Abstract
Positional identities along the anterior–posterior axis of the vertebrate nervous system are assigned during gastrulation by multiple posteriorizing signals, including retinoic acid (RA), fibroblast growth factors (Fgfs), and Wnts. Experimental evidence has suggested that RA, which is produced in paraxial mesoderm posterior to the hindbrain by aldehyde dehydrogenase 1a2 (aldh1a2/raldh2), forms a posterior-to-anterior gradient across the hindbrain field, and provides the positional information that specifies the locations and fates of rhombomeres. Recently, alternative models have been proposed in which RA plays only a permissive role, signaling wherever it is not degraded. Here we use a combination of experimental and modeling tools to address the role of RA in providing long-range positional cues in the zebrafish hindbrain. Using cell transplantation and implantation of RA-coated beads into RA-deficient zebrafish embryos, we demonstrate that RA can directly convey graded positional information over long distances. We also show that expression of Cyp26a1, the major RA-degrading enzyme during gastrulation, is under complex feedback and feedforward control by RA and Fgf signaling. The predicted consequence of such control is that RA gradients will be both robust to fluctuations in RA synthesis and adaptive to changes in embryo length during gastrulation. Such control also provides an explanation for the fact that loss of an endogenous RA gradient can be compensated for by RA that is provided in a spatially uniform manner.
Author Summary
The formation of gradients of morphogens, signaling molecules that determine cell fates in a concentration-dependent manner, is a fundamental process in developmental biology. Several morphogens pattern the anterior–posterior (head to tail) axis of the vertebrate nervous system, including the vitamin A derivative, retinoic acid (RA) and fibroblast growth factors (Fgfs). However, it remains unclear how the activities of such morphogen gradients are coordinated. We have addressed this question by combining genetic experiments in zebrafish and computational analyses. We show that RA acts as a graded signal over long distances and that its gradient is shaped, to a large extent, by local control of RA degradation. In particular, RA promotes and Fgf suppresses RA degradation, thereby linking the shapes of RA and Fgf gradients. Computational models suggest that this linkage helps make RA-mediated patterning robust to changes in the rate at which RA is synthesized (which may vary with levels of dietary vitamin A) as well as in the size and shape of the embryo during development. Analogous regulatory loops may be used for similar purposes in other tissues in which RA and Fgfs interact, as well as in other morphogen systems.
Experimental and computational studies in zebrafish reveal a complex system regulating degradation of the vitamin A derivative retinoic acid along the anterior-posterior axis, which helps explain how morphogen gradients are established and maintained.
Introduction
The anterior–posterior (A–P) axis of vertebrate embryos is patterned by multiple signals. In the gastrula-stage embryo, at least three types of diffusible, extracellular molecules—retinoic acid (RA), fibroblast growth factors (Fgfs), and Wnts—together promote posterior and suppress anterior fates [1–6]. There is evidence for both hierarchical and parallel relationships among their signaling pathways. For example, RA appears to act downstream of Fgfs and Wnts in promoting posterior identities in the neurectoderm, but not in the suppression of anterior cell fates [6].
RA, Fgfs, and Wnts are all produced at the posterior of the embryo, and might therefore be expected to form posterior-to-anterior gradients (for Fgf8 this has been demonstrated directly; [7]). It is not clear which of these gradients is ultimately responsible for providing the positional cues that specify where cell fate boundaries form. This is an important question because the signaling pathways that drive expression of cell fates need not be the same as those that convey positional information. For example, it has recently been suggested that RA acts solely as a permissive factor in hindbrain patterning, required for the adoption of posterior fates, but downstream of signals that determine where such fates are specified [8]. Support for this view comes largely from observations that, in RA-deficient mouse, rat, quail, or zebrafish embryos, hindbrain pattern can be rescued by exposure of the embryo to uniform extracellular concentrations of RA [8–15].
Despite such observations, other experiments point to a role for RA that is more like that of a classical, graded morphogen. For example, embryos deficient in vitamin A (the dietary precursor of RA), or loss-of-function mutants in the RA synthetic enzyme aldehyde dehydrogenase 1a2 (aldh1a2, also known as raldh2), show loss of posterior hindbrain segments (rhombomeres) 5–7 (r5–7), and expansion of more anterior ones (r2–4) [9–11,15,16]. Treatment of chick embryos with an RA receptor (RAR) antagonist also causes progressive, concentration-dependent anteriorization [17]. Conversely, exposure of embryos to exogenous RA leads to a concentration-dependent loss of the forebrain and eyes, and progressive posteriorization of (rhombomere) identities [12,18–21]. Loss of cyp26a1—a cytochrome p450 enzyme that oxidizes RA to more polar metabolites, promoting its removal from tissues—causes r5–7 to expand anteriorly in mice and zebrafish [22–24], and the combined depletion of Cyp26a1 and related enzymes Cyp26b1 and Cyp26c1 results in severe posteriorization of the zebrafish hindbrain [8].
Efforts to understand the role of RA in hindbrain patterning have recently been fostered by the recognition that localized degradation is critical in controlling RA signaling. Sirbu et al. [25] suggested that degradation creates shifting boundaries of RA activity and that duration of exposure to RA rather than concentration controls rhombomere fate. In contrast, Maves et al. [12] argued that pattern is determined by an RA morphogen gradient that is not fixed, but grows steeper with time, specifying rhombomeres sequentially from anterior to posterior. Recently, Hernandez et al. [8] proposed a “gradient-free” model in which discrete zones of local RA degradation are progressively induced over time, in coordination with changes in cell responsiveness to RA. Since this model does not explain how such zones are placed in their correct positions, it reverts to an essentially permissive role for RA (i.e., positional information must still come from signals other than RA, such as Fgfs or Wnts).
Here we propose a new view that reconciles both existing and new data. In it, local degradation of RA plays a central role, yet long-range RA gradients directly provide graded positional cues. This model is motivated by experimental data on the control of expression of Cyp26 enzymes, and by computational analysis of feedback and feedforward interactions between RA and Fgf signaling that are revealed by these data. We argue that this makes the RA gradient resistant to fluctuations (robust) in RA synthesis rate and stable over an expanding field of cells. We also show that one characteristic of such a morphogen gradient system based on regulated degradation is the ability to produce gradients of relatively normal shape even when RA is applied globally to an embryo. The model also suggests a mechanism by which the incorporation of retinoids into patterning systems may have taken place during chordate evolution.
Results
Long-Range, Graded Effects of RA in Zebrafish Embryos
Studies in a variety of vertebrate embryos support the view that RA diffuses through tissues [26], can act at long range, and has graded effects on patterning [27–29]. However, direct evidence that diffusible RA sets up a gradient of RA response, which in turn assigns different fates to cells at different levels of RA signaling, is still lacking. To investigate this question, we utilized a zebrafish line carrying a yellow fluorescent protein (yfp) transgene under the control of retinoic acid response elements (rare:yfp [30]). In transgenic embryos, expression of YFP was readily detectable in the developing spinal cord and posterior hindbrain at 22–24 hours post-fertilization (hpf) (up to the r6/r7 boundary; Figure 1A), and was clearly graded from posterior to anterior in the posterior hindbrain (Figure 1B). Expression was completely lost in RA-deficient embryos injected with an aldh1a2 morpholino (aldh1a2-MO; Figure 1C), and could be restored by the prior transplantation of cells overexpressing aldh1a2 and targeted to somitic mesoderm, where aldh1a2 is normally expressed (Figure 1D). In most cases, rescue spanned the width of the neural tube (approximately six cell diameters [∼70 μm]; Table 1).
Figure 1. Responses of Embryos to Sources of RA.
Long-range induction of a rare:yfp reporter. Confocal images of live embryos, dorsal view, anterior to the left.
(A) rare:yfp expression in posterior hindbrain and spinal cord at 24 hpf with an anterior boundary at r6/7.
(B) Quantification of fluorescence intensities in (A).
(C) Lack of YFP expression in an aldh1a2 morphant embryo.
(D) Rescue of YFP expression in a morphant by somitic mesoderm (yellow) transplanted during gastrulation.
(E–I) Bead implantation. RA-coated beads were implanted anterior to the first somite at 18–19 hpf and imaged at 23–24 hpf. A DMSO-coated bead (E) failed to rescue YFP expression, whereas beads soaked in either 10 μM (F) or 100 μM (G) RA induced YFP at distances of up to 200 μm.
(H and I) Inhibition of both cyp26b1 and cyp26c1 in DEAB-treated embryos leads to a symmetrical response from the rare:yfp reporter to a bead soaked in 10 μM RA (compare [H] with [I]).
