Abstract
Small noncoding RNAs have been suggested to play important roles in the regulation of gene expression across all species from plants to humans. To identify small RNAs expressed by the ovary, we generated mouse ovarian small RNA complementary DNA (srcDNA) libraries and sequenced 800 srcDNA clones. We identified 236 small RNAs including 122 microRNAs (miRNAs), 79 piwi-interacting RNAs (piRNAs), and 35 small nucleolar RNAs (snoRNAs). Among these small RNAs, 15 miRNAs, 74 piRNAs, and 21 snoRNAs are novel. Approximately 70% of the ovarian piRNAs are encoded by multicopy genes located within the repetitive regions, resembling previously identified repeat-associated small interference RNAs (rasiRNAs), whereas the remaining ∼30% of piRNA genes are located in nonrepetitive regions of the genome with characteristics similar to the majority of piRNAs originally cloned from the testis. Since these two types of piRNAs display different structural features, we categorized them into two classes: repeat-associated piRNAs (rapiRNAs, equivalent of the rasiRNAs) and non-repeat-associated piRNAs (napiRNAs). Expression profiling analyses revealed that ovarian miRNAs were either ubiquitously expressed in multiple tissues or preferentially expressed in a few tissues including the ovary. Ovaries appear to express more rapiRNAs than napiRNAs, and sequence analyses support that both may be generated through the “ping-pong” mechanism. Unique expression and structural features of these ovarian small noncoding RNAs suggest that they may play important roles in the control of folliculogenesis and female fertility.
Keywords: small noncoding RNAs, oocytes, folliculogenesis, oogenesis, miRNAs, piRNAs, snoRNAs
INTRODUCTION
The human and mouse genome sequencing projects revealed that the total number of protein-encoding genes is between ∼20,000–25,000, which is much less than what was initially predicted (Venter et al. 2001; Waterston et al. 2002). These protein-encoding genes account for only ∼1%–2% of the entire genome. The remaining portions are noncoding regions consisting of intergenic and repetitive sequences, including transposons and retrotransposons, which were once believed to be functionally insignificant. This notion is being changed with the discovery of numerous small noncoding RNA species (Lagos-Quintana et al. 2001; Lau et al. 2001, 2006; Lee and Ambros 2001; Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Saito et al. 2006; Vagin et al. 2006; Watanabe et al. 2006; Ro et al. 2007a,b), which are mostly encoded by genes located in those previously thought “noncoding” or “junk” regions of the genome. Small noncoding RNAs were previously ignored largely due to their diminutive size (20–30 nucleotides [nt]) compared to mRNAs (hundreds to thousands of nucleotides) and also to the limitation of the cloning technology available at that time. In 2001, several studies reported the identification of a class of small noncoding RNAs with sizes between 22 and 24 nt, which were named micro-RNAs (miRNAs) (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001). Subsequent studies demonstrated that miRNAs are highly conserved across almost all species from plants to mammals, and they appear to function as post-transcriptional suppressors through binding to their target mRNAs by base-pairing and subsequently inducing either translational repression or mRNA destabilization (Bartel 2004). Further studies show that miRNAs are involved in the regulation of various cellular processes, including cell differentiation, cell proliferation, development, and apoptosis (Ambros 2004). Recent data also indicate that miRNAs are directly linked to viral diseases, neuronal development, and tumorigenesis (Chang and Mendell 2007; Zhang et al. 2007). These findings suggest that miRNAs are as important as transcriptional factors in the control of gene expression in higher eukaryotes.
A new class of small RNAs called piwi-interacting RNAs (piRNAs) were recently identified in the testis of the mouse, rat, and human (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Saito et al. 2006; Watanabe et al. 2006; Ro et al. 2007b). piRNAs are 26–31 nt in length and are abundantly expressed in the testis. These small RNAs bind PIWI, a spermatogenesis-specific protein belonging to the Argonaute protein family. piRNA genes appear to exist as clusters, and genomic distribution of the piRNA gene clusters are conserved among the mouse, rat, and human genomes. However, piRNA sequences are poorly conserved between distant species, such as between the mouse and human (Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Saito et al. 2006). In the mouse, rat, and human, more than 72,000 piRNA sequences have been registered in the GenBank (http://www.ncbi.nlm.nih.gov/Genbank/). However, the structural commonality, biogenesis, expression profiles, and function of piRNAs remain unknown.
Mammalian folliculogenesis is a complex process through which primordial follicles develop into preovulatory follicles followed by ovulation that releases mature oocytes. During follicular development, oocytes increase in size, which is accompanied by proliferation and differentiation of their surrounding granulosa and thecal cells. Estrogen production by granulosa cells increases with the development from primordial to preovulatory follicles. Estrogen and follicle-stimulating hormone (FSH) induce the expression of luteinizing hormone (LH) receptors on granulosa cells from the secondary follicles onward. In response to the LH surge, fully developed oocytes are released through ovulation. After ovulation, the remaining follicular structure undergoes luteinization and the former granulosa and thecal cells are transformed into follicular and thecal lutein cells, which produce progesterone and estrogens. The coordinated development of oocytes and their supporting somatic cells suggests that close communication exists between germ cells and their supporting somatic cells. This is believed to be achieved through paracrine factors secreted by these two types of ovarian cells (Matzuk et al. 2002). The complexity of folliculogenesis indicates that tightly regulated expression and interaction of a multitude of genes are required for successful oocyte development. A total of ∼100 genes have been shown to be essential for normal folliculogenesis in mice so far (Matzuk and Lamb 2002; Roy and Matzuk 2006). Given that recently identified small RNAs may have roles in the regulation of gene expression through translational suppression, mRNA destabilization, and chromatin remodeling, there should be numerous such small RNAs expressed in the ovary. These small RNAs may play a critical role in controlling the expression of genes essential for folliculogenesis and also the endocrine function of the ovary. If these small RNAs indeed are important regulators of gene expression during folliculogenesis, mutations in these small RNA genes may lead to defects in folliculogenesis and ovarian dysfunction. Reagents that can affect the function of these small RNAs may be applicable to future nonhormonal contraceptives. Therefore, we attempted to identify small RNA species expressed by the ovary using a cloning method that we recently developed (Ro et al. 2007b). Here we report that the mouse ovary expresses all four classes of known small noncoding RNAs and some of these small RNAs display ovary-preferential expression.
