Abstract
All known guanine-sensing riboswitches regulate gene expression by specifically binding to guanine (G) or related analogs with high affinity to switch off transcription. The aptamers of this class of riboswitches are characterized by three helices (P1–P3), surrounding a central core of phylogenetically conserved nucleotides and a long-range loop–loop interaction. To gain more insight into the switching mechanism, we present here a comparison between the solution-state structures of the G-free and G-bound forms of the guanine aptamer from the xpt-pbuX operon of Bacillus subtilis, as derived from NMR chemical shifts and magnetic-field-induced residual dipolar couplings. The high-resolution NMR analysis shows the G-free aptamer is highly structured with parallel P2 and P3 helices and the long-range loop–loop interaction already present, implying that the structure is largely preformed to bind the ligand. Structural changes upon guanine binding are found to be localized to the central core. In the free state, the G-quadruple interaction and two base pairs of the P1 stem flanking the central core appear to be largely disordered. The ligand thus binds via a combined predetermined–induced fit mechanism, involving a previously unstructured five-residue loop of the J2–3 junction that folds over the ligand. These limited additional interactions within a preorganized setting possibly explain how the aptamer rapidly responds to ligand binding, which is necessary to switch the structural state of the expression platform within a narrow time frame before the RNA polymerase escapes the 5′-UTR.
Keywords: riboswitch, aptamer, RNA structure, RNA folding, NMR, mRNA, gene regulation
INTRODUCTION
Riboswitches are RNA control elements, found in the 5′- or 3′-untranslated regions (UTRs) and introns of many mRNAs of bacteria and some plant and fungi, which modulate gene expression without the need of protein cofactors (Mandal and Breaker 2004b; Nudler and Mironov 2004; Soukup and Soukup 2004). These riboswitches contain highly structured domains, which target specific metabolites, resulting in metabolite-induced conformational transitions that ultimately regulate the cellular levels of the same metabolite. So far, riboswitches have been described that control the cellular levels of purines, vitamins, coenzymes, and amino acids. Compounds for which riboswitches have been found are thiamin pyrophosphate (TPP) (Mironov et al. 2002; Winkler et al. 2002a), guanine (Mandal et al. 2003), adenine (Mandal et al. 2003; Mandal and Breaker 2004a), coenzyme B12 (Nahvi et al. 2002), flavin mononucleotide (FMN) (Mironov et al. 2002; Winkler et al. 2002b), glucosamine-6-phosphate (GlcN6P) (Winkler et al. 2004), glycine (Mandal et al. 2004), lysine (Grundy et al. 2003; Rodionov et al. 2003; Sudarsan et al. 2003), and S-adenosylmethionine (SAM) (Epshtein et al. 2003; McDaniel et al. 2003; Winkler et al. 2003; Fuchs et al. 2006). Riboswitches consist of two modular domains: an aptamer domain, which contains the high-affinity binding site for a given metabolite or ligand, connected to a downstream region termed the expression platform, which can exist in two mutually exclusive conformational states. At sufficient concentrations, binding of the ligand stabilizes the structure of the aptamer domain, which presumably results in switching the conformational state of the expression platform to regulate a certain gene expression system. While the aptamer sequences are largely conserved to ensure specific binding to a given metabolite in various organisms, the expression platforms vary in sequence to control gene expression using different mechanisms, mostly by either modulating the formation of a transcription terminator or by sequestering a Shine–Dalgarno sequence to prevent translation initiation. Besides these two dominant regulating mechanisms, riboswitches have also been found that regulate through self-cleavage of the mRNA by ribozyme activity (Winkler et al. 2004).
The purine riboswitches are among the smallest riboswitches known, contain one of the most phylogenetically conserved aptamers described to date, and hence are the ones studied most extensively by structural biology and biophysical means (Batey et al. 2004; Serganov et al. 2004; Noeske et al. 2005, 2007; Wickiser et al. 2005; Gilbert et al. 2006; Lemay et al. 2006). The aptamer part of the Bacillus subtilis xpt-pbuX-mRNA guanine riboswitch, which is the subject of this study and is representative of all known purine riboswitches, folds into a three-way junction, the arms of which are designated P1, P2, and P3 connected by three interhelical loops, J1–2, J2–3, and J3–1 (Fig. 1). In the crystal structures of the ligand–riboswitch complexes (Batey et al. 2004; Serganov et al. 2004), the P2 and P3 helices are parallel and closely packed together by two base quadruples and a noncanonical base pair between the apical P2 and P3 loops. The purine is buried within the central core by forming a base quadruple and is further closed in by two base triples from above and two base triples from below. The two upper base triples also help to anchor the P2 and P3 stems. The two lower base triples also pin down the J2–3 loop to the top of P1, closing in the guanine at the N3/N9 side.
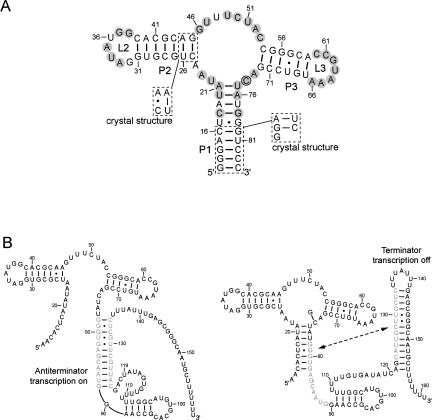
FIGURE 1.
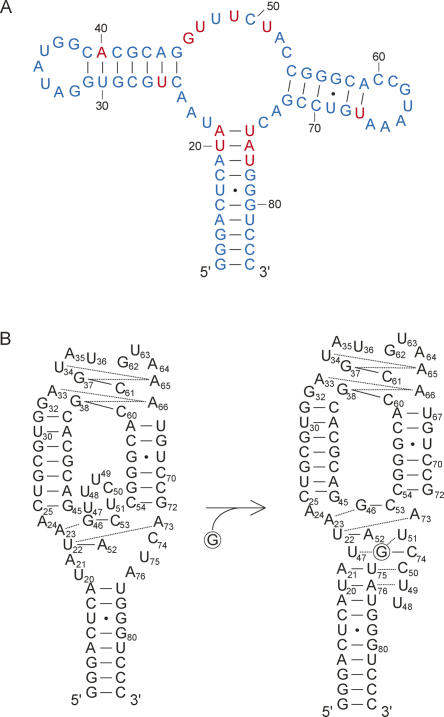
(A) Secondary structure of the G-riboswitch aptamer (GRA) used for the NMR study. Highly conserved residues are shaded in gray. Deviations from the natural sequence found in the Bacillus subtilis xpt-pbuX-mRNA gene, which have also been introduced for obtaining the crystal structure of the GRA in complex with guanine (Serganov et al. 2004), are shown in the boxed regions. C74, which confers specificity for guanine, is highlighted with a circle. (B) Gene regulation by the guanine riboswitch in the 5′-UTR of mRNA. Initial transcription generates the aptamer-binding domain, which can be preformed to bind guanine rapidly at sufficient concentration. In the absence of guanine, an antiterminator element is formed, which allows transcription to proceed (left). Binding of guanine is expected to stabilize the guanine binding domain and P1 stem, which is believed to force formation of the terminator element that stops transcription (right).