The dotted line indicates the r6/7 boundary, the dashed line, the anterior border of somite 1. nt, neural tube; s, somites.
Table 1.
Summary of Transplants and Bead Implantations
Similarly, ion-exchange beads soaked in 10–100 μM RA and implanted just anterior to the first somite at 19 hpf also strongly induced rare:yfp (Figure 1E–1G). YFP expression was observed at a greater distance—up to 300 μm—when beads were soaked in 100 μM RA (Table 1). Interestingly, responses to low (10 μM) doses of RA were markedly asymmetrical (Figure 1F and 1H), extending up to 300 μm posteriorly but truncated anteriorly near the r6/7 boundary. This asymmetry was eliminated with MOs directed against cyp26b1 and cyp26c1 (Figure 1I), which are highly expressed just anterior to the r6/7 boundary [8].
From these results, we infer that RA signaling in the neural ectoderm is graded in vivo, that signaling is dose dependent, and that long-range effects of RA can be mediated across territories in which RA cannot be synthesized (ruling out a relay-type model of RA action at a distance). In addition, asymmetries in the response to RA signaling arise in a manner consistent with localized effects of Cyp26 enzymes on RA responsiveness, as previously proposed [8]
The rare:yfp reporter appears primarily to read out sustained, high-level RA signaling. For example, YFP was not normally detected in transgenic gastrula- or early somite-stage embryos, when RA is known to act, but could be induced by beads soaked in very high levels of RA (10–25mM; Figure S1A–S1C). As a more sensitive readout of early RA signaling, we monitored two direct RA target genes, hoxb4 and hoxb5a [31]. Normally, hoxb4 has an anterior limit at the r6/7 boundary, whereas hoxb5a is expressed further posteriorly in the spinal cord (Figure 2A). Both genes require RA signaling for their early expression (Figure 2B) and both possess functional, phylogenetically conserved RAREs within their promoters [32,33]. Since hoxb4 is expressed more anteriorly, one might expect it to be activated by lower RA concentrations than hoxb5a. Consistent with this, beads coated in 100 μM RA and implanted into embryos treated with 10 μM diethylaminobenzaldehyde ([DEAB), which inhibits class 1 aldehyde dehydrogenases and thereby blocks RA synthesis [12,14,34,35], induced expression of both hox genes (Figure 2C) at early neurula stages (10–11 hpf). Induction was detectable within 60 min of bead implantation (Figure 2D–2F), and consistently extended further from the bead for hoxb4 (94.86 ± 17.18 μm; n = 4) than for hoxb5a (38.44 ± 6.27 μm; n = 5). These results suggest that there are multiple thresholds of RA response, with more anteriorly expressed target genes exhibiting lower expression thresholds. Together, the results in Figures 1 and 2 imply that RA can establish long-range gradients that directly specify the location of expression of genes that control A–P identity.
Figure 2. Concentration-Dependent Induction of hox Gene Expression by RA.
(A) In wild-type (wt) embryos, hoxb4 expression in the neural tube (nt) extends to the r6/7 boundary, whereas the limit of hoxb5a expression is level with the first somite (s). hoxb5a is also expressed in axial, lateral (lat), and cranial (cm) mesoderm.
(B and C) Beads soaked in 100 μM RA were implanted at 1–3 somites (10.3–11 hpf) into DEAB-treated embryos, fixed 3 h later, and assayed for hox expression. Dorsal views, showing in situ hybridization with probes for hoxb4 and hoxb5a at 14 hpf.
(B) DMSO-coated beads do not induce hox expression.
(C) RA-coated beads induce hoxb5a in lateral mesoderm (arrowhead), hoxb5a/hoxb4 double-positive cells near the bead, and hoxb4 single-positive cells further away (black arrow).
(D–F) Time-course of hoxb5a induction in response to a 100 μM RA bead: no induction 30 min post-implantation (D), a few cells near the bead after 60 min (arrowhead) (E), and strong expression further away after 180 min (F).
(G–L) RA-induced degradation limits the range of RA signaling. RA beads were implanted at 1–3 somites (10.3–11 hpf) into DEAB-treated embryos, fixed 3 h later, and assayed for hox expression. Dorsal views, showing in situ hybridization with probes for hoxb5a (G–I) or hoxb4 (K and L) at 14 hpf.
(G) Implantation of a DMSO bead has no effect on gene expression in DEAB–treated embryos.
(H and I) A 100 μM RA bead placed into a DEAB-treated, cyp26a1 morphant (I) induces hoxb5a over a much larger area than a DEAB-treated control (H).
(J) Graph showing an increase in the range of hox induction in cyp26a1 morphant, DEAB-treated embryos (black bar) as compared to DEAB-treated (light grey bar). The asterisk (*) indicates significantly different from DEAB-treated controls (p < 0.05), using the Student t-test.
(K and L) A 10 μM RA bead placed into a cyp26a1 morphant induces hoxb4 over a much larger area.
Effects of RA-Induced Feedback on RA Degradation and the Range of RA Signaling
In contrast to the loss of asymmetry in the RA response at 24 hpf caused by reducing cyp26b1 and cyp26c1 function (Figure 1H and 1I), the range of hoxb4 and hoxb5a responses at 13–14 hpf was more than doubled by inhibiting cyp26a1 function (Figure 2G–2L). This was unexpected because, at the stage of bead implantation for these experiments, cyp26a1 is not normally expressed within the hindbrain (Figure 3A). This suggested that RA beads were themselves inducing cyp26a1. Indeed, in situ hybridization revealed marked cyp26a1 induction up to 53 μm from the bead (Figure 3B). In contrast, RA beads only weakly induced cyp26b1 expression (in cranial mesoderm; Figure 3C, arrowhead), and did not induce cyp26c1 at all (Figure 3D), findings that are consistent with studies in chick [36]. Whole-embryo treatment with high RA levels (1 μM) also failed to induce cyp26b1 or cyp26c1, suggesting that these enzymes do not simply have lower sensitivities to RA than cyp26a1 (unpublished data). If anything, RA appeared to have a short-range inhibitory effect on cyp26c1 expression (Figure 3D).
Figure 3. Regulation of Cyp26s by RA.
(A) Loss of cyp26a1 expression in cranial mesoderm in DEAB-treated embryos and lack of induction by a DMSO-soaked bead.
(B) Induction of cyp26a1 expression by an RA bead; inset, optical section showing cyp26a1 induction in both the neural tube (nt), lying above the notochord (n), and lateral mesoderm (white arrowhead). The asterisk (*) indicates the bead.
(C) Weak induction of cyp26b1 (black arrowhead).
(D) No induction of cyp26c1.
Although it has been observed that exogenous RA induces cyp26a1 in the hindbrain [36,37], loss of RA signaling does not affect endogenous cyp26a1 expression [8,25,37]. This has led to the view that cyp26a1 induction occurs only at supra-physiological levels of RA, possibly as a form of protection against teratogenicity [8]. Yet in Figure 3, the range over which cyp26a1 is induced implies that its expression is triggered by levels of RA similar to those that induce hoxb4 and hoxb5a. This result is even more curious in view of the fact that these hox genes and cyp26a1 are not normally coexpressed in the neurectoderm.
To try to make sense of these observations, we reexamined endogenous patterns of cyp26a1 expression in embryos, starting at gastrula stage (4–9 hpf; Figure 4). At this stage, cyp26a1 is reported to be restricted anteriorly to the animal pole in the presumptive forebrain and midbrain, and posteriorly to the involuting margin [6,8]. Close inspection, however, revealed the presence of previously unrecognized, low-level cyp26a1 expression in between these two sites (Figure 4A and 4C). This was observed at mid-gastrula stages (75% epiboly), predominantly in the hypoblast (future mesendoderm; lower panel, Figure 4A) and extended approximately halfway between the anterior and marginal cyp26a1 expression domains (arrow to arrowhead on Figure 4A). By late gastrula (90% epiboly), weak cyp26a1 expression covered the entire region between the anterior and marginal domains (arrow to arrowhead on Figure 4C) and was observed in both the epiblast (future ectoderm) and hypoblast (lower panel, Figure 4C), encompassing the entire presumptive hindbrain field. Because this expression was weak, it might easily have been overlooked were it not for the fact that it completely disappeared upon treatment with 10 μM DEAB (Figure 4B and 4D). In contrast (and as previously reported), the strong expression of cyp26a1 in the anterior central nervous system and posterior margin was RA-independent [37]. Similar RA-dependent, weak cyp26a1 expression in the presumptive hindbrain can be seen in a recent study of vitamin A–deficient quails (see Figure 3B of [36]).