RESULTS
Small RNAs cloned from the mouse ovary
Using a small RNA cloning method that we recently developed (Ro et al. 2007b), we prepared two small RNA cDNA (srcDNA) libraries: one from 2-wk-old ovaries and the other from adult ovaries. Approximately 400 srcDNA clones from the 2-wk-old ovarian library and ∼400 from the adult ovarian library were sequenced. We obtained 681 reads (342 from 2-wk-old ovaries; 339 from adult ovaries). We further analyzed these sequences to determine their identity. Known miRNAs, piRNAs, and snoRNAs were identified by searching the miRBase (Ambros et al. 2003; Griffiths-Jones 2004; Griffiths-Jones et al. 2006) and GenBank (Altschul et al. 1990). Novel miRNAs were determined according to their sizes (20–24 nt), stem–loop formation in their precursor sequences, and homologs in other species in the miRBase. Novel piRNAs were determined based upon their sizes (24–40 nt), chromosomal location within the known piRNA gene clusters, and/or association with repetitive sequences. The remaining small RNAs with sizes of ∼65–200 nt were categorized as snoRNAs.
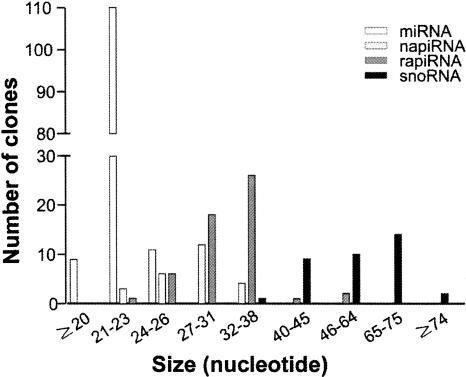
Our sequence analyses revealed that the 681 reads contained 236 small RNAs, including 122 miRNAs, 79 piRNAs, and 35 snoRNAs. By mapping the piRNAs onto the genome, we found that 54 out of the 79 piRNAs were derived from repetitive sequences, and their structural characteristics resembled those of the previously described repeat-associated small interference RNAs (rasiRNAs) (Saito et al. 2006; Vagin et al. 2006). Since rasiRNAs bind PIWI (Saito et al. 2006), they belong to the piRNA family by definition, as suggested by Lin (2007). To distinguish piRNAs that are not associated with repetitive sequences, we termed the ovarian rasiRNAs as repeat-associated piRNAs (rapiRNAs), and those derived from nonrepetitive sequences as non-repeat-associated piRNAs (napiRNAs). Among the 236 small RNAs cloned, we identified 110 novel small RNAs (15 miRNAs, 20 napiRNAs, 54 rapiRNAs, and 21 snoRNAs). Size appears to be the most visible difference among these four classes of small RNAs. By plotting small RNA size against total number of clones (Fig. 1), we found that the most common sizes are between 21 and 23 nt for miRNAs, 27 and 31 nt for napiRNAs, 32 and 38 nt for rapiRNAs, and 65 and 75 nt for snoRNAs.
FIGURE 1.
Size distribution of 236 small RNAs cloned from the mouse ovary. The most common sizes for the four classes of small RNAs identified are 21–23 nt for miRNAs, 27–31 nt for napiRNAs, 32–38 nt for rapiRNAs, and 65–75 nt for snoRNAs.
miRNAs cloned from the mouse ovary
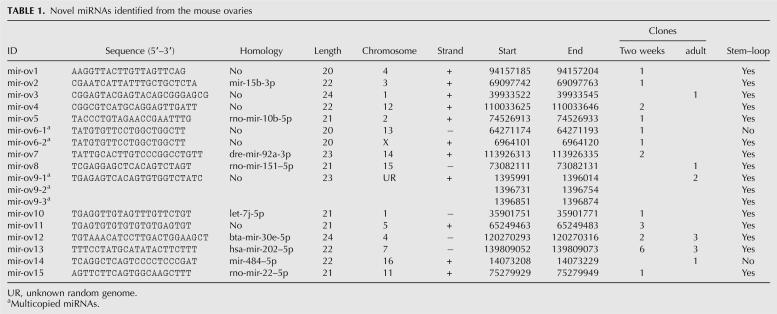
We cloned 122 miRNAs (for sequences see Supplemental Table 1) from the ovaries of 2-wk-old and adult mice, including 15 novel miRNAs (Table 1). Among the 15 novel miRNAs, seven (mir-ov5, 7, 8, 12, 13, 14, and 15) were found in other species, including rat, Zebrafish, cow, and human. mir-ov2 is the 3′ sister miRNA of the mouse mir-15b. mir-ov10 shared homology with mir-let-7i-5p (TGAGGTAGTAGTTTGTGCTGT) (Lagos-Quintana et al. 2001; Poy et al. 2004; Watanabe et al. 2006). To date, nine let-7 isoforms (a, b, c, d, e, f, g, h, and i) have been identified in various organisms and registered in the miRBase. However, since 2 nt are different in mir-ov10 (TGAGGTTGTAGTTTGTTCTGT), we believe that it is a new let-7 isoform and thus we named it “let-7j” (Table 1). Among the remaining 107 known miRNAs, 62 matched perfectly with the published sequences in the miRBase, whereas the other 45 showed 1–3-nt differences in either their 5′ end (3/45), 3′ end (41/45), or both (1/45) (Supplemental Table 1). These differences may be caused by the utilization of different 3′ and 5′ adaptors in our and the previous cloning methods (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001).
TABLE 1.
