less inhibited than cap-dependent translation by PABP depletion. Control and PABP-depleted extracts were programmed with EMCV IRES-containing poly(A+) luciferase mRNA (1 μg/mL) in the absence or presence of PABP as indicated. For panels B and C, relative luciferase units (RLU) are given for 1-μL aliquots of translation samples. Data are averages of three independent assays (with standard deviation from the mean). (D) EMCV RNA translation is slightly reduced by PABP depletion. Control and PABP-depleted extracts were programmed with the indicated concentrations of EMCV RNA in the presence of [35S]methionine as described for Fig. 2A. Labeled proteins were analyzed by SDS-PAGE and fluorography. (E) RNA synthesis in control and PABP-depleted extracts programmed with the indicated concentrations of EMCV RNA. [α-32P]CTP pulse labeling of RNA and its analysis were as described for Fig. 2B. (Arrow) Position of single-stranded EMCV RNA. (F) EMCV RNA dose response of virus yields as affected by PABP depletion. Control and PABP-depleted extracts were programmed with the indicated concentrations of EMCV RNA as described for Fig. 2C. EMCV titers were determined in triplicates. Average titer values (with standard deviation from the mean) are shown. (G) EMCV synthesis directed by the poly(A+) and poly(A−) EMCV RNA in PABP-depleted extract. The RNAs were used at a 10 μg/mL concentration; other conditions were same as described for panel F.
Next, we investigated the capacity of PABP-depleted extract to translate the full-length EMCV RNA. Strikingly, EMCV RNA was translated with comparable efficiency in the absence and presence of PABP (PABP depletion caused only ∼15% inhibition of incorporation of [35S]methionine into acid-insoluble material) (Fig. 3D; data not shown). Also, no PABP-dependent alteration in the pattern of virus-specific polypeptides was evident at any concentration of the input RNA. The possible reasons of why PABP dependence of EMCV IRES-driven translation is lower for the full-size EMCV RNA than for the reporter mRNA will be addressed in the Discussion. The next step of virus replication, RNA synthesis, was also not affected by PABP depletion, as evidenced by the amount of RNA products pulse-labeled with [α-32P]CTP (Fig. 3E). In agreement with these results, at all the concentrations of EMCV RNA, PABP depletion failed to cause a decrease in infectious virus production (Fig. 3F). Importantly, and in the support of the role of the poly(A) tail but not PABP in EMCV replication, the difference in virus synthesis directed by the poly(A+) and poly(A−) EMCV RNA (Fig. 2C) was maintained in the PABP-depleted extract (Fig. 3G).
PABP is dispensable for poliovirus synthesis in HeLa S10
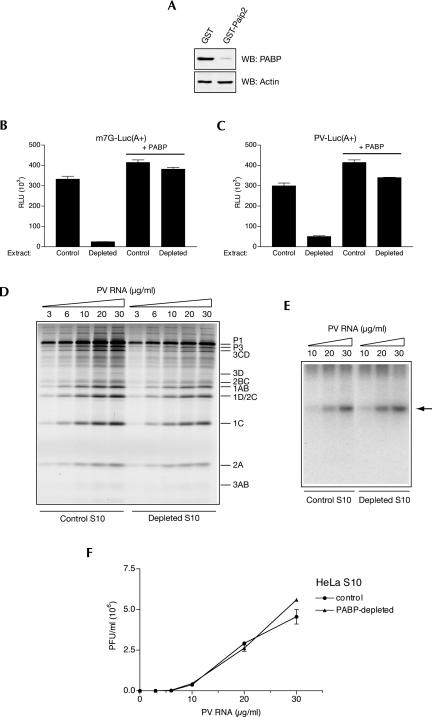
The experiments suggesting a PABP requirement for picornavirus RNA replication were performed using PV as a model (Herold and Andino 2001). It was thus possible that EMCV differs from PV in this respect. In fact, EMCV lacks the 5′ cloverleaf or similar structure presumably effecting PV genomic circularization. To determine whether PABP depletion affects PV replication in vitro, we prepared PV-replication-competent HeLa S10 extracts (Molla et al. 1991; Barton et al. 1996). We chose HeLa cells as a source of an extract because these cells are highly permissive for PV growth. As for Krebs-2 S10 extracts, the treatment of HeLa S10 extract with GST-Paip2 coupled to beads resulted in efficient (∼96%) depletion of PABP (Fig. 4A). This depletion decreased cap-dependent and PV IRES-mediated translation of poly(A+) luciferase mRNA by ∼12- and about sixfold, respectively, and the translation of both mRNAs was restored by PABP (Fig. 4B,C). We next examined full-length PV RNA dose response of translation in control and depleted extracts. Translation of PV RNA was moderately diminished (∼30%) by depleting PABP, independent of the concentration of PV RNA used (Fig. 4D). However, in spite of PABP's absence and a 30% decline in the abundance of PV proteins, PV RNA synthesis was unaffected (Fig. 4E). Finally, we analyzed PV yields in control and PABP-depleted extracts. At all PV RNA concentrations both systems generated roughly equal amounts of virus (Fig. 4F).