Biochemical studies provided evidence that the long-range loop–loop interaction forms in the absence of ligand and is essential for ligand binding. In the in-line probing experiments, the apical stem–loop regions of P2 and P3 were protected from cleavage and showed no difference in the nucleotide protection patterns in the presence or absence of ligand (Mandal et al. 2003). Destroying the loop–loop interaction by replacing the wild-type apical P2 and P3 loops by UUCG, which are incapable of interacting, abolishes the capacity of the aptamer to recognize the ligand (Batey et al. 2004). NMR studies showed the pre-existence of base pairs between P2 and P3 loop residues when no ligand is present (Noeske et al. 2007). These results are in accordance with the presence of a partially preorganized aptamer structure in the absence of ligand.
On the other hand, it has also been noted that the ligand-binding site has to undergo a substantial conformational change upon binding because the ligand cannot access a tightly preformed binding site as observed in the cocrystal structures (Batey et al. 2004; Serganov et al. 2004). This implies an induced-fit model, which is supported by in-line probing experiments, NMR studies, and thermodynamic data. In-line probing experiments of the unbound RNA showed several cleavage products mapping to the single-stranded nucleotides of the three-way junction that are protected in the presence of excess guanine, suggesting a locally disordered binding pocket in the free state (Mandal et al. 2003). NMR imino proton spectra showed additional peaks upon ligand binding without significant shifts of pre-existing peaks, indicative of a limited rearrangement of the global structure (Serganov et al. 2004). Thermodynamic measurements for ligand binding revealed unfavorable entropy values compensated by favorable enthalpy values, also in accordance with a local folding event (Gilbert et al. 2006).
Thus, given the available data, the prevailing view is that the aptamer is partially preorganized by the stems and long-range loop–loop interaction, while the junction and surrounding regions have to be disordered to allow binding of the ligand. This structural model forms the current basis for some of the remaining key questions that cannot be answered with the high-resolution crystal structures of the aptamer–ligand complexes alone: to what extent is the aptamer folded at the time of switching, and how is binding of the ligand translated into a conformational switch of the expression platform leading to transcription termination. An important aspect in evaluating this model is that riboswitch control is tightly coupled to transcription (Fig. 1B). In this process, the aptamer is synthesized first and is capable of folding independently before the expression platform is generated. Ligand binding has to be transmitted to the downstream expression platform region while it is being synthesized by the elongating polymerase before it passes through the 5′-UTR. This leaves a very narrow time frame of only a few seconds for the polymerase to choose between elongation and termination (Wickiser et al. 2005), depending on the structural state of the expression platform it encounters. For the xpt-pbuX G-riboswitch in particular, this entails residues 78–82 (Fig. 1B), which either close the P1 stem, permitting formation of the transcription terminator, or interact with a downstream region to form an antiterminator. Thus, ligand binding and subsequent closure of the core region and P1 stem has to be considerably fast, otherwise the antiterminator stem–loop structure is able to form, which is unlikely to switch back within the given time, since structural rearrangements involving unfolding/refolding stretches of RNA helices can be notably slow (e.g., Nagel et al. 2002). Overall, this model indicates the importance of a discrete thermodynamic and kinetic balance between formation of the remainder of the aptamer structure and the antiterminator. Therefore, the degree of structural organization of the aptamer is a very important consideration in understanding the mechanism of riboswitching.
To investigate the degree of structural organization, we obtained global and atomic detail structural information in solution on the G-riboswitch aptamer in its guanine bound and free states. We show that Mg2+ is required to drive the free aptamer in a single conformational state. High-quality NMR spectra were obtained for both states in the presence of Mg2+, allowing complete assignment of the imino protons' resonances, which are monitors of base hydrogen bond interactions and part of the non-exchangeable protons. In addition, by using magnetic-field-induced residual dipolar couplings (mRDCs) from imino N–H and adenine C2–H2 bonds, we show that there are no significant global differences between the structures of both states. Moreover, the changes in base pairing and short NOE-derived distances, together with the mRDCs, defined on a residue-specific basis could be used to propose a model for the extent and location of the conformational changes in the G-riboswitch aptamer upon guanine binding. This information can be used to provide further insight into the molecular mechanism of riboswitching.
RESULTS AND DISCUSSION
Mg2+ ions are required for proper folding of the guanine free G-riboswitch aptamer
The 73-mer G-riboswitch aptamer (GRA) sequence investigated in the work presented here (Fig. 1) is derived from the B. subtilis xpt-pbuX operon (Mandal et al. 2003). A few structurally silent mutations were introduced to stabilize the P2 helical region for the NMR studies (Noeske et al. 2005), i.e., P2 mutations U25C, C26U, and A45G. Additional guanosine residues were added to the 5′ end (G12–G14) needed for efficient in vitro transcription and closed by additional cytosines (C82–C84) added at the 3′ end to prevent possible RNA aggregation due to intermolecular interactions of single-stranded G-stretches.