Novel miRNAs identified from the mouse ovaries
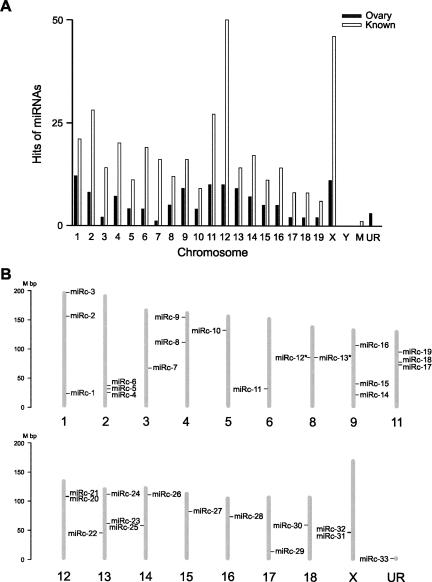
By mapping the 122 miRNAs cloned in this study and all of the known 368 mouse miRNAs registered in the miRBase onto the chromosomes, we found that all but the Y chromosome contain miRNA genes (Fig. 2A; Supplemental Table 2). Chromosomes 1, 11, 12, and X appear to contain more ovarian miRNA genes than other chromosomes, which is consistent with the chromosomal distribution of all of the known miRNA genes (Fig. 2A). The 122 miRNA genes identified in this study were derived from both the sense and antisense strands of the chromosomes (Supplemental Table 1). The sense strand appears to encode more miRNAs (64%, 77 miRNAs) than the antisense strand (36%, 45 miRNAs). By analyzing the proximity of the 122 miRNA genes in the mouse genome, we discovered 33 miRNA gene clusters (miRc) (Fig. 2B; Supplemental Table 3). Each of these miRNA gene clusters contains more than two miRNA genes, and has an average size of ∼8.0 kb. We also found two neighboring clusters, miRc-12 and miRc-13, displaying opposite orientation (Fig. 2B; Supplemental Table 3), suggesting these bidirectional miRNA gene clusters may be transcribed under the control of the same promoters.
FIGURE 2.
Genomic locations of miRNA genes and miRNA gene clusters identified in this study. (A) Chromosomal distribution of 122 miRNA genes identified in this study and 368 known miRNAs in the miRBase. The number of hits on chromosomes was calculated and graphed. M represents the mitochondrial genome and UR refers to unknown random genome. (B) Chromosomal distribution of miRNA gene clusters identified in this study. Mouse chromosomes are drawn to scale and aligned by their centromere position. Thirty-three miRNA clusters (miRcs 1–33) were located and numbered on each of the chromosomes. The minus and plus strands are located on the right and left, respectively. Bidirectional clusters located proximate on both strands are indicated by an asterisk (*).
Expression profiles of 15 novel miRNAs
To validate each of the 15 novel miRNAs cloned in this study, we examined their expression in 12 mouse tissues (Fig. 3A) using a semiquantitative PCR-based small RNA quantification method (Ro et al. 2006, 2007b). In general, all of the 15 novel miRNAs were abundantly expressed in the ovary and testis. mir-ov1, -ov5, -ov7, -ov10, -ov12, and -ov14 displayed an evenly ubiquitous expression pattern in the multiple tissues. Previously, we showed all of the nine let-7 isoforms were ubiquitously expressed in the tissues (Ro et al. 2006). The new isoform let-7j (mir-ov10) was also ubiquitously expressed in the tissues (Fig. 3A). mir-ov2, -ov4, -ov6, -ov8, -ov9, -ov11, and -ov15 were preferentially expressed in several but not all tissues examined, whereas mir-ov13 was almost exclusively expressed in the ovary, testis, and uterus (Fig. 3A).
FIGURE 3.
Expression profiles of 15 novel ovarian miRNAs. Amplicons were analyzed on 2% agarose gels. let-7a was used as a loading control. A DNA ladder on each side indicates the size of the fragments. (NTC) Nontemplate control. (A) Expression levels of 15 novel ovarian miRNAs in multiple mouse tissues. (B) Expression levels of 15 novel ovarian miRNAs in the total ovary and isolated metaphase II oocytes.
In addition to oocytes, the ovary contains multiple somatic cell types, including follicular cells, thecal cells, and lutein cells. The small RNAs cloned from the ovaries, therefore, may be derived from either oocytes or somatic cell types. To further determine the cellular origin of these novel ovarian miRNAs, we examined their expression levels in the total ovary and isolated metaphase II oocytes (Fig. 3B). Eleven out of the 15 novel ovarian miRNAs were detected in both the total ovary and oocytes, whereas the remaining 4 miRNAs (mir-ov4, -ov7, -ov9, and -ov15) were not detectable in the oocytes, suggesting these miRNAs may be expressed exclusively by ovarian somatic cells (e.g., granulosa cells and/or thecal cells).
RapiRNAs cloned from the mouse ovary
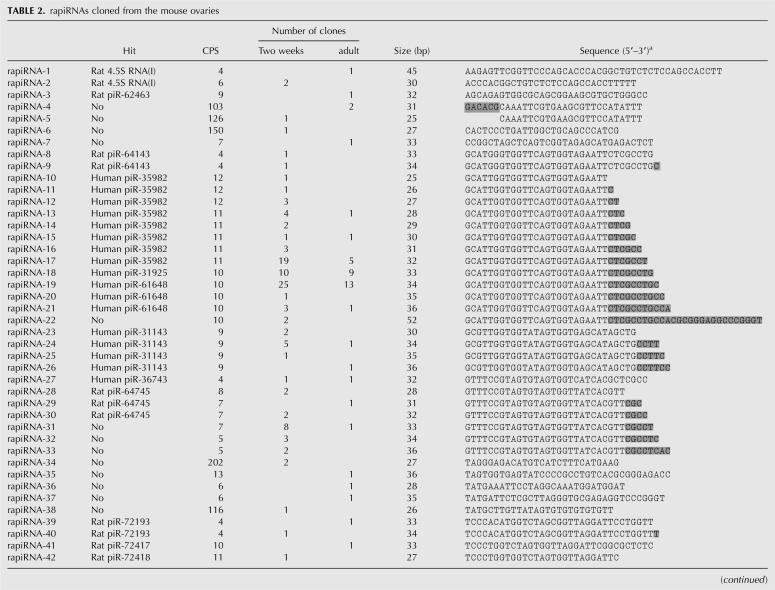
Each of the 54 rapiRNAs identified in this study was encoded by a gene with multiple copies on a single or multiple chromosomes, and the copy numbers ranged from 4 to 202 (Table 2). These rapiRNAs possess characteristics that are different from napiRNAs identified in this study (Table 3) and the majority of the piRNAs originally cloned from the testis (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Watanabe et al. 2006; Ro et al. 2007b). First, rapiRNAs appear to be longer (32–38 nt) than napiRNAs (27–31 nt) (Fig. 1). Second, rapiRNAs are mapped into repetitive regions and thus can be encoded by multicopy genes (4–202 copies) (Table 2), while genes encoding napiRNAs identified from the ovary in this study (Table 3) and the majority of the testicular piRNAs (∼84%) are located in nonrepetitive regions with one or two copies. Previously, we reported that piRNAs cloned from the mouse testis display modifications at both 5′ and 3′ ends, resulting in multiple homologous piRNAs with overlapping sequences in the middle portion (Ro et al. 2007b). Similar modifications at both 5′ and 3′ ends were observed in the rapiRNAs cloned from the mouse ovary, although the 3′ modification appears to be more common (Table 2). Five rapiRNAs (rapiRNA-22, 26, 33, 44, and 49) displayed 3′ degenerative dicing (e.g., rapiRNA-22), resulting in numerous homologous rapiRNAs (Table 2).