FIGURE 4.
Effect of PABP depletion on PV genome expression in nuclease-treated HeLa S10 extract. (A) Extracts were treated with GST or GST-Paip2 coupled to beads and their aliquots (4 μL, 50 μg of protein) were analyzed by Western blotting for PABP or actin as indicated. The degree of PABP depletion was 96%. (B) Cap-dependent translation of a reporter mRNA is strongly reduced by PABP depletion. Capped poly(A+) luciferase mRNA (1 μg/mL) was translated in control or PABP-depleted extract in the absence or presence of PABP as indicated. Translation conditions and luciferase assay are described in Fig. 3B. (C) PV IRES-mediated translation of a reporter mRNA is significantly reduced by PABP depletion. Control and PABP-depleted extracts were programmed with PV IRES-containing poly(A+) luciferase mRNA (1 μg/mL) in the absence or presence of PABP as indicated. For translation conditions and luciferase assay, see Fig. 3C. (D) PV RNA translation in PABP-depleted extracts. Control and PABP-depleted S10 extracts were programmed with the indicated concentrations of PV RNA in the presence of [35S]methionine. Labeled PV proteins were analyzed by SDS-PAGE and fluorography; their positions are indicated to the right of the gel. Endogenous mRNA translation yielded no products (data not shown). (E) RNA synthesis in control and PABP-depleted extracts programmed with the indicated concentrations of PV RNA. [α-32P]CTP pulse labeling of RNA was between 4 and 5 h of incubation. Product RNA analysis was as for Fig. 2B. (Arrow) Position of single-stranded PV RNA. (F) PV RNA-directed PV synthesis is not diminished by PABP depletion. Control and PABP-depleted extracts were programmed with the indicated concentrations of PV RNA. PV titers were determined in triplicate using HeLa R19 cell monolayers. Averages of the obtained values (with standard deviation from the mean) are shown.
DISCUSSION
The interaction of PABP with the PV-specific 5′ cloverleaf-associated proteins PCBP2 and 3CD supports a model for the formation of circular replication-competent RNP complexes (Herold and Andino 2001). In agreement with this model, a deletion mutant of PABP that is defective in binding to PCBP2 and 3CD, but not to poly(A), abrogated PV RNA replication in a coupled translation–replication system (Herold and Andino 2001). Although the 5′ UTR of EMCV RNA does not fold into a cloverleaf structure, it harbors a PCBP2 binding site (Walter et al. 1999). Thus, PABP-mediated circularization of the plus strand RNA was postulated to be of broader significance than merely to promote RNA replication of only PV (Herold and Andino 2001). A testable prediction from this model is that depleting PABP from a viral RNA-programmed cell-free system should cause a dramatic inhibition of virus replication. However, in contrast to this prediction, removing PABP from nuclease-treated S10 extracts had no effect on RNA replication and yields of both EMCV and PV (Figs. 3, 4). In addition, Paip2 and poly(A), which sequester PABP, did not adversely affect EMCV- and PV-specific translation, RNA synthesis, and virus production in vitro (Murray et al. 2001; data not shown).