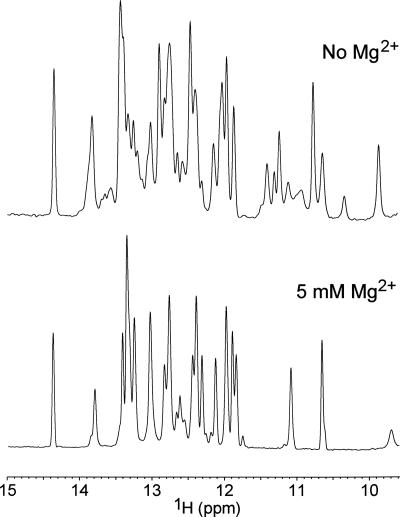
Initially the 1D imino proton spectra of the guanine free GRA were recorded in 10 mM sodium phosphate buffer, at pH 6.7. However, under these buffer conditions, the resulting spectra were rather crowded, showing many resonances with different linewidths (Fig. 2), suggesting multiple conformations that prevented complete resonance assignment. Therefore, we next tried to improve the spectra by dialyzing magnesium chloride into the sample. It is well known that magnesium ions are necessary for the tertiary structure formation of many RNA sequences (Misra and Draper 1998). Furthermore, it has been shown that Mg2+ ions are required for guanine binding (Mandal et al. 2003). Also, in the crystal structures divalent ions seem to play a stabilizing role in the interaction between the P2 and P3 helices, by neutralizing unfavorable electrostatic interactions (Batey et al. 2004; Serganov et al. 2004). Indeed, the spectra were greatly simplified, as demonstrated by a comparison of the imino proton spectra without Mg2+ and with 5 mM Mg2+ presented in Figure 2.
FIGURE 2.
One-dimensional 1H NMR spectra of the free GRA recorded in 93% H2O/7% D2O solvent with 10 mM sodium phosphate, pH 6.7, at 800 MHz, showing the imino proton region. The spectra were measured without magnesium (top) and with 5 mM Mg2+ (bottom).
In the absence of Mg2+ ions, the Watson–Crick base pair region between 12 and 15 ppm shows many overlapping resonances with different linewidths and intensities, which changes into a well-resolved spectrum with many fewer resonances and of similar width after dialysis into 5 mM Mg2+. In the non-Watson–Crick base pair region of the spectrum, i.e., between 9 and 12 ppm, the difference is even more profound, showing at least six out of eight resonances to disappear between 10 and 11.5 ppm. The crowded spectrum of the free GRA in the absence of Mg2+ ions yielding many additional resonances with different linewidths is indicative of multiple species with different dynamic properties. This likely represents a mixture between folded and unfolded states, lacking the long-range loop–loop interaction as demonstrated for the adenine-sensing riboswitch aptamer by a single-molecule FRET study (Gilbert and Batey 2006; Lemay et al. 2006). Apparently magnesium ions are required to force the free GRA into a single stable conformation, illustrating their structural and biological importance.
Resonance assignment and solution structure of the GRA–guanine complex
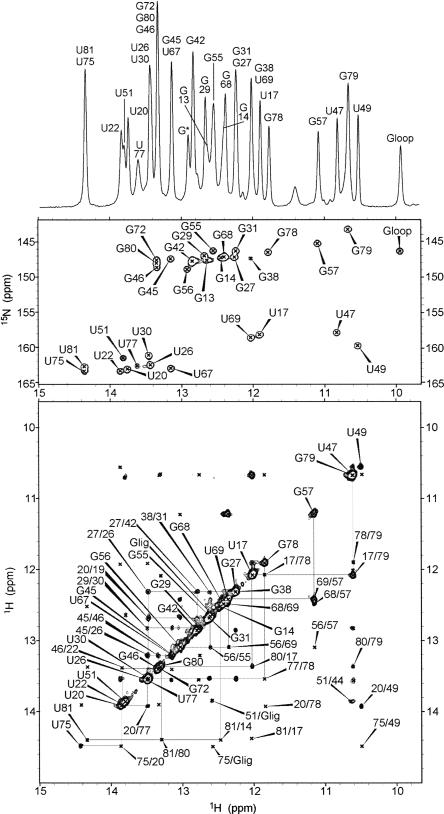
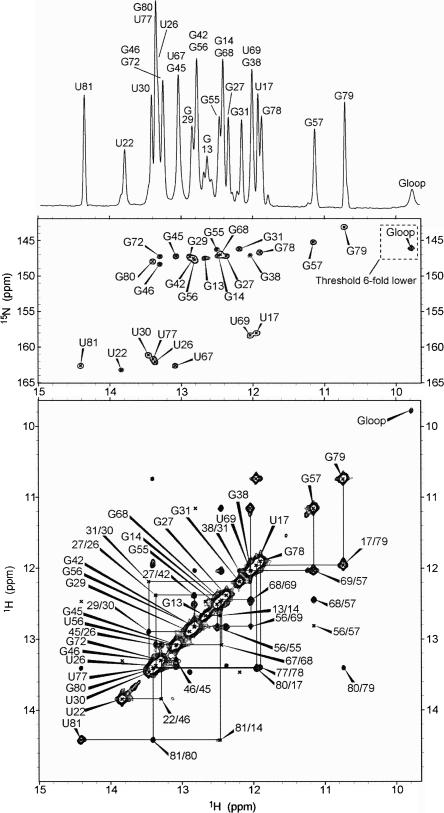
We next recorded NMR spectra of the GRA–guanine complex in the presence of 5 mM Mg2+. The quality of the NMR spectra is excellent for a molecule of this size, showing many well-resolved imino proton resonances with small linewidths that allow for nearly complete resonance assignment. Imino protons were assigned (Fig. 3) using standard assignment protocols (Wijmenga and van Buuren 1998), involving sequential imino–imino NOE contacts assigned to residue type by separation of U and G imino resonances in 15N HMQC spectra. To resolve some ambiguous NOE contacts involving overlapping resonances, both the P2 and P3 stem–loops were also synthesized and investigated by NMR separately (O.M. Ottink, S.M. Rampersad, M. Tessari, H.A. Heus, and S.S. Wijmenga, unpubl.).
FIGURE 3.
NMR spectra of the GRA–guanine complex recorded in 93% H2O/7% D2O solvent with 10 mM sodium phosphate, 5 mM Mg2+Cl2, pH, 6.7 at 800 MHz. 1D imino proton spectrum of the complex (top), with resonance assignments indicated by residue type and number. The corresponding 15N–1H HMQC (middle) spectrum and 2D 1H–1H NOESY spectrum (bottom), showing the same imino proton region. Assignments of the crosspeaks in the HMQC and diagonal peaks in the NOESY are indicated by residue type and number. Crosspeaks in the NOESY are indicated by residue numbers involved. Sequential NOE connectivities are indicated by lines.