TABLE 2.
rapiRNAs cloned from the mouse ovaries
TABLE 3.
napiRNAs identified from the mouse ovaries
Chromosomal distribution of rapiRNAs
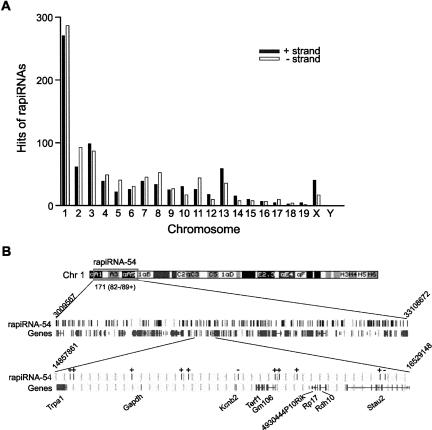
Since rapiRNA genes are located within repetitive regions, many could have hundreds of copies distributed in single or multiple chromosomes (Supplemental Table 4). For example, rapiRNA-4 and -5 genes had 103 and 126 copies, respectively, located on all chromosomes except the Y, whereas the rapiRNA-54 gene had 171 copies, all of which were located on chromosome 1 (Supplemental Table 4). The rapiRNAs had multiple hits (4–202) on both the antisense and sense strands of the chromosomes. The total number of hits by the 54 rapiRNAs was 1674, of which 817 (49%) and 857 (51%) were on the sense and antisense strands, respectively, suggesting that rapiRNAs are produced from both strands during their biogenesis (Supplemental Table 5). Additionally, we analyzed the number of rapiRNA hits for each chromosome (Fig. 4A). Interestingly, 33% of all hits (556/1674) were on chromosome 1, while only 1 hit was on chromosome Y.
FIGURE 4.
Chromosomal distribution of rapiRNAs. (A) A total of 54 rapiRNAs cloned in this study have multiple hits (4–202) on both “+” and “−” strands of all chromosomes. The total number of hits by the 54 rapiRNAs was 1674, of which 817 (49%) and 857 (51%) were on the “+” and “−” strand, respectively. (B) Genomic locations of 171 copies of the rapiRNA-54 gene on chromosome 1. The 171 copies of the rapiRNA-54 gene, including 82 copies on the “−” strand and 89 on the “+” strand are located within a region between A1 (nucleotide 3009567) and 1qB (nucleotide 33108672). The same region contains 147 protein-coding genes. Locations of all rapiRNA-54 hits are shown along with the exons of their neighboring protein-coding genes. Black bars on the rapiRNA-54 map represent hits on the genome and black bars under the rapiRNA-54 map represent exons of the genes. “+” or “−”on the map indicates plus or minus strand of the chromosome. On each side, nucleotide numbers are shown. A region in A3 (nucleotides 14857861–16529148) was scaled up to show the locations of rapiRNA-54 hits and the exons of the protein-coding genes.
Characteristics of rapiRNA-54
Since chromosome 1 possessed the most rapiRNA hits, we chose the rapiRNA-54 gene, which has 171 copies exclusively on chromosome 1, for further analysis. The 171 copies of the rapiRNA-54 gene were distributed within a region of ∼30 Mb (nucleotides 3009567–33108672) on chromosome 1, which contains a total of 147 protein-encoding genes (Fig. 4B). We mapped all the rapiRNA gene copies and protein-encoding genes in this region and found that rapiRNA gene copies were located exclusively in intergenic or intronic (no exonic) sequences within the repetitive regions (Fig. 4B; Supplemental Table 6). rapiRNA-54 displayed 75% intergenic hits (129/171) and 25% intronic hits (42/171) within the region. A region encompassing nucleotides 14857861–16529148 was enlarged to show relative genomic locations of the rapiRNA-54 gene copies and their surrounding protein-encoding genes (Fig. 4B). Eleven copies of the rapiRNA-54 and nine protein-encoding genes were found in this region. Eight copies were located in the intergenic regions and three were found in the intronic regions, which was consistent with the overall distribution of rapiRNA-54 gene copies. We also found that 48% of the hits (82/171) were from the antisense strand, and 52% (89/171) were from the sense strand (Supplemental Table 6). This is consistent with the strand origins observed for all of the 54 rapiRNAs described above (Fig. 4A).