Figure 4. RA-Dependent Expression of cyp26a1 .
Wild-type (wt) and DEAB-treated embryos were stained for extended periods of time for cyp26a1 expression to investigate low-level expression. Top, dorsal view; bottom, lateral view.
(A and C) Weak expression posterior to the presumptive midbrain (arrow to arrowhead).
(B and D) This expression is lost in DEAB-treated embryos.
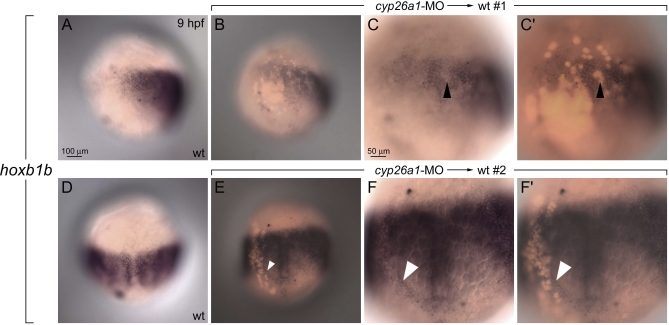
These data indicate that expression of the major RA-degrading enzyme, cyp26a1, in cells within or adjacent to the presumptive hindbrain during gastrulation is under the control of RA signaling. This suggests a more subtle role for RA degradation than simply creating boundaries beyond which RA cannot spread. Given that expression at such locations is weak, the question arises as to whether it is of any physiological consequence. To address this, we used mosaic analysis, transplanting cyp26a1 morphant cells at 5 hpf (50% epiboly) into wild-type embryos and assaying the expression of hoxb1b at 9–10 hpf (90% epiboly–tailbud; Figure 5A and 5D). When located within the endogenous hoxb1b domain, Cyp26a1-deficient cells down-regulated hoxb1b (Figure 5B–5C′), Cyp26a1-deficient cells also often caused an increase in expression in adjacent cells (Figure 5E–5F′). These results indicate that the low-level expression of cyp26a1 within and near the presumptive hindbrain is, indeed, functionally relevant. Moreover, because the observed effects were limited either to the transplanted cells themselves or cells nearby, the results imply that cyp26a1 normally influences RA signaling only over very short ranges (the significance of this point will be discussed later).
Figure 5. cyp26a1 Is Required Outside of the Anterior Neural Domain.
(A and D) hoxb1b expression in wild-type (wt) embryos.
(A–C′) Lateral view, anterior to top, dorsal to right.
(D–F′) Dorsal view, anterior to top.
(B, C′, E, and F′) Merged brightfield and fluorescent images show positions of transplanted cyp26a1-deficient cells labeled with lysine-fixable rhodamine dextran (red).
(B–C′) Transplants of cyp26a1-deficient cells located within the endogenous hoxb1b domain cause a nearly cell-autonomous decrease in hoxb1b expression (black arrowheads).
(E–F′) Transplants of cyp26a1-deficient cells also cause changes in hoxb1b expression in adjacent host cells (white arrowheads).
Fgf Regulates cyp26a1 Expression
If low-level cyp26a1 expression in the future hindbrain of the gastrula-stage embryo is induced by RA signaling, why does such expression decline from anterior to posterior (Figure 4A–4D), a direction along which RA signaling presumably increases? This observation suggests that there must be additional, position-specific inputs into cyp26a1.
To clarify the nature of such inputs, we implanted RA beads into different A–P locations in DEAB-treated embryos at mid-gastrula (8 hpf). This rapidly induced cyp26a1 expression (≤30 min), but the pattern of induction depended on the precise bead position (Figure 6A–6D). Beads placed within 100 μm of the anterior domain of high-level cyp26a1 expression in the brain induced cyp26a1 anterior to the bead—in a pattern that merged with endogenous cyp26a1—but not posteriorly (Figure 6C). Beads placed further posteriorly at mid-trunk levels (Figure 6D), failed to induce cyp26a1 expression near the bead at all (where RA levels should be highest), but only far anteriorly (white arrowhead) and near the posterior margin (white arrow). These results suggest that the capacity of cells to induce cyp26a1 in response to RA declines from anterior to posterior, rising again near the posterior end of the embryo. A similar phenomenon can be seen if DEAB-treated embryos are treated with increasing concentrations of exogenous RA (Figure S2A–S2D).
Figure 6. Regulation of cyp26a1 by RA and Fgfs.
(A and B) Anterior and marginal expression domains of cyp26a1 expression are unchanged by 10 μM DEAB. Anterior is to the left in all panels.
(C and D) RA beads (black arrowheads) induce cyp26a1 expression in DEAB-treated embryos far from (white arrowhead and arrow in [D]), but not adjacent to the bead.
(E–H) Fgf signaling sets the posterior boundary of cyp26a1 expression.
(E) cyp26a1 expression in wild type (wt) (lateral view, anterior to the left).
(F and G) Expression in SU5402-treated embryos shifts posteriorly in a concentration-dependent manner (compare brackets in [E], [F] and [G]).
(H) The posterior shift caused by blocking Fgf signaling is abolished by DEAB treatment.
An attractive candidate for a factor controlling such positional effects is Fgf. Eliminating all Fgf signaling is known to lead to cyp26a1 expression along the entire A–P axis [6], which suggests that one role of Fgfs may be to suppress cyp26a1 expression. We examined this possibility by treating embryos with various concentrations of the Fgf receptor tyrosine kinase inhibitor SU5402. As shown in Figure 6E–6G, in the presence of SU5402, the border of high-level cyp26a1 in the neurectoderm shifted, in a concentration-dependent fashion, progressively toward the posterior. Interestingly, the expansion of cyp26a1 expression upon blockade of Fgf signaling was prevented by simultaneous administration of DEAB (Figure 6H). Thus, Fgf appears to act indirectly, by inhibiting RA-mediated activation of cyp26a1 expression.
Modeling the RA Gradient System
From the information presented above, we infer that RA gradients, at least during gastrulation, should be influenced by interacting feedback (RA signaling inducing RA degradation) and feedforward (Fgf signaling repressing RA degradation) effects. To identify the consequences of such effects, we turned to quantitative modeling, using partial differential equations to describe the spatial dynamics of RA diffusion and degradation (see Materials and Methods). Because RA is a lipophilic molecule that can pass directly through cell membranes, we initially ignored cell boundaries, modeling RA as a substance that moves freely through tissue, being degraded at any location in proportion to the amount of Cyp26 enzyme expressed by cells there. However, the observations in Figure 5 made us realize that this could not be the case. The fact that the effects of blockade of cyp26a1 in transplanted cells were nearly cell autonomous tells us that RA molecules that enter cells with even low levels of cyp26a1 expression get degraded before they have an opportunity to exit the cell. The only way that RA can act over long distances, yet effectively never leave cells, is if transport precedes cell entry, i.e., RA moves a long way within the extracellular space before it is taken up.
In fact, this makes sense when one considers that RA binds proteins tightly, and must dissociate from them to cross a plasma membrane. It has been shown in cell cultures that the presence of physiological levels of albumin results in very slow RA uptake by cells [38]. Given that RA bound to a soluble protein carrier could be expected to diffuse several hundred micrometers in a matter of minutes, we reasoned that a realistic model of RA dynamics needs to include separate, interconverting pools of extracellular protein-bound and intracellular RA, with RA signaling being solely a function of the latter.
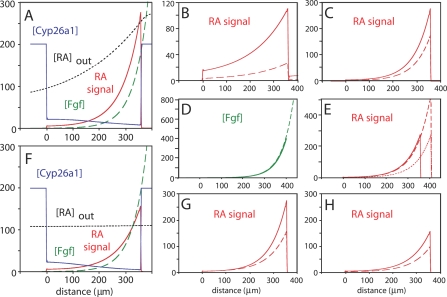
We developed such a model for the early gastrula-stage embryo (see Materials and Methods), when RA is known to be required for hindbrain patterning, and its behavior was analyzed over a wide range of parameter choices (Figure 7). The model readily generates patterns of RA signaling and cyp26a1 expression (Figure 7A) that, for reasonable parameter choices, fit experimental expectations. As a result of exploration of the model, three observations were made:
Figure 7. Modeling the RA Morphogen Gradient.
A one-dimensional mathematical model of the gastrula-stage zebrafish embryo was developed that incorporates RA synthesis, diffusion, cell permeation, degradation, and signaling; a stable Fgf gradient; and expression of cyp26a1 under the control of RA and Fgf signaling, as well as other position-specific cues (see Materials and Methods). In each panel, anterior is to the left and posterior to the right. On the abscissa, zero represents the posterior boundary of the domain of high, anterior cyp26a1 expression (approximately the r1/r2 border), which is set independently of RA. Values on the ordinate are in arbitrary units. Fgf concentration is shown in green, Cyp26a1 in blue, extracellular RA ([RA]out) in black, and RA signaling (a function of intracellular RA) is shown in red. Parameter values common to all panels are given in Materials and Methods.