All stem imino protons could be assigned, indicating formation of all base pairs in the three stems with the exception of the closing G12C84 pair, in which the imino proton is not observable due to terminal fraying. The other resonances thus originate from hydrogen-bonded imino protons outside the stem regions, of which several could be assigned to residues within the junction loops. U22 and G46, which form U22A52 and G46C53 base pairs in the cocrystal structure, are also base paired in the solution structure and form a continuous stack on P1, as shown by the chemical shift positions and sequential NOE walk. Using the crystal structure as basis (Batey et al. 2004; Serganov et al. 2004), which helps in achieving self-consistent assignment, the imino proton resonances of the J2–3 loop residues U47, U49, and U51, as well as the imino proton of the bound guanine could be assigned, and together with the observed NOEs is in accordance with a folded J2–3 loop structure enclosing the ligand. U47 and U51 are nearby, and crucial NOE contacts position the ligand surrounded along its periphery by U51 and U75 and the J2–3 loop close to the upper base pairs of P1. Interestingly, the observation of an NOE between the U47 and U49 imino protons, which are 7.4 Å apart in the crystal structure, suggests enhanced stacking and a tighter fold of the central core in the solution structure. In support of the long-range L2–L3 tertiary interaction, we were able to assign L2 residue G38 by an NOE contact with the G31 imino proton. The chemical shift position of the G38 imino proton agrees with formation of a Watson–Crick base pair, corresponding to the L2 G38–L3 C60 base pair, indicative of the presence of the L2–L3 loop–loop interaction. The near-complete assignment leaves only a few imino protons of loop nucleotides unassigned, i.e., the remainder of L2, J2–3 U48, and L3 G62 and U63. However, when we again take the crystal structure (Batey et al. 2004; Serganov et al. 2004) as basis, complete exposure to solvent of the bases of L2 U36 and J2–3 U48 as well as the imino protons of G62 and U63 is unlikely to give an imino proton signal for these residues due to exchange with water. The broad guanosine resonance at 11.4 ppm, which only shows up in the 1D spectrum, can thus be assigned to either L2 G32 or G37. Judged from its resonance position, this could well be G32, which is unpaired but stacked within the L2–L3 kissing loop region in the crystal structure. In summary, the imino proton assignment and observed NOE contacts show the presence of the P1–P3 stems and also reveal many contacts involving interactions of junction loop residues and bound ligand that compare to the tightly packed central core observed in the crystal structures. Besides these, we were also able to identify a crucial contact that is consistent with formation of the long-range L2–L3 interaction. Thus the NMR structural data, inferred from the imino proton patterns and the many NOEs involved, perfectly match the crystal structure of the bound GRA, indicating a high structural homology with the solution structure.
The GRA–guanine complex in solution closely resembles the crystal structure
To further define the resemblance of the solution and crystal structures of the GRA-guanine complex, we also measured magnetic-field-induced RDCs (mRDCs) and compared those with calculated values using the crystal structure as model. RDCs provide long-range spatial information that is orientational rather than distance-based, and in general they are used in NMR structure determination for refinement and domain orientation by fitting RDC values to calculated structures. Thus the degree of correlation between the experimental and calculated mRDCs can be taken as a measure for the structural comparison of the solution and the target crystal structure.
This comparison is legitimate, provided structurally equivalent N–H and C–H bonds are used, and the differences between the sequence of the NMR construct and the natural xpt-pbuX sequence (see Fig. 1) also relate to the crystal structures. Thus, the H–N mRDC for C25G45 was compared with the corresponding U25A45 base pair of the crystal structure. Similarly, due to the insertion of the C16–G80 base pair, the value for C16–G80 was compared to A16U80 in the crystal structure and A15U81 to G15C81. The G14C82 and U26A44 base pairs could not be used for comparison since either the corresponding base pair is absent or does not contain an imino proton in the crystal structure. Finally, the mRDC for G56 could not reliably be derived due to resonance overlap. Using the NOESY spectrum in H2O, the imino proton assignment was extended to the H2 protons of the eight AU base-paired adenosine residues in the three stems, yielding a total of 31 mRDCs.
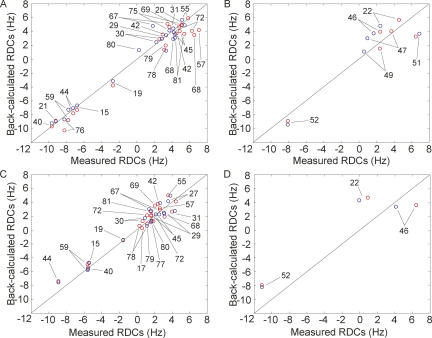
The asymmetric elongated shape of the GRA–ligand complex suggests susceptibility for magnetic field alignment, and indeed the mRDCs could be derived by measuring one-bond N–H or C–H splittings at three different field strengths (see Materials and Methods) or by using a J-coupling/chemical shift relationship (Grzesiek et al. 2004). Figure 4A shows the comparison between the 25 mRDCs measured for residues located in the three stems and those predicted from the crystal structure. The stem mRDCs are well spread out over the three stems (P1: 10; P2: 8; P3: 7), providing a reliable comparison of the stem orientations as only a small set of (m)RDCs is required to determine the global structure of branched nucleic acids (Mollova et al. 2000; van Buuren et al. 2004). The measured mRDCs correlate quite well with the predicted values, calculated from the crystal structure model. The mean deviation (0.8 Hz), which is on the order of the experimental error, corresponds to a difference of ∼4°–8°, depending on the N–H or C–H bond orientation, and the highest deviation of 3 Hz corresponds to a difference of ∼11°–18°. Assuming no local differences in the helical regions between the solution structure and the structure in the crystal, these small deviations indicate that the stem orientations of the solution and crystal structures are highly similar and adopt the same fork-like structures, i.e., with parallel P2 and P3 stems pointing in one direction and P1 pointing in the opposite direction.
FIGURE 4.
Comparison of the measured and predicted RDCs values from the crystal structure model (Serganov et al. 2004) of the GRA–guanine complex. (A) Stem residues of the GRA–guanine complex. (B) Residues located in the three-way junction of the guanine-bound complex. (C) Stem residues of the free GRA aptamer. (D) Residues located in the three-way junction of the free GRA. The numbers represent the G-riboswitch aptamer's residues. Numbers indicate residues of the GRA. Red circles indicate RDCs derived using the magnetic field strength dependence (500, 600, and 800 MHz), and blue circles indicate RDCs derived using the 15N J-coupling/chemical shift relation (Grzesiek et al. 2004).