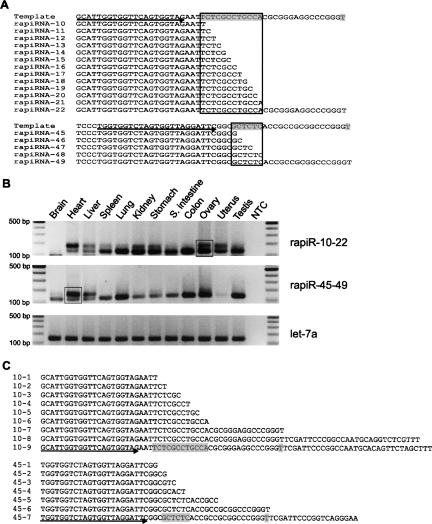
As mentioned above, one of the structural characteristics of rapiRNAs is the 3′ end degenerative dicing, which results in numerous rapiRNA homologs with differential ending nucleotides in a staggered fashion (Table 2). It is likely that these homologous rapiRNAs are initially encoded by the same genomic sequence because they differ only at their distal 3′ ends. Our expression profiling for two groups of such rapiRNA homologs (rapiRNAs 10–22 and rapiRNAs 45–49) (Fig. 5A) using semiquantitative PCR demonstrated that these rapiRNAs are expressed in many organs, including the ovary (Fig. 5B). The primers used could amplify all the homologs within each of the two groups (Fig. 5A). Since these homologs differ by 1–17 nt at their 3′ ends, the PCR products were expected to show a small, but appreciable, difference in size. Using high-resolution agarose gel electrophoresis, we could roughly distinguish between the abundance of the shorter rapiRNAs and their slightly larger homologs. For rapiRNAs 10–22, the longer forms (e.g., rapiRNAs-22) appeared to be dominant in heart, whereas the shorter rapiRNAs (e.g., rapiRNAs-10) were the major forms in the majority of the tissues analyzed including spleen, lung, kidney, stomach, small intestine, colon, ovary, and testis (Fig. 5B, upper panel). In liver and uterus, both the shorter and longer forms appeared to be equally abundant. Among RapiRNAs 45–49, the shorter forms (e.g., rapiRNA-45) were expressed in most organs, whereas the longer forms (e.g., rapiRNA-49) were predominant in heart and liver (Fig. 5B, middle panel). Interestingly, rapiRNAs 10–22 were absent in brain, while rapiRNAs 45–49 were barely detectable in the uterus (Fig. 5B), suggesting that rapiRNA expression displays tissue specificity, although the majority of rapiRNAs are expressed in every tissue analyzed. To confirm the cloned rapiRNAs and PCR data, we subcloned the rapiR-10–22 PCR amplicons derived from the ovary and the rapiR-45–49 amplicons from the heart into the pcDNA3.1 TOPO vector (see boxes in the gel images, Fig. 5B) and sequenced 30 clones randomly selected from each group. Sequence analysis revealed that the PCR amplicons contained most of the cloned homologs shown in Figure 5A (Fig. 5C). In addition, we identified even longer forms (PCR clones, 10-8, 10-9, 45-7) from the PCR products (Fig. 5C). The PCR clone 10-9 was 32 nt longer in size than rapiRNA-22, whereas 45-7 was 19 nt longer than rapiRNA-49. The clone 10-8 was 3 nt shorter than 10-9 and contained divergent 10 nt at the 3′ end, which did not match those of 10-9. In fact, the clones 10-9 and 10-8 were derived from different chromosomes, 11 and 13, respectively, suggesting rapiRNA-10–22 were derived from precursors encoded by different gene copies located on the two chromosomes.
FIGURE 5.
rapiRNA homologs and expression profiles. (A) Alignment of two groups of rapiRNA homologs. Multiple rapiRNA homologs were generated from a single precursor template. rapiRNAs-10–22 (top) and rapiRNAs-45–49 (bottom) are aligned along with their precursor templates. The diced nucleotides in the precursor templates are highlighted in gray, and the different ends within the diced region are framed. Primers used for PCR are indicated by arrows under the template DNA sequences. (B) Expression levels of the homologous rapiRNAs in multiple tissues. let-7a was used as a loading control. Amplicons were analyzed on 2% agarose gels. A DNA ladder on each side indicates the size of the fragments. (NTC) Nontemplate control. Two PCR amplicons (rapiR-10–22 and rapi-45–49), boxed in the gel images, were excised, purified, and subcloned into a pcDNA3.1 vector for sequencing analyses. (C) Sequencing results of the two amplicons. Nine clones (10-1 to 10-9) were obtained from the rapiR-10–22 amplicons of the ovary and seven clones (45-1 to 45-7) were obtained from the rapiR-45–49 amplicons of the heart. The diced nucleotides in the precursor templates are highlighted in gray, and primers used for PCR are indicated by arrows under the template DNA sequences.
napiRNAs cloned from the mouse ovary
Our cloning efforts also identified 25 napiRNAs (Table 3). Five out of the 25 napiRNAs matched known piRNAs registered in the miRBase and the remaining 20 were novel napiRNAs (Table 3). The most common sizes of these napiRNAs were between 27 and 31 nt, and the majority (20 napiRNAs) of these piRNA precursor sequences fail to form stem–loop structures (Table 3). piRNAs were once believed to be exclusively expressed in the testis because their binding partner Piwi is a testis-specific protein (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Watanabe et al. 2006). In a recent study, however, we found that ∼16% of testicular piRNA are actually expressed in multiple tissues in addition to the testis (Ro et al. 2007b). Cloning of 20 more novel napiRNAs from the mouse ovary further supports the notion that these piRNAs are not exclusively expressed in the testis. Their functions, therefore, may not be limited to the male germ line.
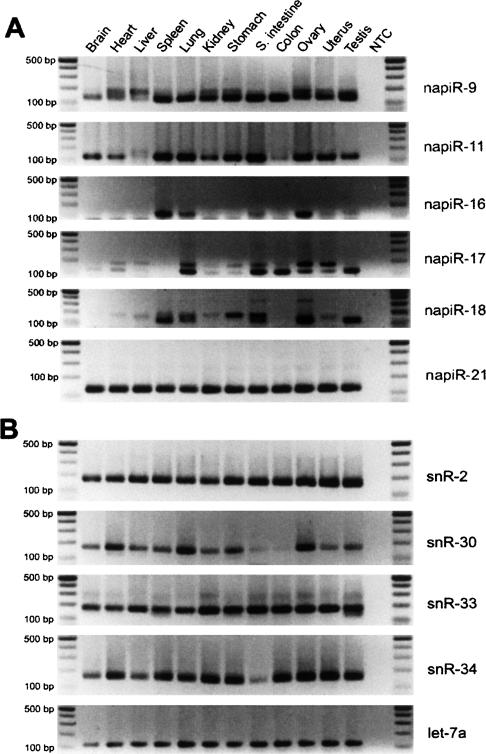
To validate these cloned novel napiRNAs, we chose six (napiR-9, -11, -16, -17, -18, and -21) to analyze their expression levels in multiple mouse tissues (Fig. 6A). All of the six napiRNAs were detected in multiple tissues. napiR-11 was absent or weakly detectable in liver and colon, but abundantly expressed in spleen, lung, gastrointestinal tract, ovary, uterus, and testis. napiR-16 was only expressed in spleen, lung, and ovary. napiR-17 and napiR-18 were preferentially expressed in several tissues. These diverse expression patterns of napiRNAs suggest that they may have tissue-dependent roles. More interestingly, napiR-17 and napiR-18 were amplified with multiple PCR products. Sequencing of these products (data not shown) revealed that they were homologs with variably staggered 3′ ends, suggesting that these napiRNAs, similar to their rapiRNA counterparts, were subjected to degenerative dicing events as well during their biogenesis.