(A) Typical patterns of RA signaling and cyp26a1 expression generated by the model. Note the presence of low-level cyp26a1 expression that declines from anterior to posterior.
(B and C) Feedback and feedforward regulation of cyp26a1 expression leads to robustness to RA synthesis rates. In both panels, the dashed curves show the effects of a 2-fold decrease in RA production. In (B), cyp26a1 is taken to be constant over the interval 0 < x < 360 μm, whereas in (C), it is regulated as prescribed by the model. Note the dramatic increase in robustness in (C).
(D and E) Effect of embryo elongation on Fgf and RA gradients. In both panels, solid lines are calculated for a posterior margin at 400 μm; dashed lines for a posterior margin at 450 μm. In (D), the dashed line is calculated using a rate of Fgf synthesis 2.75 times higher than for the solid line; this is to the degree of increase that would be required to compensate for the rearward movement of the Fgf source. (E) shows that, using the same sets of parameters, the RA gradient automatically compensates for the rearward movement of the RA source, without any need for readjustment of RA synthesis. For comparison, the dotted line shows what would happen to the RA gradient were there no such compensation.
(F and G) RA supplied in a completely delocalized manner can produce a relatively normal morphogen gradient. (F) is a modification of (A), in which the endogenous posterior source of RA (from 360 < x < 400) has replaced by a constant influx of RA at all locations. For an appropriate rate of exogenous RA influx, relatively normal patterns of RA signaling and cyp26a1 expression ensue, despite the lack of an extracellular RA gradient. This is particularly true at anterior locations, as shown in (G), in which the RA signaling gradients from (A) and (F) are compared (solid and dashed curves, respectively).
(H) RA signaling gradients produced by delocalized RA are robust to rates of RA influx. Like gradients produced by endogenous RA (C), gradients produced by exogenous, delocalized RA are also predicted to be robust (solid curve is RA signaling from [F]; dashed curve shows the effect of a 2-fold decrease the in the rate of RA influx). Values of parameters that differed between panels were β = 1 ([A–E] and solid curve in [G]) or 0.5 ([F], [H], and dashed curve in [G]); x f = 400 μm ([A–C] and [F–H]) or 450 μm (D–E); f 0 = 0 (B), 400 ([A], [C], [F–H], and solid curves in [D] and [E]), or 1,100 (dashed curves in [D] and [E]); k deg = 500 sec−1 ([A] and [C–G]) or 0.03 sec−1 (B); k max = 0.001 ([A] and [C–G]) or 4 (B); V(x) = 5 for x > x f – 40, zero otherwise ([A], [D], [E], and solid curves in [B], [C], and [G]), 2.5 for x > x f – 40, zero otherwise (dashed curves in [B] and [C]), 0.2 for all x ([F], dashed curve in [G], and solid curve in [H]), or 0.1 for all x (dashed curve in [H]).
First, feedback regulation of RA degradation makes the intracellular RA gradient robust to changes in the rate of RA synthesis. In the absence of any controls on cyp26a1 expression, a change in the rate of RA synthesis would be expected to produce a similar change in the amplitude of the RA gradient. Figure 7B illustrates the effect of a 2-fold decrease in RA synthesis rate on the (intracellular) RA signaling gradient. Note the large shift in the locations at which RA signaling levels cross threshold values. In contrast, by allowing for strong feedback regulation of cyp26a1, the model produces a much more robust gradient (Figure 7C). The idea that “self-enhanced degradation” could render morphogen gradients robust to fluctuations in synthesis rates was first shown in theoretical work on Wingless and Hedgehog gradients in Drosophila [39]. Since rates of RA synthesis in vivo are likely to be highly labile to environmental factors (e.g., the availability of its dietary precursor, vitamin A), exploitation of such a mechanism would seem especially advantageous for RA gradients.
Second, feedforward effects of Fgfs couple the shape of the RA gradient to that of the Fgf gradient. Previous work has shown that the Fgf gradient is formed by diffusion and receptor-mediated destruction [7], so its shape should be determined by a balance between Fgf synthesis and receptor binding and dynamics [40,41]. During gastrulation, the source of Fgf expression in the posterior marginal zone moves continuously away from the anterior of the embryo, so the Fgf gradient itself would need to undergo continual change in order to maintain stable levels of Fgf at constant positions. The easiest way to accomplish this would be for Fgf expression to increase as gastrulation proceeds (Figure 7D). Were the RA gradient not coupled to the Fgf gradient through the latter's effects on RA degradation, elongation of the embryo would continually shift the RA gradient toward the posterior (Figure 7E, dotted line), unless RA synthesis were continually increased by just the right amount. In contrast, because of Fgf's influence on cyp26a1 induction, increased Fgf production automatically results in an increase in RA signaling that compensates for axis elongation (Figure 7E, dashed line). In effect, the RA gradient becomes entrained to the Fgf gradient.
Third, even an embryo that makes no RA and is bathed in uniform RA will generate a relatively normal RA signaling gradient. A remarkable consequence of the entrainment of the RA gradient to the Fgf gradient is that replacement of the endogenous RA source at the posterior with a spatially uniform influx of RA into the extracellular space still produces a marked posterior-to-anterior gradient of intracellular RA (Figure 7F). At appropriate doses of exogenous RA, the RA signaling gradient will closely resemble the endogenous one, especially at anterior sites (Figure 7G). This effect nicely reconciles data showing that RA acts in a concentration-dependent manner [12,17–21] with observations that uniform administration of RA can effectively rescue pattern in RA-deficient embryos [8–12].
Furthermore, because of the feedback effect of RA on its own degradation, the exogenous gradient can be expected to be as robust to RA dosage as the endogenous one (compare Figure 7H and 7C). Indeed, marked robustness of rescue by exogenous RA can be demonstrated experimentally (Figure S3): when DEAB-treated embryos were exposed to exogenous RA starting at 4 hpf, relatively normal hindbrain patterning was restored over an 8-fold range (0.625–5 nM) of RA concentrations. Hernandez et al. [8] reported similar rescue over a 20-fold RA range (0.5–10 nM), using embryos treated at slightly later stages (5.3 hpf). We interpret such robustness not as an indication that RA acts in a concentration-independent manner, but rather that the distribution and concentration of RA within cells is controlled through RA- and Fgf-signaling–dependent control processes, such as those described here.
Discussion
RA and Fgf Collaborate to Produce a Robust Patterning System
The classical view of a morphogen is a molecule with a graded distribution that acts in a concentration-dependent fashion to assign positional identities to a field of cells [42]. The question of whether RA is a morphogen has been controversial in a number of contexts, including patterning of the limb, heart, and brain [8,12,43–47]. Because RA so often acts together with other signaling molecules—such as Fgfs, Hedgehogs, and Bmps—to influence patterning, the issue of whether RA plays an instructive or merely permissive role in conveying positional information is a continual source of debate. Here, we suggest that the key to resolving this debate—in the hindbrain at least—is to treat RA and Fgf as a single, integrated morphogen system, in which an RA gradient assigns positional identities, but the shape of the RA gradient is established collaboratively by RA and Fgf through their control of cyp26a1 expression (Figure 8).
Figure 8. Model of Hindbrain Patterning by RA.
(A) Diagram of interactions between RA, Fgf, and cyp26a1. Only those shown in blue are included in the mathematical model; dotted lines are extrapolated from other animal models.
(B) Schematics illustrate dorsal views of gastrulating embryos and corresponding gradients, anterior to the left. RA is produced by Aldh1a2 in somitic mesoderm (red) and diffuses through the neurectoderm. Cyp26a1 (blue) degrades RA at differing rates to produce a gradient across the hindbrain that specifies rhombomere fates. The regulation of cyp26a1 by both Fgf and RA signaling creates a robust RA gradient that grows appropriately as the A–P axis of the embryo elongates.
This view reconciles several observations: It agrees with experimental evidence that RA and Fgf both promote posterior identity, with RA acting downstream of Fgf [6]. It accommodates data that RA acts over long range to produce graded outputs (Figures 1 and 2), and that both Fgf and RA control the distribution of RA-degradative enzymes (Figures 3–5). Perhaps most importantly, it explains how administration of uniform, exogenous RA can rescue patterning that is normally driven by RA synthesized in a localized fashion (Figure 7). This view also provides a mechanism by which the RA gradient can grow over time during gastrulation (Figure 8B)—a key element of the hindbrain patterning model of Maves and Kimmel [12]—without the need for complex regulation of RA synthesis. Although this still requires Fgf signaling to increase during gastrulation, the known coupling of Fgf signaling to gastrulation movements provides a means by which such increases may be coordinated [48,49].