Poly(A) stimulates PV IRES, coxsackievirus IRES, and, to a significantly lesser degree, EMCV IRES-mediated translation of reporter mRNAs in vitro (Bergamini et al. 2000; Michel et al. 2001; Svitkin et al. 2001; Bradrick et al. 2007). Similarly, PABP enhances the translation from EMCV and PV IRESs, presumably by increasing the affinity of eIF4G for these IRESs (Figs. 3C, 4C; Michel et al. 2001; Svitkin et al. 2001). It is therefore surprising that stimulation of translation by the PABP/poly(A) tail complex is not recapitulated in the context of the genomic PV and EMCV RNAs (Figs. 3D, 4D; Spector et al. 1975; Barton et al. 1996; Silvestri et al. 2006). A plausible explanation is that initiation is not the rate-limiting step of translation of the long picornavirus genomes. In fact, in the course of translation of EMCV RNA in vitro, elongating ribosomes slow down their advance somewhere at the border between the coding regions for the structural and nonstructural proteins (Shih et al. 1979; Svitkin and Agol 1983). Data suggesting that polyprotein elongation, rather than initiation, is limiting for the synthesis of EMCV proteins in virus-infected cells were also reported (Jen et al. 1978). Alternatively, the interaction between eIF4G and PABP may not function efficiently in the context of the full-length picornavirus RNAs where the IRES and poly(A) tail structures are positioned very far apart.
Although PABP-depleted cytoplasmic extracts are fully competent in supporting EMCV and PV syntheses, the possibility that PABP modulates some virus-specific processes in virus-infected cells could not be excluded. We attempted to address this issue by studying PV and EMCV replication in HeLa cells in which PABP expression had been silenced using RNA interference. Although the amount of PABP in these cells was decreased by ∼80% of the control level by siRNA treatment, there was no down-regulation of either EMCV or PV replication (data not shown). These negative data, although consistent with our in vitro results, cannot be readily interpreted because Paip2 degrades in PABP knockdown cells, thereby ensuring PABP homeostasis (Yoshida et al. 2006). However, the fact that in PV-infected cells, a substantial fraction of PABP and PCBP2 is cleaved at a time when virus-specific RNA synthesis occurs most efficiently is hardly compatible with the proposed role of these proteins in PV RNA circularization (Kuyumcu-Martinez et al. 2002, 2004; B. Semler, pers. comm.).
If PABP is not essential for picornavirus replication, then what kind of advantage to the virus is conferred by poly(A), which is conserved in all picornaviruses? Why does poly(A) removal dramatically diminish the infectivity of picornavirus RNAs as well as its capacity to direct virus synthesis in vitro? Translation of viral genomes is not likely to be the step critically affected by deadenylation, at least not in vitro (Figs. 1B, 2A; Spector et al. 1975; Barton et al. 1996; Silvestri et al. 2006). Similarly, poly(A) tail length does not appear to be very important for the stability of EMCV and PV RNA (Fig. 1C; Silvestri et al. 2006). Thus, the only essential function that might be severely compromised by the lack of poly(A) is the synthesis of a VPg-poly(U) primer by 3Dpol as originally suggested (Paul et al. 1998). The 3′ UTR of PV and EMCV apparently increases the efficiency of this reaction by specifically recruiting 3Dpol and probably other components of the replication complex. Interactions between the proteins bound to the 3′ UTR and those bound to the 5′ cloverleaf of PV may assist in forming a circular mRNP complex. It is possible that the protein-bridging role is played by 3CD alone, provided that this protein can bind to the 5′ and 3′ UTRs and also form a homodimer, as suggested by several studies (Andino et al. 1993; Harris et al. 1994; Xiang et al. 1995; Barton et al. 2001; Murray and Barton 2003). In addition, a cellular protein(s) that has been modified by PV 2Apro might enhance the formation of the circular RNP complex (Jurgens et al. 2006). RNP circularization should block the loading of ribosomes onto the 5′ IRES structure, thereby promoting a switch from translation to replication (Gamarnik and Andino 1998; Barton et al. 1999).
It is puzzling that the effect of poly(A) tail removal on EMCV synthesis is much more profound in the untreated than in nuclease-treated extract (cf. Figs. 1E and 2C). A relatively high virus synthesis directed by poly(A−) EMCV RNA in the nuclease-treated extract could result from incomplete digestion of the poly(A) tail with RNAse H. This may generate a fraction of EMCV RNA with poly(A) tails that while being too short for hybridization with oligo(dT) are nevertheless sufficient for promoting RNA synthesis and virus production. It is also possible that competition from cellular mRNAs for one or more components of the RNA replication complex in the untreated extract impose greater restriction on the replication of viral RNAs with short than with long poly(A) tails.