FIGURE 6.
Expression profiling analyses for novel napiRNAs and snoRNAs identified in this study. Expression levels in 12 mouse tissues were examined by semi-quantitative PCR. let-7a was used as a loading control. Amplicons were analyzed on 2% agarose gels. A DNA ladder on each side indicates the size of the fragments. (NTC) Nontemplate control. (A) Expression profiles of six novel napiRNAs (napiR-9, -11, -16, -17, -18 and -21). (B) Expression profiles of four novel snoRNAs (snR-2, -30, -33, and -34).
snoRNAs cloned from the mouse ovary
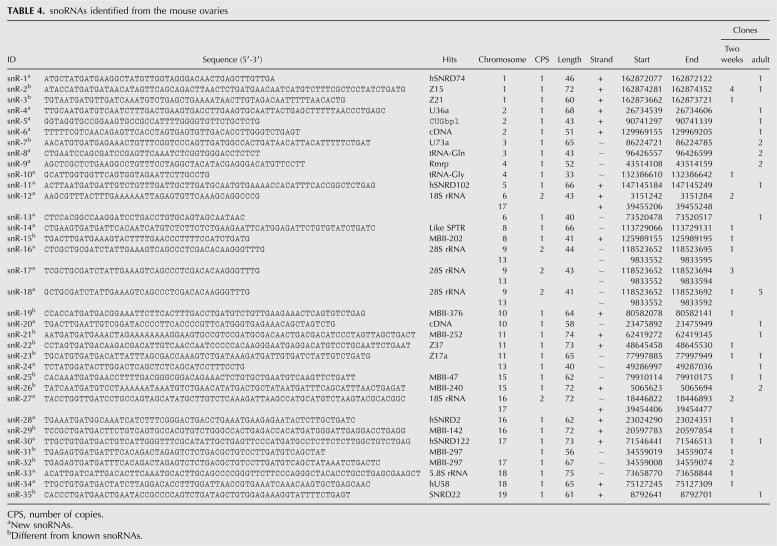
SnoRNAs range from 45 to 100 nt in size and have been implicated in the modification of pre-mRNAs (Terns and Terns 2002). In this study, we identified 35 snoRNAs from the ovarian srcDNA libraries, 21 of which were novel (Table 4). Size analyses of these 35 snoRNAs showed that the most common size of ovarian snoRNAs is between 65 and 75 nt (Fig. 1). The chromosomal origin, size, and genes that these snoRNAs perfectly or partially match are summarized in Table 4. Expression profiling for four novel snoRNAs (snR-2, -30, -33, and -34) revealed that snR-2 and snR-33 were ubiquitously expressed in all tissues examined, while snR-30 and snR-34 were expressed preferentially in most but not all of the tissues analyzed (Fig. 6B).
TABLE 4.
snoRNAs identified from the mouse ovaries
DISCUSSION
Mammalian oogenesis is a complex process involving the regulation of a multitude of genes at the transcriptional and post-transcriptional levels. Discovery of miRNAs and their roles in post-transcriptional regulation suggest that small regulatory RNAs may be important regulators of gene expression. Recent identification of rasiRNAs and piRNAs further support this notion. We attempted to clone small RNAs from the mouse ovary as the first step toward understanding the regulatory roles of small RNAs in oogenesis. By using a novel small RNA cloning method developed by our laboratory, we identified a total of 236 small RNAs, including 122 miRNAs, 25 napiRNAs, 54 rapiRNAs, and 35 snoRNAs. By analyzing the structural features and expression profiles of these ovarian small RNAs, we identified many characteristics unique to each of the four classes, suggesting that these small RNAs are generated through different mechanisms and may have differential roles in the regulation of gene expression during folliculogenesis.
Structural features of the four classes of ovarian small RNAs
Listed in ascending order of their major sizes, the four classes of small RNAs are miRNAs (21–23 nt), <napiRNAs (27–31 nt), <rapiRNAs (32–38 nt), and <snoRNAs (65–75 nt). Although the majority of small RNAs can be distinguished based upon their sizes, other structural features can help determine their identity, including stem–loop formation of the precursor sequences (for miRNAs), genes derived from highly repetitive regions (rapiRNAs), and numerous homologs with degenerative dicing at both the 5′ and 3′ ends (rapiRNAs and napiRNAs). It appears that the majority of the rapiRNA (∼53%) genes are located on chromosomes 1–3 (Fig. 4A). The significance of the chromosomal preference of rapiRNA genes remains unclear. It has been shown that ∼17% of the testicular piRNAs are derived from exonic regions (Ro et al. 2007b), and these piRNAs belong to the napiRNAs population. Unlike those piRNAs, rapiRNAs are exclusively derived from intergenic and intronic sequences (Table 2), which may be due to the fact that repetitive sequences tend to exist in these regions.
The degenerative dicing at both the 5′ and 3′ ends is a unique feature to the biogenesis of rapiRNAs and napiRNAs. These degenerative dicing events result in multiple homologous piRNAs with overlapping sequences in the middle portions, but differential sequences at either their 5′ or 3′ ends, or both. Our PCR-based small RNA detection method is superior to conventional PAGE-based Northern blot analysis because it is much more sensitive and can also simultaneously detect levels of the longer and shorter forms of the multiple homologous piRNAs displaying degenerative dicing. Ratios of the longer and shorter forms of rapiRNAs vary in a tissue-dependent manner. The physiological significance of differential ratios of the longer-to-shorter forms of piRNAs in specific tissues requires further study.