Central to the mechanism proposed in this study is the regulation of RA degradation in a manner that provides for robustness to uncertain levels of RA synthesis. Evidence that RA synthesis is highly susceptible to fluctuations in levels of dietary precursors includes the fact that high doses of vitamin A readily produce retinoid teratogenicity in mammals [50]. Moreover, in the zebrafish embryo, even modest increases in intracellular levels of the RA precursor retinal lead to strong posteriorization if Cyp26a1 function is blocked [8]. Although the latter effect has been interpreted as evidence that Cyp26a1 exists to protect embryos from teratogenicity, it is also a consequence of our model in which Cyp26a1 plays a central role in controlling the shape of an RA gradient. Surprisingly, this role is not one in which Cyp26a1 simply acts as a sink for extracellular RA, since removing Cyp26a1 function in transplanted cells has nearly cell autonomous effects (Figure 5). This suggests that RA degradation within a cell does not greatly affect the concentration of RA within a neighboring cell and that RA must act over long distances by traveling within the extracellular space before it enters cells.
Indeed, our results agree with those of Sirbu et al. [25], Hernandez et al. [8], and others in emphasizing the importance of localized degradation in controlling RA signaling in the developing hindbrain. Unlike Hernandez et al. [8], however, we find no need to reject the idea of an RA gradient in favor of a model driven by the sequential appearance of sharp boundaries of cyp26b1 and cyp26c1 expression. In fact, an RA gradient primarily controlled by early, smoothly graded Cyp26a1 activity better explains the relatively mild hindbrain phenotypes of cyp26b1/cyp26c1 double morphants, and the requirement for cyp26a1 to be functional for global RA treatments to rescue RA-deficient embryos.
Towards a More Complete Understanding of RA Patterning Systems
In the present study, a mathematical model was used to explore the consequences of feedforward and feedback effects of Fgf and RA on hindbrain patterning. Such a model is not intended to represent a complete or accurate picture of the early embryo, but simply to elucidate how interacting signaling pathways might work together to achieve useful ends. In fact, there is evidence that the regulatory interactions in this system go well beyond those explicitly modeled here (Figure 8A). For example, there is evidence that RA signaling inhibits expression of the RA biosynthetic enzyme aldh1a2 [37], which is likely to further decrease the sensitivity to RA synthesis to RA precursor levels. RA may affect the expression of Fgfs, and Fgf signaling is required for the expression of aldh1a2 [51]. RA signaling also appears to have complex effects on the expression of Fgf receptors [51]. Determining whether these effects contribute to robustness, to the coupling between RA and Fgf gradients, or to the performance of other tasks, awaits further experimental and computational work.
It should also be noted that, whereas the present model treats the RA gradient as a system operating near its steady state, the rapidity with which gastrulation proceeds in the zebrafish raises the possibility that patterning occurs under pre–steady-state conditions. Interestingly, recent theoretical work suggests that responses to morphogen gradients in the pre–steady-state regime can be much more robust than those at steady state [52], and preliminary calculations confirm that this is true for the RA gradient model described here (unpublished data). Thus, the sources of robustness in RA gradients may be many and varied.
Collaboration between RA and Fgfs appears to be emerging as a common motif in vertebrate pattern formation, not only in the nervous system (e.g., hindbrain, forebrain, ventral retina, and tailbud), but elsewhere (e.g., somites, heart, pancreas, and limbs). Intriguingly, the details of the interactions between RA and Fgfs are often quite different in different systems. For example, RA and Fgf gradients with opposite, rather than parallel, orientation are thought to play key roles in the patterning of somites, and recent modeling studies have provided insights into the tasks such an arrangement can accomplish [53,54]. It will be interesting to see whether or not there are common regulatory loops and control mechanisms in all RA/Fgf systems. It will also be interesting to see whether or not analogous collaborative relationships exist among other morphogens that pattern animal embryos.
Evolution of the RA Gradient System
The roles of Fgf and RA in A–P patterning appear to have arisen in the deuterostome lineage, as protostomes (e.g., nematodes and arthropods) apparently lack both RARs and the enzymes that synthesize RA [55]. Yet nuclear hormone receptors, including Retinoid X receptors (RXR)s, are expressed throughout the animal kingdom and serve in the detection of both endogenous hormones and environmental compounds [56,57]. Invertebrates also use cytochrome p450 enzymes to oxidize a wide variety of endogenous and environmental compounds [58,59]. Our results suggest that the initial step in the evolution of the RA gradient system may have occurred when one of these enzymes happened to fall under the control of an ancestral A–P patterning system, so that its expression in embryos became graded from posterior to anterior. As a result, any environmental substance that it degraded would automatically form a gradient within the embryo, the shape of which would be read out at the transcriptional level through RXR-mediated signaling. In this way, a primitive RA-like gradient system could be established that was later refined, e.g., by developing ways to store the precursor of the compound, and produce an active form of it at one end of the embryo.
Although we can only speculate as to the advantage gained by incorporating RA into a preexisting A–P patterning system, we note that Kerszberg [60] showed that gradients of nuclear hormone receptor ligands have an intrinsic ability to form domains of gene expression with multiple sharp boundaries. Bringing RA into A–P patterning may thus have been critical in the evolution of the highly subdivided, rhombomeric organization of the vertebrate hindbrain.
Materials and Methods
Animals.
Embryos were obtained in natural crosses and staged according to Kimmel et al. [61]. rare:yfp (Tg(RARE-gata2:NTD-eYFP)—ZDB-LOCUS-051209–1) fish [30] were a kind gift of E. Linney.
Morpholinos.
Antisense MOs were designed to translation start sites in cyp26b1 (5′-TCAAAACTCTCGAAGAGCATGGCTG-3′) and cyp26c1/d1 (5′-AAATCGTGCCCGAACATCTCGAACG-3′). aldh1a2- and cyp26a1-MOs have been described [11,24]. MOs were dissolved in Danieau buffer; 0.5 nl of solution was injected at the one-cell stage.
Cell transplantation.
rare:yfp experiments. Wild-type donor embryos were injected at the one-cell stage with a mixture of 1.5% TRITC-dextran (neutral, 10,000 M r), 1.5% biotinylated-dextran (lysine-fixable, 10,000 M r), and 2 ng of aldh1a2 RNA, and treated with 10 nM retinol (vitamin A). rare:yfp host embryos were injected with 1 ng of aldh1a2-MO. At sphere stage, embryos were mounted in 3% methylcellulose, and 30–50 cells were transplanted from the margins of labeled donor embryos to the margins of unlabeled hosts. Post-gastrula transplants were performed by mounting embryos in 0.75% low-melt agarose, and cells were transplanted from the somites of donor embryos into the forming somites of host embryos. Transplanted embryos were mounted in 1.5% agarose for analysis by confocal microscopy (using a Zeiss LSM510Meta). Images were processed, and the fluorescence intensity graph was produced using ImageJ software (http://rsb.info.nih.gov/ij/).
cyp26a1-MO experiments. Wild-type donor embryos were injected at the one-cell stage with a mixture of 1.5% Fluor-ruby-dextran (lysine-fixable, 10,000 M r), 1.5% biotinylated-dextran (lysine-fixable, 10,000 M r), and 2 ng of cyp26a1-MO. At 30%–50% epiboly (4.7–5.3 hpf), 30–50 cells were transplanted from the margins of labeled donor embryos to the margins of unlabeled wild-type hosts. Transplanted embryos were fixed at 90% epiboly (9 hpf) and stained for hoxb1b expression by in situ hybridization.
Pharmacological treatments.
The following stock solutions were stored in DMSO at −80 °C: 10 mM 4-(diethylamino)-benzaldehyde (DEAB; Acros organics), 10 mM all-trans retinoic acid (RA; Sigma), and 10 mM SU5402 (CalBiochem). These were diluted in Embryo Medium (EM) for treatments. Sibling controls were incubated in corresponding dilutions of DMSO. All incubations were done in the dark.
Bead implantation.
AG 1-X8 ion-exchange beads (formate form; BIORAD) were soaked in different concentrations of all-trans RA in EM for 1 h (control beads were incubated in similar amounts of DMSO in EM). Beads were rinsed in EM for 5 min before implantation. Embryos were mounted in 3% methylcellulose, a small slit was cut in the epidermis with a tungsten needle, and an RA/DMSO-coated bead (40–60 μm in diameter) was inserted through the slit. Embryos were then incubated at 28.5 °C in the dark prior to fixation.