MATERIALS AND METHODS
Viral RNA preparations
PV and EMCV were propagated in HeLa and Krebs-2 cells, respectively. Viruses were purified by centrifugation through a sucrose cushion and precipitation with polyethylene glycol (Svitkin et al. 1998). RNA was extracted with a mixture of phenol and chloroform. For poly(A) tail removal, EMCV RNA (40 μg) was hybridized to oligo(dT)16 (2 μg) in 39 μL of 50 mM KCl by slow cooling the reaction mixture from 40°C to room temperature in a water bath. Ten microliters of 5× RNase H buffer (100 mM Tris-HCl, pH 7.8, 40 mM MgCl2, 5 mM DTT, and 10% glycerol) and 1 μL (2 U) of RNase H (Sigma) was added at room temperature, and the reaction mixture was incubated for 20 min at 37°C. After incubation, RNA was phenol–chloroform extracted and ethanol precipitated. To circumvent the possibility of the regeneration of the poly(A) tail in cell-free extracts (van Ooij et al. 2006), the 3′ end of deadenylated RNA was blocked with cordycepin-5′-triphosphate using yeast poly(A) polymerase (USB) (Herold and Andino 2001). Control poly(A+) EMCV RNA was blocked with cordycepin-5′-triphosphate in a similar way. Poly(A+) and poly(A−) EMCV RNAs were purified by CHROMA SPIN-1000 column (BD Biosciences Clontech) chromatography. The integrity of RNAs and efficiency of deadenylation was verified by formaldehyde–agarose gel electrophoresis and Northern blotting as described below. Plasmids encoding luciferase, T3luc(A+), T7EMCVluc(A+), and T7PVluc(A+), their linearization, and transcription with T3 or T7 RNA polymerase were described previously (Svitkin et al. 2001).
Preparation and PABP depletion of Krebs-2 and HeLa cell extracts
Untreated or micrococcal nuclease-treated Krebs-2 and HeLa S3 cell extracts (S10 fractions) were prepared as previously described (Barton et al. 1996; Svitkin and Sonenberg 2004, 2007). For the depletion of endogenous PABP, nuclease-treated extracts were incubated with the GST-Paip2 protein that was immobilized onto glutathione-Sepharose beads (Svitkin and Sonenberg 2004). Mock-depleted (control) extracts were incubated with GST alone. Western blotting assessment of PABP depletion was carried out on 2, 4, and 6 μL aliquots of mock- and PABP-depleted extracts using an anti-PABP polyclonal antibody and Western Lightning chemiluminescence kit (Perkin-Elmer Life Sciences) (Svitkin and Sonenberg 2004). Densitometric analysis of band intensities was performed using NIH Image version 1.63 software.
In vitro translation, RNA replication, and virus synthesis
EMCV and PV RNAs were translated in 20 μL reaction mixtures containing Krebs-2 or HeLa S3 cell extracts, respectively (Barton et al. 1996; Svitkin and Sonenberg 2003, 2007). Where indicated, nuclease-treated extracts depleted of PABP were used. [35S]methionine labeling of proteins was for 3 h at 32°C. Protein products were resolved by SDS-PAGE (15% gels) and detected by fluorography. The unlabeled reaction mixtures (40 μL) used to measure RNA replication were incubated for 4 h at 32°C. Tracer [α-32P]CTP was then added for a period of 1 h. Labeled product RNA was deproteinized and analyzed by native 1% agarose gel electrophoresis essentially as described previously (Svitkin and Sonenberg 2003, 2007). To measure virus synthesis, the unlabeled reaction mixtures (40 μL) were incubated for 18 h at 32°C. The input RNA was then destroyed by RNase A/T1 treatment (Molla et al. 1991). Virus titers were determined in serially diluted samples using confluent monolayers of BHK-21 (EMCV) or HeLa R19 (PV) cells in 60-mm-diameter plates (Rueckert and Pallansch 1981; Svitkin et al. 2005; Svitkin and Sonenberg 2007). Cap- and IRES-containing reporter mRNAs were translated at their optimal potassium ion concentrations (75 and 150 mM, respectively) in a 10-μL total reaction volume. Recombinant human PABP (Svitkin and Sonenberg 2004) was included in the reaction mixtures at a 10 μg/mL final concentration where indicated. After incubation for 60 min at 32°C, luciferase levels were measured by enzymatic assay (Svitkin and Sonenberg 2004).