Chromosomal mapping of rapiRNAs supports the “ping-pong” mechanism of piRNA biogenesis
rapiRNAs were originally identified from fly testes and embryos (Aravin et al. 2003). rapiRNAs have been suggested to play a role in regulating transposon activity and in defining chromatin structures based upon the fact that they are cognate to transposable elements and other repetitive sequences in the fly genome (Saito et al. 2006; O'Donnell and Boeke 2007). Consistent with previous studies (Saito et al. 2006; Vagin et al. 2006), our data show that rapiRNAs are in both the antisense and sense orientations and the 5′ ends of the rapiRNAs display a uridine preference, suggesting that they are generated by an RNase III domain enzyme. Recent data suggest that piRNAs are generated via a self-reinforcing amplification cycle, also called the “ping-pong” mechanism (Brennecke et al. 2007; Lin 2007; O'Donnell and Boeke 2007). In this model, the cycle starts with processing the primary piRNAs, which are long transcripts derived from defective transposon copies in regions of heterochromatin and are antisense to the expressed transposons. The primary piRNAs bind either Piwi or Aubergine, and the Piwi/Aubergine-piRNA complexes recognize and cleave their active transposon targets, generating new sense piRNAs that bind Ago3. Subsequently, the sense piRNA-Ago3 complex directs another cleavage event of a piRNA cluster transcript, creating a new antisense piRNA capable of binding Piwi or Aubergine. The cycle will be reinforced by producing additional sense piRNAs when the secondary piRNA-Piwi/Aubergine complexes recognize and cleave their target transposon elements. Our finding appears to support this model because the numbers of rapiRNAs derived from the sense (52%) strand and the antisense (48%) strand are nearly equivalent (Supplemental Table 5). Since this self-amplification cycle is theoretically indefinite, there must be a yet-to-be-defined mechanism regulating the rate of the amplification.
Potential functions of ovarian small RNAs
miRNAs mainly function as post-transcriptional regulators of gene expression through binding to their target sequences generally located in the 3′ UTR, controlling mRNA stability and translation (Bartel 2004). Several of the novel miRNAs identified in this study are preferentially expressed in the ovary, suggesting that they may have a role limited to ovarian functions. Further identification of the target genes of these ovarian miRNAs will shed light on their regulatory roles in folliculogenesis.
DNA transposons and retrotransposons are mobile elements, which can insert themselves at new locations in the host genome, thus modifying gene structure and altering gene expression. Therefore, their transposition must be regulated to avoid endangering the host. Since rapiRNAs share 100% complementarity with transposons or repetitive sequences, it has been suggested that they play a role in silencing transposon activity (O'Donnell and Boeke 2007). Our expression profiling studies demonstrate that expression of rapiRNAs and napiRNAs is not limited to the testis. In fact, several other tissues, including the ovary, also express these piRNAs, although the number is much less than that expressed in the testis.
Extensive studies on the function of snoRNAs has revealed that they participate in the site-specific modification of nucleotides in target RNAs, including the 2′O-ribose methylation and pseudouridylation through base-pairing with their target sequences and recruiting associated proteins to achieve modifications (Terns and Terns 2002). Further analyses of the ovarian snoRNAs should reveal their RNA targets and physiological role in follicular development.
Taken together, ovarian expression of all of the four known classes of small RNAs species implies that folliculogenesis involves complex regulation of gene expression. Further studies on the functions of these small regulatory RNAs will help us understand the complex regulatory machinery in the control of oogenesis and female fertility.
MATERIALS AND METHODS
Oocyte collection
To collect oocytes (metaphase II), CD-1 female mice (Charles River Laboratories) were treated with 5 IU of PMSG (Sigma), followed by 5 IU of hCG (Sigma) to induce superovulation as described previously (Yan et al. 2005). Oocytes were recovered from oviducts 18–24 h after hCG treatment in M2 medium (Sigma) containing 1 mg/mL hyaluronidase (Sigma). Complete removal of cumulus cells was verified by observation under a stereomicroscope. Oocytes free of cumulus cells were collected and snap frozen in liquid nitrogen until used for small RNA isolation. A total of 2000 metaphase II oocytes were collected for small RNA isolation.
Small RNA isolation
Small RNA samples from 12 different mouse organs (brain, heart, liver, spleen, lung, kidney, stomach, small intestine, colon, uterus, testis, and ovary) and oocytes were isolated using mirVanaPTMP miRNA isolation kit according to the manufacturer's instructions (Ambion). Briefly, 50–250 mg of tissue were homogenized in 10 volumes of Lysis/Binding buffer. A 1/10 volume of miRNA Homogenate Additive was added and incubated on ice for 10 min. Total RNA was extracted by adding an equal volume of Acid-Phenol:Chloroform. Small RNA was extracted from the total RNA using a filter cartridge with 100 μL of preheated (95°C) elution solution. The concentration of small RNA was measured using a NanoDrop, ND-1000 spectrophotometer (NanoDrop Technologies).
Cloning of small RNAs
Small RNA cloning was performed as described (Ro et al. 2007b). Briefly, small RNAs isolated from the ovaries of 2-wk-old and adult (2–4 mo) mice were polyadenylated and a 5′ RNA adapter was ligated with the poly(A)-tailed RNA. The ligation products were reverse transcribed to make small RNA cDNAs (srcDNAs). The srcDNAs were amplified using PCR, and PCR amplicons were subcloned into a pcDNA3.1 TOPO vector (Invitrogen) for sequencing. Primers for cloning can be found in Supplemental Table 7.
Bioinformatic analysis
DNA sequences were analyzed by Vector NTI Suite v.6.0 (InforMax) to locate small RNA sequences in the cloning vector. Each small RNA was then located in the mouse genome using the BLAST program in the UCSC Genome Browser (Karolchik et al. 2003). The strand, as well as its start and end nucleotides, in the genome were determined according to the BLAST results. To identify miRNAs, all small RNAs were initially analyzed using the miRBase search tool (http://microrna.sanger.ac.uk/sequences/search.shtml) (Ambros et al. 2003; Griffiths-Jones 2004; Griffiths-Jones et al. 2006). If a small RNA completely or partially matched any registered miRNA from different organisms and its size ranged from 20 to 24 nt, we classified it as a miRNA. To further confirm its identity as a miRNA, we selected a fragment of ∼110 bp from each of the two sides flanking the small RNA for predicting the secondary structure of the miRNA precursor (stem–loop formation) using the Mfold program (Zuker 2003). If a small RNA was 20–24 nt long, its precursor sequence could form a stem–loop structure, and it was not registered in the miRBase (Ambros et al. 2003; Griffiths-Jones 2004; Griffiths-Jones et al. 2006), we classified it as a novel miRNA. miRNA gene clusters were identified according to proximity of genomic locations of all of the miRNAs cloned in this study. The size and cluster distributions were calculated using Microsoft Excel. The data were plotted and graphed using GraphPad Prism (GraphPad Software).