To assess the structural comparison of the central core, six N–H and C2–H2 mRDCs, representing the majority of junction nucleotides, could be used for the correlation shown in Figure 4B. The residual deviations of these mRDCs are similar to those of the mRDCs of the stem residues. The N–H mRDCs of U22, G46, and A52 match quite well. These mRDCs correlate with formation of the two base triples, A73:U22–A52 and A23:G46–C53, closing in the ligand from above. In agreement with formation of the base quadruple (C74–G:U47,U51) surrounding the bound ligand, the N–H mRDC of U51 shows a deviation of ∼3 Hz, and only a small deviation is seen for U47. Finally, further evidence for existence of the two base triples, A21–U75:C50 and U20–A76:U49, closing in the ligand from below and bringing the J2–3 loop and P1-stem together is provided by the N–H mRDC of U49 that matches also quite well, as do the values for U22 and U75. Thus, the mRDC values also match the crystal structure for the region encompassing the binding pocket.
Structural organization of the free GRA
In order to gain insight into the structural rearrangements that takes place upon ligand binding, we compared the NMR spectra of the free guanine riboswitch with the bound aptamer and performed a similar mRDC analysis. The 1D imino proton and 15N-HMQC spectra of the free and the guanine-bound states are very similar, suggesting largely similar structures. Only a limited number of resonances are not observed for the unbound state, and small chemical shift changes are observed for the common resonances (Figs. 3, 5). Nearly all stem imino proton resonances observed for the bound state are also present in the spectra of the free state, with the exception of U20 and U75 of the upper two P1 stem base pairs adjacent to the three-way junction. Importantly, the mRDC-based analysis of the free GRA indicates a global fold with stem orientations similar to the bound form in the crystal structure. The 17 N–H imino mRDCs and five C2–H2 mRDCs of the stem residues (P1, 7; P2, 8; P3, 7) correlate quite well, as is observed for the GRA–guanine complex (Fig. 4).
FIGURE 5.
NMR spectra of the free GRA in water, with conditions as indicated in the legend of Figure 3. (Top) The 1D imino proton spectrum. (Middle) 15N–1H HMQC spectrum. (Bottom) 2D NOESY spectrum. Assignment indication is as in Figure 3.
Besides the majority of P1–P3 stem-imino-proton resonances, some resonances remain which originate from residual tertiary interactions in the central domain. U22 and G46 are still discernible in the spectra of the unbound state, and the mRDC values confirm presence of the U22–A52 and G46–C50 base pairs. On the other hand, in addition to the two P1 residues, U20 and U75 mentioned above, the majority of junction nucleotides in the free state do not yield a detectable imino proton resonance, as a result of enhanced imino proton exchange with water, suggesting a more disordered central core in the absence of bound ligand. It seems that the structural organization of the central core in the free state we observe goes beyond that proposed by Schwalbe and co-workers (Noeske et al. 2007). This research group proposed that all central core interactions and adjacent stem base pairs are not involved in any stabilizing interaction, based on the absence of imino proton resonances (Noeske et al. 2007). This difference might originate from different buffer conditions, as these studies were conducted in the absence of Mg2+, which most likely further destabilizes the central core, thus emphasizing the importance of Mg2+ ions in RNA folding.
In summary, the resonance assignment and mRDC comparison essentially show that the solution structure of the free GRA closely resembles the bound state in the GRA–guanine complex. The structure is preorganized in the free state with the three stems in the fork-like structure surrounding a partially unfolded central core.
Structural rearrangements upon ligand binding
Besides the presence or absence of imino proton resonances and a mRDC comparison as described above, proton chemical shifts, which are highly sensitive for structural variation due to large aromatic ring current effects (Cromsigt et al. 2001), also provide a powerful tool to monitor conformational changes. The chemical shift perturbation, the 1H-chemical shift difference between the free and guanine-bound form, is mapped onto the secondary structure of the GRA, shown in Figure 6A. The average 1H-chemical shift difference of the imino and adenine C2–H2 groups is only 0.16 ppm, again illustrating the great overall structural resemblance. Also, the imino proton resonances of U22 and G46, which are involved in the A73:U22–A52 and A23:G46–C53 base triples, do not shift significantly. The largest changes map around the binding pocket, although some of the P2 and P3 stem residues near the L2 and L3 loop are affected as well. The latter changes might represent tighter folding of the bound state, with the P2 and P3 arms somewhat closer together in the bound state than in the free RNA. As described above, the imino proton resonances, which are present in the spectra of the bound state but absent in the NMR spectra of the free state, originate from U20 and U75 of the P1 stem; the J2–3 loop nucleotides U47, U49, and U51; and the guanine ligand.
FIGURE 6.
(A) Chemical shift perturbation map indicating structural differences between the free and ligand-bound states of the GRA. Blue letters indicate residues that experience chemical shift changes of imino protons resonances <0.25 ppm or for which no NMR data are available; red letters indicate residues with chemical shift changes >0.25 ppm or those that show a peak in the spectrum of the G-bound state, which is absent in the free state. (B) Schematic representation of the solution structures derived by NMR of the GRA in the free and guanine-bound forms.
The chemical shift perturbation together with the NMR data described in the earlier paragraphs provide a detailed account of the nucleotides involved in the conformational changes that occur in the GRA upon ligand binding. One key observation is pre-existence of the three stems and two central core base triples, A73:U22–A52 and A23:G46–C53, in the free state, which close in the ligand from above in the bound state. These two base triples also anchor the P2–P3 stems at the junction interface, which together with pre-existence of the L2–L3 interaction accounts for the identical P1 and P2 stem orientations we observe for the free and bound states. Another key observation is the absence of the other two base triples, A21–U75:C50 and U20–A76:U49,U47, closing in the ligand from below in the bound state and disordered nucleotides surrounding the guanine in the base quadruple (C74–G:U47,U51). Thus, prior to ligand binding, the free GRA appears to be largely organized, with preformation of the three stems and base triples facing P2 and P3, but disordered in the lower part of the central core and the upper part of P1.