Northern blot analysis of RNA degradation
The unlabeled reaction mixtures (100 μL) assembled to assay mRNA decay contained 10 μg/mL poly(A+) or poly(A−) EMCV RNA, Krebs-2 S10 extract, and other translation components. After incubation for the times indicated at 32°C, 15 μL aliquots were withdrawn into 185 μL of 0.4 mg/mL proteinase K (made up in buffer containing 100 mM NaCl, 20 mM Tris-HCl, pH 8.0, 1 mM EDTA, 1% SDS, and 100 μg/mL Escherichia coli tRNA). Following incubation for 15 min at 37°C, total RNA was phenol–chloroform extracted, separated on a formaldehyde–0.8% agarose gel, and transferred onto a nylon membrane (Hybond-N; GE-Healthcare) (Sambrook et al. 1989). To confirm equal loading, blots were stained with Blot Stain Blue (Sigma), and the intensities of the bands of rRNA were compared. Hybridization with a fragment of the randomly primed 32P-labeled EMCV cDNA (pE-C9) (Hahn and Palmenberg 1995) was performed using ExpressHyb hybridization solution (BD Biosciences Clontech) according to the company's protocol. For analysis of EMCV RNA deadenylation after RNase H/oligo(dT) treatment (Fig. 1A), hybridization was conducted with 32P-5′-phosphorylated oligo(dT)16. Band intensities were determined using the BAS-2000 phosphorimaging system (FUJI Medical Systems USA).
ACKNOWLEDGMENTS
We thank Ann Palmenberg for EMCV cDNA, Vadim Agol and Bert Semler for critical reading of the manuscript, Amy Rosenfeld for helpful discussions, and Colin Lister for excellent technical assistance. This research was supported by a Public Health Service Research Grant from NIH to N.S. (Grant #5R01 GM66157-03). N.S. is a Howard Hughes Medical Institute International Scholar.
Footnotes
Note: Reported in part at the 23rd Annual Meeting of the American Society for Virology (Montreal, Quebec, 2004).
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.606407.
REFERENCES
- Agol, V.I. Picornavirus genome: An overview. In: Semler B.L., Wimmer E., editors. Molecular biology of picornaviruses. ASM Press; Washington, DC: 2002. pp. 127–148. [Google Scholar]
- Andino, R., Rieckhof, G.E., Baltimore, D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- Andino, R., Rieckhof, G.E., Achacoso, P.L., Baltimore, D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, D.J., Morasco, B.J., Flanegan, J.B. Assays for poliovirus polymerase, 3DPol, and authentic RNA replication in HeLa S10 extracts. Methods Enzymol. 1996;275:35–57. doi: 10.1016/s0076-6879(96)75005-x. [DOI] [PubMed] [Google Scholar]
- Barton, D.J., Morasco, B.J., Flanegan, J.B. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol. 1999;73:10104–10112. doi: 10.1128/jvi.73.12.10104-10112.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, D.J., O'Donnell, B.J., Flanegan, J.B. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 2001;20:1439–1448. doi: 10.1093/emboj/20.6.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamini, G., Preiss, T., Hentze, M.W. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA. 2000;6:1781–1790. doi: 10.1017/s1355838200001679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradrick, S.S., Dobrikova, E.Y., Kaiser, C., Shveygert, M., Gromeier, M. Poly(A)-binding protein is differentially required for translation mediated by viral internal ribosome entry sites. RNA. 2007;13:1–12. doi: 10.1261/rna.556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumakov, K.M., Agol, V.I. Poly(C) sequence is located near the 5′-end of encephalomyocarditis virus RNA. Biochem. Biophys. Res. Commun. 1976;71:551–557. doi: 10.1016/0006-291x(76)90822-6. [DOI] [PubMed] [Google Scholar]