Whole-mount in situ hybridization.
In situ hybridization was carried out as described previously [62]. Probes included hoxb4, hoxb5a, and hoxd4a [31], cyp26a1 [6], cyp26b1 (IMAGEclone3722563; EcoRI, SP6), cyp26c1/d1 [63], hoxb1b [64], krox-20/egr2b [65], and pax2a [66] . Images were taken and measurements were made using Openlab software (http://www.improvision.com/products/openlab/). Ranges of hox gene induction (e.g., assayed as the distance from the edge of the bead to the most distant hox-expressing cell) were compared using a two-tailed Student t-test.
Modeling.
The RA gradient in the gastrula was modeled as a one-dimensional reaction diffusion system (cf. [41,67,68]) represented by the following equations.
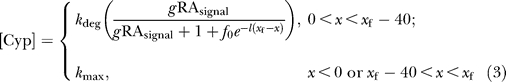 |
[RA]out and [RA]in represent extracellular and intracellular concentrations, respectively, of RA; RAsignal represents the strength of the RA signal (which may or may not vary linearly with [RA]in, in accordance with exponent n); and [Cyp] represents the intracellular concentration of Cyp26a1. D is the effective extracellular diffusion coefficient of RA; V(x) is the rate of production of RA at position x, typically taken to be a constant in a fixed posterior domain and zero elsewhere; k p is a first-order permeability coefficient that lumps together the processes of RA dissociation from protein, association with and diffusion through the plasma membrane, and reassociation with protein on the other side of the membrane; and k deg is a degradation constant for intracellular RA. Effects of [RA]in and Fgf on cyp26a1 expression are captured through multiplication of the parameter k deg by the expression containing γ, f 0, λ, and x f. Here γ is sensitivity to RAsignal-mediated feedback; f 0 represents the sensitivity of feedback to Fgf; λ is the length constant of the Fgf gradient (the inverse of the distance over which it drops to e −1 of its initial value); and x f is the location along the Fgf gradient where [Fgf] = f 0. The parameter β is introduced to allow the effective rate constant for flux of RA out of the extracellular space to be higher than for flux into the intracellular space (to capture the fact that some endogenous RA should leave the extracellular space entirely, e.g., by diffusing out of the embryo, or into long-term capture in yolk).
The x-axis is taken to represent the A–P axis of the gastrula-stage embryo, with x = 0 corresponding to the posterior border of an anterior domain of high cyp26a1 expression (approximately the r2/r3 boundary), and x = x f at the posterior margin. The boundary of a second domain of high cyp26a1 expression is placed 40 μm from the posterior margin. The value of [Cyp] in both of these domains is fixed at k max. A no-flux boundary condition is placed at x = −200 (to represent the fact that the problem could be viewed as symmetrical about the anterior-most point of the embryo). At the posterior margin, it was assumed that extracellular RA could only leave the embryo by diffusing through cells, so a “leaky” posterior boundary condition was used, in which the parameter k p captured the rate constant of leakage.
Steady-state solutions to the equations were obtained numerically using Mathematica software by solving the time-dependent equations for sufficiently long times that results no longer changed significantly (typically after a few hours). In some cases, steady-state calculations were done directly using a Matlab boundary value ODE Solver. Parameter values were manually explored over several orders of magnitude. In all panels of Figure 7, parameter values were D = 18 μm2sec−1, n = 2, λ = 0.019 μm−1, k p = 10−4sec−1, and γ = 2.5 × 10−5. (Other parameter values are given in the figure legend.) The choice of modest cooperativity in signaling (n = 2) acknowledges the intrinsic capabilities of nuclear hormone receptors [60], but is not, in fact, required to produce the qualitative findings of this study. It should be noted that the length scales of the RA gradients produced in Figure 7 are determined largely by the length scale of the Fgf gradient (1/λ), which was chosen arbitrarily, and the diffusion coefficient D of protein-bound RA, which is not known accurately. Accordingly the micrometer values on the abscissa should be taken as essentially arbitrary and easily adjustable to be several fold smaller or larger through modest changes in these parameter values.
Supporting Information
(A–C) Implantation of a bead soaked in 10 mM RA at 8 hpf induces YFP expression after 2 h. Single confocal sections show nuclear YFP expression (A) and cells labeled with BODIPY-TR (B). (C) Merged image. dashed circle indicates the position of the implanted bead.
(5.7 MB TIF)
(A–D) cyp26a1 is up-regulated in a concentration-dependent manner that also depends on a cell's position in the embryo (compare [B], [C], and [D] to [A]).
(3.1 MB TIF)
Embryos were treated at 4 hpf with DEAB either with or without a low concentration of RA between 0.625–20 nM, fixed at 19 hpf, and pattern was assessed by in situ hybridization for hoxd4a (blue; r6/7 boundary), krox-20 (egr2b, red; r3 and r5), and pax2a (red; midbrain–hindbrain boundary).
(A) Wild-type expression.
(B) DEAB-treated embryos show loss of r5–r7.
(C–F) Concentrations from 0.625–5 nM rescue wild-type patterning to the same extent.
(G–H) Higher concentrations produce anterior defects.
(9.5 MB TIF)
Accession Numbers
The GenBank (http://www.ncbi.nlm.nih.gov/Genbank) accession numbers for the genes discussed in this paper are aldh1a2 (NM_131850), cyp26a1 (NM_131146), cyp26b1 (NM_212666), cyp26c1 (NM_001029951), hoxb1b (NM_131142) hoxb4 (NM_131118), hoxb5a (NM_131101), hoxd4a (NM_130757), krox-20/egr2b (NM_130997), and pax2a (NM_131184).
The ZFIN (Zebrafish Model Organism database; http://www.zfin.org) ID numbers are aldh1a2 (ZDB-GENE-011010–3), cyp26a1 (ZDB-GENE-990415–44), cyp26b1 (ZDB-GENE-990415–44), cyp26c1 (ZDB-GENE-050714–2), hoxb1b (ZDB-GENE-980526–290), hoxb4 (ZDB-GENE-990415–105), hoxb5a (ZDB-GENE-980526–70), hoxd4a (ZDB-GENE-980526–214) krox-20/egr2b (ZDB-GENE-980526–283), and pax2a (ZDB-GENE-990415–8).
Acknowledgments
We are grateful to Elwood Linney for the rare:yfp fish, and Yasuhiko Kawakami for the cyp26c1 cDNA. We thank Frederic Wan for insights into the dynamics of gradient formation, and Ira Blitz and members of the Schilling lab for critical reading of the manuscript.
Abbreviations
- A–P
anterior–posterior
- Fgf
fibroblast growth factor
- hpf
hours post-fertilization
- MO
Morpholino
- RA
retinoic acid
- RARE
retinoic acid response element
- YFP
yellow fluorescent protein
Footnotes
Author contributions. RJW, ADL and TFS conceived and designed the experiments and wrote the paper. RJW performed the experiments. RJW, QN, ADL, and TFS analyzed the data. QN and ADL contributed reagents/materials/analysis tools.
Funding. This work was supported by the National Institutes of Health grants NS-41353 to TFS and GM67247 and GM76516 to QN and ADL.
Competing interests. The authors have declared that no competing interests exist.