Mechanism of riboswitching
A wide body of structural and biophysical evidence is available suggesting the GRA is partially preorganized by the stems and long-range loop–loop interaction, while the junction and surrounding regions have to be partially disordered to allow binding of the ligand. Our NMR study can be used to evaluate this model and further specify the conformational changes on a residue-specific basis. The NMR structural data confirm existence of a preorganized structure in the free state containing the P1–P3 stem–loops including the L2–L3 interaction surrounding a disordered central core, as previously suggested by in-line probing experiments and earlier NMR studies (Mandal et al. 2003; Serganov et al. 2004; Noeske et al. 2007). The in-line probing studies revealed that nearly all central core residues and adjacent base pairs become protected from cleavage upon binding of the ligand, while nearly all P1–P3 stem as well as the L2 and L3 residues are protected, whether the ligand is present or not. Only U36, U48, and U63 are susceptible for cleavage both in the free and bound states. A profound difference between our study and the in-line probing and earlier NMR studies is that we observe pre-existence of all base pairs facing the central core and the two base triples that assist in anchoring the P2 and P3 helices. However, the in-line probing studies (Mandal et al. 2003) were conducted in different buffer conditions (e.g., an elevated pH of 8.5) to promote phosphodiester bond cleavage, and the earlier NMR studies (Noeske et al. 2007) were conducted in the absence of Mg2+, which, as we mentioned above, might further destabilize the central core and surrounding stem base pairs. We should also point out that we modified the two base pairs of the P2 arm facing the binding core, in order to stabilize this helix. These may assist in preorganization of the adjacent core residues to an extent greater than for the natural RNA. There are, however, examples of guanine riboswitches where the P2 arm has similar stabilizing base pairs adjacent to the core (Mandal et al. 2003).
Using the key observations described in the last paragraph, we can propose a multistep model for the sequential folding pathway induced by ligand binding (Fig. 6B). In the first step, the ligand is able to access the preorganized GRA and enters a partially disordered binding pocket by forming a base pair with C74, which in the initial binding event can be further stabilized by stacking interactions with the existing base triple above, i.e., A73:U22–A52. Subsequently, formation of the base quadruple surrounds the ligand, and finally the ligand is closed in by formation of the two base triples, which close in the ligand from below. Preformation of the two base triples implies that the site of entry of the ligand is on the J2–3 side of the binding pocket. In the last folding step, after formation of the two upper Watson–Crick base pairs in P1, the J2–3 loop can thus be viewed as a lid that closes in the ligand on the N9/N3 side and simultaneously fastens the J2–3 loop to the P1 stem by forming the tertiary interactions. A salient feature of this proposed predetermined–induced fit model of ligand binding is that only limited conformational changes in the central core are required to drive the GRA to its stable bound-state configuration, which is an important aspect for the mechanism of riboswitching. In the structure we observe for the free GRA, only C50 and C74 are accessible to form a Watson–Crick base pair with the incoming ligand, which possibly promotes fast recognition without the need for further proofreading. This suggests an additional role for a preformed GRA in the free state, since more disorder would render more cytidines accessible, which might lead to unproductive binding. While this consideration seems less important for the GRA, with only two accessible acceptors to form the Watson–Crick base pair, it might be more important for the structurally related adenine riboswitch where the many conserved uridines in the central core might further enhance unproductive binding. Besides the intrinsic property of a A–U base pair to form one less hydrogen bond, this might contribute to the apparent lower affinity of the adenine aptamer for its cognate ligand, with a dissociation constant (K d) of 0.5 mM, i.e., 100-fold higher than the K d value reported for the GRA–guanine interaction (5 nM) (Gilbert et al. 2006).
The limited structural rearrangement upon ligand binding that we propose nicely corresponds with the thermodynamic data reported by Batey and co-workers (Gilbert et al. 2006). Assuming temperature independent enthalpy and entropy values, the reported values for guanine binding correspond with a ΔG 37 of −11.0 kcal/mol in accordance with formation of roughly five to six Watson–Crick base pairs (Serra and Turner 1995), which matches well to formation of the remainder of the central core interactions and the two upper base pairs in P1. Assuming that we are correct in modeling the upper two base pairs of P1 as disordered in the unbound state, a total energy gain of −19.5 kcal/mol at 37°C can be predicted for locking the aptamer structure around guanine by adding the energy gain (Serra and Turner 1995) for formation of the lower six base pairs of P1, i.e., nucleotides 14–19 with 77–82 (Fig. 1B). This value is comparable to the predicted thermodynamic stability of the antiterminator, i.e., the structure with which ligand binding to the aptamer has to compete, which in case of the xpt-pbuX riboswitch amounts to −17.5 kcal/mol.
Increasing evidence is emerging that RNA folding proceeds via a hierarchic folding pathway in which highly structured RNAs form through compaction of preformed secondary-structure elements (Brion and Westhof 1997). In this view secondary structure elements, such as helices or hairpin–loops are formed first, which subsequently interact locally through coaxial stacking or pseudoknot or triple-helix interactions. Finally, these independently folded subdomains interact cooperatively by loop–loop and numerous contacts involving loops, bulges, and helices. Preorganization of the free GRA, followed by the multistep sequential folding pathway that we propose occurs upon ligand binding, provides a classic example of such a hierarchic folding pathway on the one hand, but on the other hand shows how the unique folding capacity of RNA can be adapted for a specific purpose. By making effective use of the folding landscape of RNA, the aptamer is folded to such an extent that binding specificity is secured, but at the same time a sufficient stretch is left unstructured to ensure quick response to ligand binding for transcription control. We expect other riboswitches to operate via this combined predetermined–induced fit mechanism as well, and it will for instance be interesting to see how this mechanism is adapted by structurally dissimilar riboswitches, such as the S-adenosylmethionine riboswitch, which also displays a high affinity for its cognate metabolite (K d=4 nM) (Soukup and Soukup 2004).
RNAs that change their shape in response to external stimuli, such as the riboswitch–metabolite interaction, are ideal tools for molecular design, for instance, in the development of nanomechanical devices and synthetic RNA circuits (e.g., Davidson and Ellington 2007), due to its great predictive power in constructing secondary- and tertiary-structure elements in combination with functional diversity. To be able to build such devices, prior knowledge of the three-dimensional structures and concomitant RNA folding processes remains a necessity. The detailed structural description of the GRA–guanine interaction we have presented provides such a structural framework for riboswitch utilization. It will, for instance, be interesting to see if this framework can be used to engineer an adenine-responsive aptamer (ARA) with high adenine affinity by mutating the uridine-rich core while preserving the compact three-dimensional structure.