- Cui, T., Porter, A.G. Localization of binding site for encephalomyocarditis virus RNA polymerase in the 3′-noncoding region of the viral RNA. Nucleic Acids Res. 1995;23:377–382. doi: 10.1093/nar/23.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsch-Hasler, K., Yogo, Y., Wimmer, E. Replication of picornaviruses. I. Evidence from in vitro RNA synthesis that poly(A) of the poliovirus genome is genetically coded. J. Virol. 1975;16:1512–1517. doi: 10.1128/jvi.16.6.1512-1517.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik, A.V., Andino, R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes & Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow, I., Chaudhry, Y., Richardson, A., Meredith, J., Almond, J.W., Barclay, W., Evans, D.J. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 2000;74:4590–4600. doi: 10.1128/jvi.74.10.4590-4600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow, I.G., Polacek, C., Andino, R., Evans, D.J. The poliovirus 2C cis-acting replication element-mediated uridylylation of VPg is not required for synthesis of negative-sense genomes. J. Gen. Virol. 2003;84:2359–2363. doi: 10.1099/vir.0.19132-0. [DOI] [PubMed] [Google Scholar]
- Hahn, H., Palmenberg, A.C. Encephalomyocarditis viruses with short poly(C) tracts are more virulent than their mengovirus counterparts. J. Virol. 1995;69:2697–2699. doi: 10.1128/jvi.69.4.2697-2699.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, K.S., Xiang, W., Alexander, L., Lane, W.S., Paul, A.V., Wimmer, E. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem. 1994;269:27004–27014. [PubMed] [Google Scholar]
- Herold, J., Andino, R. Poliovirus RNA replication requires genome circularization through a protein–protein bridge. Mol. Cell. 2001;7:581–591. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruby, D.E., Roberts, W.K. Encephalomyocarditis virus RNA. II. Polyadenylic acid requirement for efficient translation. J. Virol. 1977;23:338–344. doi: 10.1128/jvi.23.2.338-344.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson, A. Poly(A) metabolism and translation: The closed-loop model. In: Hershey J.W.B., et al., editors. Translational control. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1996. pp. 451–480. [Google Scholar]
- Jen, G., Birge, C.H., Thach, R.E. Comparison of initiation rates of encephalomyocarditis virus and host protein synthesis in infected cells. J. Virol. 1978;27:640–647. doi: 10.1128/jvi.27.3.640-647.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens, C.K., Barton, D.J., Sharma, N., Morasco, B.J., Ogram, S.A., Flanegan, J.B. 2Apro is a multifunctional protein that regulates the stability, translation, and replication of poliovirus RNA. Virology. 2006;345:346–357. doi: 10.1016/j.virol.2005.09.067. [DOI] [PubMed] [Google Scholar]
- Kahvejian, A., Svitkin, Y.V., Sukarieh, R., M'Boutchou, M.N., Sonenberg, N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes & Dev. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim, M.M., Svitkin, Y.V., Kahvejian, A., De Crescenzo, G., Costa-Mattioli, M., Sonenberg, N. A mechanism of translational repression by competition of Paip2 with eIF4G for poly(A) binding protein (PABP) binding. Proc. Natl. Acad. Sci. 2006;103:9494–9499. doi: 10.1073/pnas.0603701103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleghpour, K., Svitkin, Y.V., Craig, A.W., DeMaria, C.T., Deo, R.C., Burley, S.K., Sonenberg, N. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol. Cell. 2001;7:205–216. doi: 10.1016/s1097-2765(01)00168-x. [DOI] [PubMed] [Google Scholar]
- Kuyumcu-Martinez, N.M., Joachims, M., Lloyd, R.E. Efficient cleavage of ribosome-associated poly(A)-binding protein by enterovirus 3C protease. J. Virol. 2002;76:2062–2074. doi: 10.1128/jvi.76.5.2062-2074.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyumcu-Martinez, N.M., Van Eden, M.E., Younan, P., Lloyd, R.E. Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: A novel mechanism for host translation shutoff. Mol. Cell. Biol. 2004;24:1779–1790. doi: 10.1128/MCB.24.4.1779-1790.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons, T., Murray, K.E., Roberts, A.W., Barton, D.J. Poliovirus 5′-terminal cloverleaf RNA is required in cis for VPg uridylylation and the initiation of negative-strand RNA synthesis. J. Virol. 2001;75:10696–10708. doi: 10.1128/JVI.75.22.10696-10708.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus, D.A., Evans, M.C., Jacobson, A. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, Y.M., Borman, A.M., Paulous, S., Kean, K.M. Eukaryotic initiation factor 4G-poly(A) binding protein interaction is required for poly(A) tail-mediated stimulation of picornavirus internal ribosome entry segment-driven translation but not for X-mediated stimulation of hepatitis C virus translation. Mol. Cell. Biol. 2001;21:4097–4109. doi: 10.1128/MCB.21.13.4097-4109.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla, A., Paul, A.V., Wimmer, E. Cell-free, de novo synthesis of poliovirus. Science. 1991;254:1647–1651. doi: 10.1126/science.1661029. [DOI] [PubMed] [Google Scholar]
- Morasco, B.J., Sharma, N., Parilla, J., Flanegan, J.B. Poliovirus cre(2C)-dependent synthesis of VPgpUpU is required for positive- but not negative-strand RNA synthesis. J. Virol. 2003;77:5136–5144. doi: 10.1128/JVI.77.9.5136-5144.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, K.E., Barton, D.J. Poliovirus CRE-dependent VPg uridylylation is required for positive-strand RNA synthesis but not for negative-strand RNA synthesis. J. Virol. 2003;77:4739–4750. doi: 10.1128/JVI.77.8.4739-4750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, K.E., Roberts, A.W., Barton, D.J. Poly(rC) binding proteins mediate poliovirus mRNA stability. RNA. 2001;7:1126–1141. doi: 10.1017/s1355838201010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsley, T.B., Towner, J.S., Blyn, L.B., Ehrenfeld, E., Semler, B.L. Poly(rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- Paul, A.V. Possible unifying mechanism of picornavirus genome replication. In: Semler B.L., Wimmer E., editors. Molecular biology of picornaviruses. ASM Press; Washington, DC: 2002. pp. 227–246. [Google Scholar]
- Paul, A.V., van Boom, J.H., Filippov, D., Wimmer, E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- Paul, A.V., Rieder, E., Kim, D.W., van Boom, J.H., Wimmer, E. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 2000;74:10359–10370. doi: 10.1128/jvi.74.22.10359-10370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Kolupaeva, V.G., Lomakin, I.B., Pilipenko, E.V., Shatsky, I.N., Agol, V.I., Hellen, C.U. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko, E.V., Poperechny, K.V., Maslova, S.V., Melchers, W.J., Slot, H.J., Agol, V.I. Cis-element, oriR, involved in the initiation of (−) strand poliovirus RNA: A quasiglobular multidomain RNA structure maintained by tertiary (“kissing”) interactions. EMBO J. 1996;15:5428–5436. [PMC free article] [PubMed] [Google Scholar]
- Racaniello, V.R. Picornaviridae: The viruses and their replication. In: Knipe D.M., Howley P.M., editors. Fields virology. Lippincott Williams & Wilkins; Philadelphia, PA: 2001. pp. 685–722. [Google Scholar]
- Romanova, L.I., Agol, V.I. Interconversion of linear and circular forms of double-stranded RNA of encephalomyocarditis virus. Virology. 1979;93:574–577. doi: 10.1016/0042-6822(79)90260-5. [DOI] [PubMed] [Google Scholar]
- Rueckert, R.R., Pallansch, M.A. Preparation and characterization of encephalomyocarditis (EMC) virus. Methods Enzymol. 1981;78:315–325. [PubMed] [Google Scholar]
- Sachs, A. Physical and functional interactions between the mRNA cap structure and the poly(A) tail. In: Sonenberg N., et al., editors. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 447–465. [Google Scholar]
- Sambrook, J., Fritsch, E.F., Maniatis, T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Sarnow, P. Role of 3′-end sequences in infectivity of poliovirus transcripts made in vitro. J. Virol. 1989;63:467–470. doi: 10.1128/jvi.63.1.467-470.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih, D.S., Shih, C.T., Zimmern, D., Rueckert, R.R., Kaesberg, P. Translation of encephalomyocarditis virus RNA in reticulocyte lysates: Kinetic analysis of the formation of virion proteins and a protein required for processing. J. Virol. 1979;30:472–480. doi: 10.1128/jvi.30.2.472-480.