References
- Cox WG, Hemmati-Brivanlou A. Caudalization of neural fate by tissue recombination and bFGF. Development. 1995;121:4349–4358. doi: 10.1242/dev.121.12.4349. [DOI] [PubMed] [Google Scholar]
- Lamb TM, Harland RM. Fibroblast growth factor is a direct neural inducer, which combined with noggin generates anterior-posterior neural pattern. Development. 1995;121:3627–3636. doi: 10.1242/dev.121.11.3627. [DOI] [PubMed] [Google Scholar]
- McGrew LL, Lai CJ, Moon RT. Specification of the anteroposterior neural axis through synergistic interaction of the Wnt signaling cascade with noggin and follistatin. Dev Biol. 1995;172:337–342. doi: 10.1006/dbio.1995.0027. [DOI] [PubMed] [Google Scholar]
- Koshida S, Shinya M, Mizuno T, Kuroiwa A, Takeda H. Initial anteroposterior pattern of the zebrafish central nervous system is determined by differential competence of the epiblast. Development. 1998;125:1957–1966. doi: 10.1242/dev.125.10.1957. [DOI] [PubMed] [Google Scholar]
- Kiecker C, Niehrs C. A morphogen gradient of Wnt/beta-catenin signalling regulates anteroposterior neural patterning in Xenopus. Development. 2001;128:4189–4201. doi: 10.1242/dev.128.21.4189. [DOI] [PubMed] [Google Scholar]
- Kudoh T, Wilson SW, Dawid IB. Distinct roles for Fgf, Wnt and retinoic acid in posteriorizing the neural ectoderm. Development. 2002;129:4335–4346. doi: 10.1242/dev.129.18.4335. [DOI] [PubMed] [Google Scholar]
- Scholpp S, Brand M. Endocytosis controls spreading and effective signaling range of Fgf8 protein. Curr Biol. 2004;14:1834–1841. doi: 10.1016/j.cub.2004.09.084. [DOI] [PubMed] [Google Scholar]
- Hernandez RE, Putzke AP, Myers JP, Margaretha L, Moens CB. Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development. Development. 2007;134:177–187. doi: 10.1242/dev.02706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederreither K, Vermot J, Schuhbaur B, Chambon P, Dolle P. Retinoic acid synthesis and hindbrain patterning in the mouse embryo. Development. 2000;127:75–85. doi: 10.1242/dev.127.1.75. [DOI] [PubMed] [Google Scholar]
- White JC, Highland M, Kaiser M, Clagett-Dame M. Vitamin A deficiency results in the dose-dependent acquisition of anterior character and shortening of the caudal hindbrain of the rat embryo. Dev Biol. 2000;220:263–284. doi: 10.1006/dbio.2000.9635. [DOI] [PubMed] [Google Scholar]
- Begemann G, Schilling TF, Rauch GJ, Geisler R, Ingham PW. The zebrafish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development. 2001;128:3081–3094. doi: 10.1242/dev.128.16.3081. [DOI] [PubMed] [Google Scholar]
- Maves L, Kimmel CB. Dynamic and sequential patterning of the zebrafish posterior hindbrain by retinoic acid. Dev Biol. 2005;285:593–605. doi: 10.1016/j.ydbio.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Molotkova N, Molotkov A, Sirbu IO, Duester G. Requirement of mesodermal retinoic acid generated by Raldh2 for posterior neural transformation. Mech Dev. 2005;122:145–155. doi: 10.1016/j.mod.2004.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begemann G, Marx M, Mebus K, Meyer A, Bastmeyer M. Beyond the neckless phenotype: influence of reduced retinoic acid signaling on motor neuron development in the zebrafish hindbrain. Dev Biol. 2004;271:119–129. doi: 10.1016/j.ydbio.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Gale E, Zile M, Maden M. Hindbrain respecification in the retinoid-deficient quail. Mech Dev. 1999;89:43–54. doi: 10.1016/s0925-4773(99)00202-6. [DOI] [PubMed] [Google Scholar]
- Maden M, Gale E, Kostetskii I, Zile M. Vitamin A-deficient quail embryos have half a hindbrain and other neural defects. Curr Biol. 1996;6:417–426. doi: 10.1016/s0960-9822(02)00509-2. [DOI] [PubMed] [Google Scholar]
- Dupe V, Lumsden A. Hindbrain patterning involves graded responses to retinoic acid signalling. Development. 2001;128:2199–2208. doi: 10.1242/dev.128.12.2199. [DOI] [PubMed] [Google Scholar]
- Durston AJ, Timmermans JP, Hage WJ, Hendriks HF, de Vries NJ, et al. Retinoic acid causes an anteroposterior transformation in the developing central nervous system. Nature. 1989;340:140–144. doi: 10.1038/340140a0. [DOI] [PubMed] [Google Scholar]
- Sive HL, Draper BW, Harland RM, Weintraub H. Identification of a retinoic acid-sensitive period during primary axis formation in Xenopus laevis. Genes Dev. 1990;4:932–942. doi: 10.1101/gad.4.6.932. [DOI] [PubMed] [Google Scholar]
- Marshall H, Nonchev S, Sham MH, Muchamore I, Lumsden A, et al. Retinoic acid alters hindbrain Hox code and induces transformation of rhombomeres 2/3 into a 4/5 identity. Nature. 1992;360:737–741. doi: 10.1038/360737a0. [DOI] [PubMed] [Google Scholar]
- Godsave SF, Koster CH, Getahun A, Mathu M, Hooiveld M, et al. Graded retinoid responses in the developing hindbrain. Dev Dyn. 1998;213:39–49. doi: 10.1002/(SICI)1097-0177(199809)213:1<39::AID-AJA4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Abu-Abed S, Dolle P, Metzger D, Beckett B, Chambon P, et al. The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures. Genes Dev. 2001;15:226–240. doi: 10.1101/gad.855001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y, Meno C, Fujii H, Nishino J, Shiratori H, et al. The retinoic acid-inactivating enzyme CYP26 is essential for establishing an uneven distribution of retinoic acid along the anterio-posterior axis within the mouse embryo. Genes Dev. 2001;15:213–225. doi: 10.1101/gad.851501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto Y, Wada H, Okamoto H, Kudo A, Imai Y. Retinoic acid-metabolizing enzyme Cyp26a1 is essential for determining territories of hindbrain and spinal cord in zebrafish. Dev Biol. 2005;278:415–427. doi: 10.1016/j.ydbio.2004.11.023. [DOI] [PubMed] [Google Scholar]
- Sirbu IO, Gresh L, Barra J, Duester G. Shifting boundaries of retinoic acid activity control hindbrain segmental gene expression. Development. 2005;132:2611–2622. doi: 10.1242/dev.01845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichele G, Thaller C. Characterization of concentration gradients of a morphogenetically active retinoid in the chick limb bud. J Cell Biol. 1987;105:1917–1923. doi: 10.1083/jcb.105.4.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpert L. Positional information revisited. Development. 1989;107(Suppl):3–12. doi: 10.1242/dev.107.Supplement.3. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Krumlauf R. Patterning the vertebrate neuraxis. Science. 1996;274:1109–1115. doi: 10.1126/science.274.5290.1109. [DOI] [PubMed] [Google Scholar]
- Marshall H, Morrison A, Studer M, Popperl HRK. Retinoids and Hox genes. FASEB J. 1996;10:969–978. [PubMed] [Google Scholar]
- Perz-Edwards A, Hardison NL, Linney E. Retinoic acid-mediated gene expression in transgenic reporter zebrafish. Dev Biol. 2001;229:89–101. doi: 10.1006/dbio.2000.9979. [DOI] [PubMed] [Google Scholar]
- Prince VE, Joly L, Ekker M, Ho RK. Zebrafish hox genes: genomic organization and modified colinear expression patterns in the trunk. Development. 1998;125:407–420. doi: 10.1242/dev.125.3.407. [DOI] [PubMed] [Google Scholar]
- Gould A, Itasaki N, Krumlauf R. Initiation of rhombomeric Hoxb4 expression requires induction by somites and a retinoid pathway. Neuron. 1998;21:39–51. doi: 10.1016/s0896-6273(00)80513-9. [DOI] [PubMed] [Google Scholar]
- Oosterveen T, Niederreither K, Dolle P, Chambon P, Meijlink F, et al. Retinoids regulate the anterior expression boundaries of 5' Hoxb genes in posterior hindbrain. EMBO J. 2003;22:262–269. doi: 10.1093/emboj/cdg029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo JE. Inhibition of mouse and human class 1 aldehyde dehydrogenase by 4-(N,N-dialkylamino)benzaldehyde compounds. Adv Exp Med Biol. 1997;414:217–224. doi: 10.1007/978-1-4615-5871-2_25. [DOI] [PubMed] [Google Scholar]
- Russo JE, Hauguitz D, Hilton J. Inhibition of mouse cytosolic aldehyde dehydrogenase by 4-(diethylamino)benzaldehyde. Biochem Pharmacol. 1988;37:1639–1642. doi: 10.1016/0006-2952(88)90030-5. [DOI] [PubMed] [Google Scholar]