MATERIALS AND METHODS
RNA sample preparation
A double-stranded DNA fragment coding for the GRA (Fig. 1) and flanked by a T7 promoter sequence was cloned into a pUC18 vector. Unlabeled and uniformly 13C/15N-labeled G-riboswitch aptamer samples were prepared by in vitro run-off transcription using the linearized plasmid and T7 RNA polymerase (Milligan and Uhlenbeck 1989). The products were purified using denaturing PAGE, collected by electroelution, and dialyzed against NMR buffer (10 mM sodium phosphate, pH 6.7). Magnesium was added to the solution by dialyzing the samples against the NMR buffer containing the appropriate concentration of MgCl2. The final concentrations of the NMR samples were ∼0.2 and 0.7 mM for the unlabeled G-riboswitch and uniformly labeled G-riboswitch, respectively.
Complex formation
Because of the low solubility of guanine in H2O, the GRA–guanine complexes were prepared at low concentration. For the unlabeled GRA–guanine complex, this was achieved by mixing together 1 mL of a 60 μM G-riboswitch solution with 3.5 mL of a 21 μM guanine solution, both dissolved in NMR buffer containing 5 mM MgCl2. The sample was subsequently concentrated to 300 μL using a 10-kDa cut-off Centricon tube (Amicon). The final concentration of the NMR sample was 0.2 mM. Using the same protocol, the uniformly 13C/15N-labeled GRA-guanine complex was prepared in a concentration of 0.7 mM.
NMR experiments
All spectra were recorded at 5 and 15°C on 500- and 800-MHz Varian Inova spectrometers equipped with cryoprobes and on a 600-MHz Varian Inova spectrometer. NOESY spectra in water (93% H2O, 7% D2O) were recorded with 100-, 200-, and 300-msec mixing times. Nitrogen-decoupled 15N-HMQC spectra were also recorded in 93% H2O/7% D2O. The water signal was suppressed either by a selective 270° sine-shaped pulse in combination with a Watergate water suppression scheme (Piotto et al. 1992) or with a jump–return pulse (Plateau and Gueron 1982) in combination with a Watergate sequence. 15N-HMQC-IPAP spectra (Ottiger et al. 1998) were measured to derive the 1H–15N splittings. Adenine C2–H2 correlations were measured in water by 13C-HSQC experiments using a constant time evolution period to refocus carbon–carbon couplings. 1H–13C splittings were measured by recording the 13C-HSQC spectra in an IPAP fashion. All spectra were processed with NMRpipe (Delaglio et al. 1995) and assigned using the Sparky software (Kneller and Kuntz 1993).
RDC measurements
The asymmetric elongated shape of the 25-kDa GRA created sufficient susceptibility for magnetic field alignment for derivation of mRDCs by measuring one-bond N–H or C–H splittings at three different field strengths or by using a J-coupling/chemical shift relationship (Grzesiek et al. 2004). In the first approach, the imino 15N–H RDCs of the aptamer are derived by measuring the splittings at 500-, 600-, and 800-MHz proton frequencies. The measured splitting relates to the magnetic field strength, following the equation J(Hz) =J 0(Hz)+k(1/MHz) ×B 2(MHz), with J the total splitting, J 0 the isotropic J coupling, B the magnetic field strength, and k the RDC per magnetic-field-strength unit. Thus, by plotting the measured splittings against the square of the magnetic field strength, the RDC values can be derived from the slope of the fitted straight line. Multiplying the value for the slope with the square of a certain magnetic field strength yields the RDC value at that field strength. The C2–H2 RDCs were derived by calculating the difference between the measured splitting and the theoretical value of 203.2 Hz for AMP. The other approach makes use of the linear correlation between the strength of the imino 15N–H scalar couplings involved in N–H–N hydrogen bonds and their chemical shift value following the equation 1 J NH = (1.016 Hz/ppm)δHN − 100.58 Hz (Grzesiek et al. 2004). Using this equation, the theoretical J couplings are calculated. The RDC is then the difference between the measured splitting and the calculated scalar coupling.
Module software (Dosset et al. 2001) was used to derive the alignment tensor and back-calculated RDCs from the PDB coordinate file (1Y27) of the guanine-bound GRA crystal structure (Serganov et al. 2004) and our measured values. The entire system was considered as one structural entity. For obtaining A a and A r values given by Module, only N–H RDCs from residues located in the three stem regions were used. From the A a and A r values, the D a NH and D a CH values were obtained by multiplying A a with (μ0 hγHγN,C)/16(πr NH,CH)3, assuming N–H and C–H bond lengths of 1.02 and 1.09 Å, respectively. The R values were obtained by dividing A r by A a. A a and A r are the unitless axial and rhombic components of the molecular alignment tensor A, respectively. The back-calculated RDCs were directly given by Module. All values were obtained assuming an order parameter of 1. The D a NH,CH and R values, derived for the bound and unbound states of the GRA are listed in Table 1.
TABLE 1.
A a, A r, D a, and R values for the bound and unbound states of the G-riboswitch aptamer
SUPPLEMENTAL DATA
Figures showing the 1D imino proton, 2D NOESY spectra, and imino proton assignments of the P2 and P3 arms, and bond-angle deviation–RDC deviation correlation plots of the guanine-bound GRA and tables with measured and back-calculated mRDCs for the guanine-bound and guanine-free GRA are available upon request from the authors (send an e-mail message to h.heus@science.ru.nl).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.635307.