A semi-quantitative PCR-based quantification of small RNAs
Construction of small RNA cDNA (srcDNA) libraries and the semi-quantitative PCR were performed as previously described (Ro et al. 2006). Primer sequences used for expression profiling are shown in Supplemental Table 7. For each of the small RNAs analyzed, the cycle number was empirically determined such that the PCR was in the exponential range (generally between 20 and 30 cycles).
SUPPLEMENTAL DATA
All Supplemental Materials can be found at http://www.medicine.nevada.edu/physio/facyan.html, and are also available upon request.
ACKNOWLEDGMENTS
The authors thank Anita Batra and Jason Michaels for editing the text. The Nevada Genomic Center (NIH Grant Number P20 RR-016464) is acknowledged for sequencing services. This work was supported by grants from the National Institute of Health (HD048855 and HD050281), and, in part, by a start-up funds from the University of Nevada, Reno, to W.Y.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.754207.
REFERENCES
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ambros, V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Ambros, V., Bartel, B., Bartel, D.P., Burge, C.B., Carrington, J.C., Chen, X., Dreyfuss, G., Eddy, S.R., Griffiths-Jones, S., Marshall, M., et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin, A.A., Lagos-Quintana, M., Yalcin, A., Zavolan, M., Marks, D., Snyder, B., Gaasterland, T., Meyer, J., Tuschl, T. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Aravin, A., Gaidatzis, D., Pfeffer, S., Lagos-Quintana, M., Landgraf, P., Iovino, N., Morris, P., Brownstein, M.J., Kuramochi-Miyagawa, S., Nakano, T., et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Brennecke, J., Aravin, A.A., Stark, A., Dus, M., Kellis, M., Sachidanandam, R., Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila . Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Chang, T.C., Mendell, J.T. The roles of microRNAs in vertebrate physiology and human disease. Annu. Rev. Genomics Hum. Genet. 2007 doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- Girard, A., Sachidanandam, R., Hannon, G.J., Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones, S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones, S., Grocock, R.J., van Dongen, S., Bateman, A., Enright, A.J. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna, S.T., Beyret, E., Wang, Z., Lin, H. A novel class of small RNAs in mouse spermatogenic cells. Genes & Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik, D., Baertsch, R., Diekhans, M., Furey, T.S., Hinrichs, A., Lu, Y.T., Roskin, K.M., Schwartz, M., Sugnet, C.W., Thomas, D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Lendeckel, W., Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lau, N.C., Lim, L.P., Weinstein, E.G., Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans . Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lau, N.C., Seto, A.G., Kim, J., Kuramochi-Miyagawa, S., Nakano, T., Bartel, D.P., Kingston, R.E. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., Ambros, V. An extensive class of small RNAs in Caenorhabditis elegans . Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lin, H. piRNAs in the germ line. Science. 2007;316:397. doi: 10.1126/science.1137543. [DOI] [PubMed] [Google Scholar]
- Matzuk, M.M., Lamb, D.J. Genetic dissection of mammalian fertility pathways. Nat. Cell Biol. 2002;4(Suppl):s41–s49. doi: 10.1038/ncb-nm-fertilityS41. [DOI] [PubMed] [Google Scholar]
- Matzuk, M.M., Burns, K.H., Viveiros, M.M., Eppig, J.J. Intercellular communication in the mammalian ovary: Oocytes carry the conversation. Science. 2002;296:2178–2180. doi: 10.1126/science.1071965. [DOI] [PubMed] [Google Scholar]
- O'Donnell, K.A., Boeke, J.D. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poy, M.N., Eliasson, L., Krutzfeldt, J., Kuwajima, S., Ma, X., Macdonald, P.E., Pfeffer, S., Tuschl, T., Rajewsky, N., Rorsman, P., et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- Ro, S., Park, C., Jin, J., Sanders, K.M., Yan, W. A PCR-based method for detection and quantification of small RNAs. Biochem. Biophys. Res. Commun. 2006;351:756–763. doi: 10.1016/j.bbrc.2006.10.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro, S., Park, C., Sanders, K.M., McCarrey, J.R., Yan, W. Cloning and expression profiling of testis-expressed microRNAs. Dev. Biol. 2007a doi: 10.1016/j.ydbio.2007.09.009. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro, S., Park, C., Song, R., Nguyen, D., Jin, J., Sanders, K.M., McCarrey, J.R., Yan, W. Cloning and expression profiling of testis-expressed piRNA-like RNAs. RNA. 2007b;13:1693–1702. doi: 10.1261/rna.640307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, A., Matzuk, M.M. Deconstructing mammalian reproduction: Using knockouts to define fertility pathways. Reproduction. 2006;131:207–219. doi: 10.1530/rep.1.00530. [DOI] [PubMed] [Google Scholar]
- Saito, K., Nishida, K.M., Mori, T., Kawamura, Y., Miyoshi, K., Nagami, T., Siomi, H., Siomi, M.C. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes & Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns, M.P., Terns, R.M. Small nucleolar RNAs: Versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 2002;10:17–39. [PMC free article] [PubMed] [Google Scholar]
- Vagin, V.V., Sigova, A., Li, C., Seitz, H., Gvozdev, V., Zamore, P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Watanabe, T., Takeda, A., Tsukiyama, T., Mise, K., Okuno, T., Sasaki, H., Minami, N., Imai, H. Identification and characterization of two novel classes of small RNAs in the mouse germline: Retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes & Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Yan, W., Ma, L., Stein, P., Pangas, S.A., Burns, K.H., Bai, Y., Schultz, R.M., Matzuk, M.M. Mice deficient in oocyte-specific oligoadenylate synthetase-like protein OAS1D display reduced fertility. Mol. Cell. Biol. 2005;25:4615–4624. doi: 10.1128/MCB.25.11.4615-4624.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B., Pan, X., Cobb, G.P., Anderson, T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]