- Reijntjes S, Blentic A, Gale E, Maden M. The control of morphogen signalling: regulation of the synthesis and catabolism of retinoic acid in the developing embryo. Dev Biol. 2005;285:224–237. doi: 10.1016/j.ydbio.2005.06.019. [DOI] [PubMed] [Google Scholar]
- Dobbs-McAuliffe B, Zhao Q, Linney E. Feedback mechanisms regulate retinoic acid production and degradation in the zebrafish embryo. Mech Dev. 2004;121:339–350. doi: 10.1016/j.mod.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Tsukada M, Schroder M, Seltmann H, Orfanos CE, Zouboulis CC. High albumin levels restrict the kinetics of 13-cis retinoic acid uptake and intracellular isomerization to all-trans retinoic acid and inhibit its anti-proliferative effect on SZ95 sebocytes. J Invest Dermatol. 2002;119:182–185. doi: 10.1046/j.1523-1747.2002.01816.x. [DOI] [PubMed] [Google Scholar]
- Eldar A, Rosin D, Shilo BZ, Barkai N. Self-enhanced ligand degradation underlies robustness of morphogen gradients. Dev Cell. 2003;5:635–646. doi: 10.1016/s1534-5807(03)00292-2. [DOI] [PubMed] [Google Scholar]
- Lander AD, Nie Q, Wan FY. Do morphogen gradients arise by diffusion? Dev Cell. 2002;2:785–796. doi: 10.1016/s1534-5807(02)00179-x. [DOI] [PubMed] [Google Scholar]
- Lander AD, Nie Q, Vargas B, Wan FY. Aggregation of a distributed source in morphogen gradient formation. SIAM Stud Appl Math. 2005;114:343–374. doi: 10.1111/j.0022-2526.2005.01556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpert L. Positional information and pattern formation. Curr Top Dev Biol. 1971;6:183–224. doi: 10.1016/s0070-2153(08)60641-9. [DOI] [PubMed] [Google Scholar]
- Thaller C, Eichele G. Retinoid signaling in vertebrate limb development. Ann N Y Acad Sci. 1996;785:1–11. doi: 10.1111/j.1749-6632.1996.tb56237.x. [DOI] [PubMed] [Google Scholar]
- Moss JB, Xavier-Neto J, Shapiro MD, Nayeem SM, McCaffery P, et al. Dynamic patterns of retinoic acid synthesis and response in the developing mammalian heart. Dev Biol. 1998;199:55–71. doi: 10.1006/dbio.1998.8911. [DOI] [PubMed] [Google Scholar]
- Tickle C. Morphogen gradients in vertebrate limb development. Semin Cell Dev Biol. 1999;10:345. doi: 10.1006/scdb.1999.0294. [DOI] [PubMed] [Google Scholar]
- Hochgreb T, Linhares VL, Menezes DC, Sampaio AC, Yan CY, et al. A caudorostral wave of RALDH2 conveys anteroposterior information to the cardiac field. Development. 2003;130:5363–5374. doi: 10.1242/dev.00750. [DOI] [PubMed] [Google Scholar]
- Yashiro K, Zhao X, Uehara M, Yamashita K, Nishijima M, et al. Regulation of retinoic acid distribution is required for proximodistal patterning and outgrowth of the developing mouse limb. Dev Cell. 2004;6:411–422. doi: 10.1016/s1534-5807(04)00062-0. [DOI] [PubMed] [Google Scholar]
- Nutt SL, Dingwell KS, Holt CE, Amaya E. Xenopus Sprouty2 inhibits FGF-mediated gastrulation movements but does not affect mesoderm induction and patterning. Genes Dev. 2001;15:1152–1166. doi: 10.1101/gad.191301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HA, Hyodo-Miura J, Nagamune T, Ueno N. FGF signal regulates gastrulation cell movements and morphology through its target NRH. Dev Biol. 2005;282:95–110. doi: 10.1016/j.ydbio.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Adams J. Structure-activity and dose-response relationships in the neural and behavioral teratogenesis of retinoids. Neurotoxicol Teratol. 1993;15:193–202. doi: 10.1016/0892-0362(93)90015-g. [DOI] [PubMed] [Google Scholar]
- Shiotsugu J, Katsuyama Y, Arima K, Baxter A, Koide T, et al. Multiple points of interaction between retinoic acid and FGF signaling during embryonic axis formation. Development. 2004;131:2653–2667. doi: 10.1242/dev.01129. [DOI] [PubMed] [Google Scholar]
- Bergmann S, Sandler O, Sberro H, Shnider S, Schejter E, et al. Pre-steady-state decoding of the Bicoid morphogen gradient. PLoS Biol. 2007;5:e46. doi: 10.1371/journal.pbio.0050046. doi: 10.1371/journal.pbio.0050046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbeter A, Gonze D, Pourquie O. Sharp developmental thresholds defined through bistability by antagonistic gradients of retinoic acid and FGF signaling. Dev Dyn. 2007;236:1495–1508. doi: 10.1002/dvdy.21193. [DOI] [PubMed] [Google Scholar]
- Baker RE, Maini PK. Travelling gradients in interacting morphogen systems. Math Biosci. 2007;209:30–50. doi: 10.1016/j.mbs.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Marletaz F, Holland LZ, Laudet V, Schubert M. Retinoic acid signaling and the evolution of chordates. Int J Biol Sci. 2006;2:38–47. doi: 10.7150/ijbs.2.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szanto A, Narkar V, Shen Q, Uray IP, Davies PJ, et al. Retinoid X receptors: X-ploring their (patho)physiological functions. Cell Death Differ. 2004;11(Suppl 2):S126–S143. doi: 10.1038/sj.cdd.4401533. [DOI] [PubMed] [Google Scholar]
- King-Jones K, Horner MA, Lam G, Thummel CS. The DHR96 nuclear receptor regulates xenobiotic responses in Drosophila. Cell Metab. 2006;4:37–48. doi: 10.1016/j.cmet.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Niwa R, Matsuda T, Yoshiyama T, Namiki T, Mita K, et al. CYP306A1, a cytochrome P450 enzyme, is essential for ecdysteroid biosynthesis in the prothoracic glands of Bombyx and Drosophila. J Biol Chem. 2004;279:35942–35949. doi: 10.1074/jbc.M404514200. [DOI] [PubMed] [Google Scholar]
- Bogwitz MR, Chung H, Magoc L, Rigby S, Wong W, et al. Cyp12a4 confers lufenuron resistance in a natural population of Drosophila melanogaster. Proc Natl Acad Sci U S A. 2005;102:12807–12812. doi: 10.1073/pnas.0503709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerszberg M. Accurate reading of morphogen concentrations by nuclear receptors: a formal model of complex transduction pathways. J Theor Biol. 1996;183:95–104. doi: 10.1006/jtbi.1996.0205. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Raya A, Raya RM, Rodriguez-Esteban C, Belmonte JC. Retinoic acid signalling links left-right asymmetric patterning and bilaterally symmetric somitogenesis in the zebrafish embryo. Nature. 2005;435:165–171. doi: 10.1038/nature03512. [DOI] [PubMed] [Google Scholar]
- McClintock JM, Carlson R, Mann DM, Prince VE. Consequences of Hox gene duplication in the vertebrates: an investigation of the zebrafish Hox paralogue group 1 genes. Development. 2001;128:2471–2484. doi: 10.1242/dev.128.13.2471. [DOI] [PubMed] [Google Scholar]
- Oxtoby E, Jowett T. Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res. 1993;21:1087–1095. doi: 10.1093/nar/21.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss S, Johansen T, Korzh V, Fjose A. Expression of the zebrafish paired box gene pax[zf-b] during early neurogenesis. Development. 1991;113:1193–1206. doi: 10.1242/dev.113.4.1193. [DOI] [PubMed] [Google Scholar]
- von Dassow G, Meir E, Munro EM, Odell GM. The segment polarity network is a robust developmental module. Nature. 2000;406:188–192. doi: 10.1038/35018085. [DOI] [PubMed] [Google Scholar]
- Eldar A, Dorfman R, Weiss D, Ashe H, Shilo BZ, et al. Robustness of the BMP morphogen gradient in Drosophila embryonic patterning. Nature. 2002;419:304–308. doi: 10.1038/nature01061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A–C) Implantation of a bead soaked in 10 mM RA at 8 hpf induces YFP expression after 2 h. Single confocal sections show nuclear YFP expression (A) and cells labeled with BODIPY-TR (B). (C) Merged image. dashed circle indicates the position of the implanted bead.
(5.7 MB TIF)
(A–D) cyp26a1 is up-regulated in a concentration-dependent manner that also depends on a cell's position in the embryo (compare [B], [C], and [D] to [A]).
(3.1 MB TIF)
Embryos were treated at 4 hpf with DEAB either with or without a low concentration of RA between 0.625–20 nM, fixed at 19 hpf, and pattern was assessed by in situ hybridization for hoxd4a (blue; r6/7 boundary), krox-20 (egr2b, red; r3 and r5), and pax2a (red; midbrain–hindbrain boundary).
(A) Wild-type expression.
(B) DEAB-treated embryos show loss of r5–r7.
(C–F) Concentrations from 0.625–5 nM rescue wild-type patterning to the same extent.
(G–H) Higher concentrations produce anterior defects.
(9.5 MB TIF)