REFERENCES
- Batey, R.T., Gilbert, S.D., Montange, R.K. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- Brion, P., Westhof, P. Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- Cromsigt, J.A.M.T.C., Hilbers, C.W., Wijmenga, S.S. Prediction of proton chemical shifts in RNA. Their use in structure refinement and validation. J. Biomol. NMR. 2001;21:11–29. doi: 10.1023/a:1011914132531. [DOI] [PubMed] [Google Scholar]
- Davidson, E.A., Ellington, A.D. Synthetic RNA circuits. Nat. Chem. Biol. 2007;3:23–28. doi: 10.1038/nchembio846. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., Bax, A. NMRpipe: A multidimensional spectral processing system based on Unix pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dosset, P., Hus, J.C., Marion, D., Blackledge, M. A novel interactive tool for rigid-body modeling of multidomain macromolecules using residual dipolar couplings. J. Biomol. NMR. 2001;20:223–231. doi: 10.1023/a:1011206132740. [DOI] [PubMed] [Google Scholar]
- Epshtein, V., Mironov, A.S., Nudler, E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl. Acad. Sci. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, R.T., Grundy, F.J., Henkin, T.M. The S(MK) box is a new SAM- binding RNA for translational regulation of SAM synthetase. Nat. Struct. Mol. Biol. 2006;13:226–233. doi: 10.1038/nsmb1059. [DOI] [PubMed] [Google Scholar]
- Gilbert, S.D., Batey, R.T. Riboswitches: Fold and function. Chem. Biol. 2006;13:805–807. doi: 10.1016/j.chembiol.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Gilbert, S.D., Stoddard, C.D., Wise, S.J., Batey, R.T. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J. Mol. Biol. 2006;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Grundy, F.J., Lehman, S.C., Henkin, T.M. The L box regulon: Lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc. Natl. Acad. Sci. 2003;100:12057–12062. doi: 10.1073/pnas.2133705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzesiek, S., Cordier, F., Jaravine, V., Barfield, M. Insights into biomolecular hydrogen bonds from hydrogen bond scalar couplings. Prog. Nucl. Magn. Reson. Spectrosc. 2004;45:275–300. [Google Scholar]
- Kneller, D.G., Kuntz, I.D. UCSF Sparky: An NMR display, annotation and assignment tool. J. Cell. Biochem. 1993;53:254. [Google Scholar]
- Lemay, J.F., Penedo, J.C., Tremblay, R., Lilley, D.M., Lafontaine, D.A. Folding of the adenine riboswitch. Chem. Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Mandal, M., Breaker, R.R. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004a;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- Mandal, M., Breaker, R.R. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 2004b;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- Mandal, M., Boese, B., Barrick, J.E., Winkler, W.C., Breaker, R.R. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- Mandal, M., Boese, B., Barrick, J.E., Winkler, W.C., Breaker, R.R. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- McDaniel, B.A., Grundy, F.J., Artsimovitch, I., Henkin, T.M. Transcription termination control of the S box system: Direct measurement of S-adenosylmethionine by the leader RNA. Proc. Natl. Acad. Sci. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan, J.F., Uhlenbeck, O.C. Synthesis of small RNAs using T7 RNA-polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- Mironov, A.S., Gusarov, I., Rafikov, R., Lopez, L.E., Shatalin, K., Kreneva, R.A., Perumov, D.A., Nudler, E. Sensing small molecules by nascent RNA: A mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- Misra, V.K., Draper, D.E. On the role of magnesium ions in RNA stability. Biopolymers. 1998;48:113–135. doi: 10.1002/(SICI)1097-0282(1998)48:2<113::AID-BIP3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Mollova, E.T., Hansen, M.R., Pardi, A. Global structure of RNA determined with residual dipolar couplings. J. Am. Chem. Soc. 2000;122:11561–11562. [Google Scholar]
- Nagel, J.H., Gultyaev, A.P., Oistamo, K.J., Gerdes, K., Pleij, C.W. A pH-jump approach for investigating secondary structure refolding kinetics in RNA. Nucleic Acids Res. 2002;30:e63. doi: 10.1093/nar/gnf057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahvi, A., Sudarsan, N., Ebert, M.S., Zou, X., Brown, K.L., Breaker, R.R. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002;9:1043–1049. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- Noeske, J., Richter, C., Grundl, M.A., Nasiri, H.R., Schwalbe, H., Wohnert, J. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc. Natl. Acad. Sci. 2005;102:1372–1377. doi: 10.1073/pnas.0406347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noeske, J., Buck, J., Furtig, B., Nasiri, H.R., Schwalbe, H., Wohnert, J. Interplay of “induced fit” and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic Acids Res. 2007;35:572–583. doi: 10.1093/nar/gkl1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler, E., Mironov, A.S. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Ottiger, M., Delaglio, F., Bax, A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J. Magn. Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- Piotto, M., Saudek, V., Sklenar, V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- Plateau, P., Gueron, M. Exchangeable proton NMR without base-line distortion, using new strong-pulse sequences. J. Am. Chem. Soc. 1982;104:7310–7311. [Google Scholar]
- Rodionov, D.A., Vitreschak, A.G., Mironov, A.A., Gelfand, M.S. Regulation of lysine biosynthesis and transport genes in bacteria: Yet another RNA riboswitch? Nucleic Acids Res. 2003;31:6748–6757. doi: 10.1093/nar/gkg900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov, A., Yuan, Y.R., Pikovskaya, O., Polonskaia, A., Malinina, L., Phan, A.T., Hobartner, C., Micura, R., Breaker, R.R., Patel, D.J. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra, M.J., Turner, D.H. Predicting thermodynamic properties of RNA. Methods Enzymol. 1995;259:242–261. doi: 10.1016/0076-6879(95)59047-1. [DOI] [PubMed] [Google Scholar]
- Soukup, J.K., Soukup, G.A. Riboswitches exert genetic control through metabolite-induced conformational change. Curr. Opin. Struct. Biol. 2004;14:344–349. doi: 10.1016/j.sbi.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Sudarsan, N., Nahvi, A., Roth, A., Collins, J.A., Breaker, R.R. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes & Dev. 2003;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Buuren, B.N.M., Schleucher, J., Wittmann, V., Griesinger, C., Schwalbe, H., Wijmenga, S.S. NMR spectroscopic determination of the solution structure of a branched nucleic acid from residual dipolar couplings by using isotopically labeled nucleotides. Angew. Chem. 2004;43:187–192. doi: 10.1002/anie.200351632. [DOI] [PubMed] [Google Scholar]
- Wickiser, J.K., Cheah, M.T., Breaker, R.R., Crothers, D.M. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- Wijmenga, S.S., van Buuren, B.N.M. The use of NMR methods for conformational studies of nucleic acids. Prog. Nucl. Magn. Reson. Spectrosc. 1998;32:287–387. [Google Scholar]
- Winkler, W., Nahvi, A., Breaker, R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002a;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- Winkler, W.C., Cohen-Chalamish, S., Breaker, R.R. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. 2002b;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, W.C., Nahvi, A., Sudarsan, N., Barrick, J.E., Breaker, R.R. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Mol. Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- Winkler, W.C., Nahvi, A., Roth, A., Collins, J.A., Breaker, R.R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]