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestri, L.S., Parilla, J.M., Morasco, B.J., Ogram, S.A., Flanegan, J.B. Relationship between poliovirus negative-strand RNA synthesis and the length of the 3′ poly(A) tail. Virology. 2006;345:509–519. doi: 10.1016/j.virol.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Spector, D.H., Baltimore, D. Requirement of 3′-terminal poly(adenylic acid) for the infectivity of poliovirus RNA. Proc. Natl. Acad. Sci. 1974;71:2983–2987. doi: 10.1073/pnas.71.8.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector, D.H., Villa-Komaroff, L., Baltimore, D. Studies on the function of polyadenylic acid on poliovirus RNA. Cell. 1975;6:41–44. doi: 10.1016/0092-8674(75)90071-9. [DOI] [PubMed] [Google Scholar]
- Svitkin, Y.V., Agol, V.I. Translational barrier in central region of encephalomyocarditis virus genome. Modulation by elongation factor 2 (eEF-2) Eur. J. Biochem. 1983;133:145–154. doi: 10.1111/j.1432-1033.1983.tb07440.x. [DOI] [PubMed] [Google Scholar]
- Svitkin, Y.V., Sonenberg, N. Cell-free synthesis of encephalomyocarditis virus. J. Virol. 2003;77:6551–6555. doi: 10.1128/JVI.77.11.6551-6555.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin, Y.V., Sonenberg, N. An efficient system for cap- and poly(A)-dependent translation in vitro. Methods Mol. Biol. 2004;257:155–170. doi: 10.1385/1-59259-750-5:155. [DOI] [PubMed] [Google Scholar]
- Svitkin, Y.V., Sonenberg, N. Translational control by the poly(A) binding protein: A check for mRNA integrity. Mol. Biol. (Mosk.) 2006;40:611–619. [PubMed] [Google Scholar]
- Svitkin, Y.V., Sonenberg, N. A highly efficient and robust in vitro translation system for expression of picornavirus and hepatitis C virus RNA genomes. Methods Enzymol. 2007;429:53–82. doi: 10.1016/S0076-6879(07)29004-4. [DOI] [PubMed] [Google Scholar]
- Svitkin, Y.V., Hahn, H., Gingras, A.C., Palmenberg, A.C., Sonenberg, N. Rapamycin and wortmannin enhance replication of a defective encephalomyocarditis virus. J. Virol. 1998;72:5811–5819. doi: 10.1128/jvi.72.7.5811-5819.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin, Y.V., Imataka, H., Khaleghpour, K., Kahvejian, A., Liebig, H.D., Sonenberg, N. Poly(A)-binding protein interaction with eIF4G stimulates picornavirus IRES-dependent translation. RNA. 2001;7:1743–1752. [PMC free article] [PubMed] [Google Scholar]
- Svitkin, Y.V., Herdy, B., Costa-Mattioli, M., Gingras, A.C., Raught, B., Sonenberg, N. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol. Cell. Biol. 2005;25:10556–10565. doi: 10.1128/MCB.25.23.10556-10565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teterina, N.L., Egger, D., Bienz, K., Brown, D.M., Semler, B.L., Ehrenfeld, E. Requirements for assembly of poliovirus replication complexes and negative-strand RNA synthesis. J. Virol. 2001;75:3841–3850. doi: 10.1128/JVI.75.8.3841-3850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooij, M.J., Polacek, C., Glaudemans, D.H., Kuijpers, J., van Kuppeveld, F.J., Andino, R., Agol, V.I., Melchers, W.J. Polyadenylation of genomic RNA and initiation of antigenomic RNA in a positive-strand RNA virus are controlled by the same cis-element. Nucleic Acids Res. 2006;34:2953–2965. doi: 10.1093/nar/gkl349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, B.L., Nguyen, J.H., Ehrenfeld, E., Semler, B.L. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA. 1999;5:1570–1585. doi: 10.1017/s1355838299991483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, W., Harris, K.S., Alexander, L., Wimmer, E. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J. Virol. 1995;69:3658–3667. doi: 10.1128/jvi.69.6.3658-3667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, M., Yoshida, K., Kozlov, G., Lim, N.S., De Crescenzo, G., Pang, Z., Berlanga, J.J., Kahvejian, A., Gehring, K., Wing, S.S., et al. Poly(A) binding protein (PABP) homeostasis is mediated by the stability of its inhibitor, Paip2. EMBO J. 2006;25:1934–1944. doi: 10.1038/sj.emboj.7601079. [DOI] [PMC free article] [PubMed] [Google Scholar]