Abstract
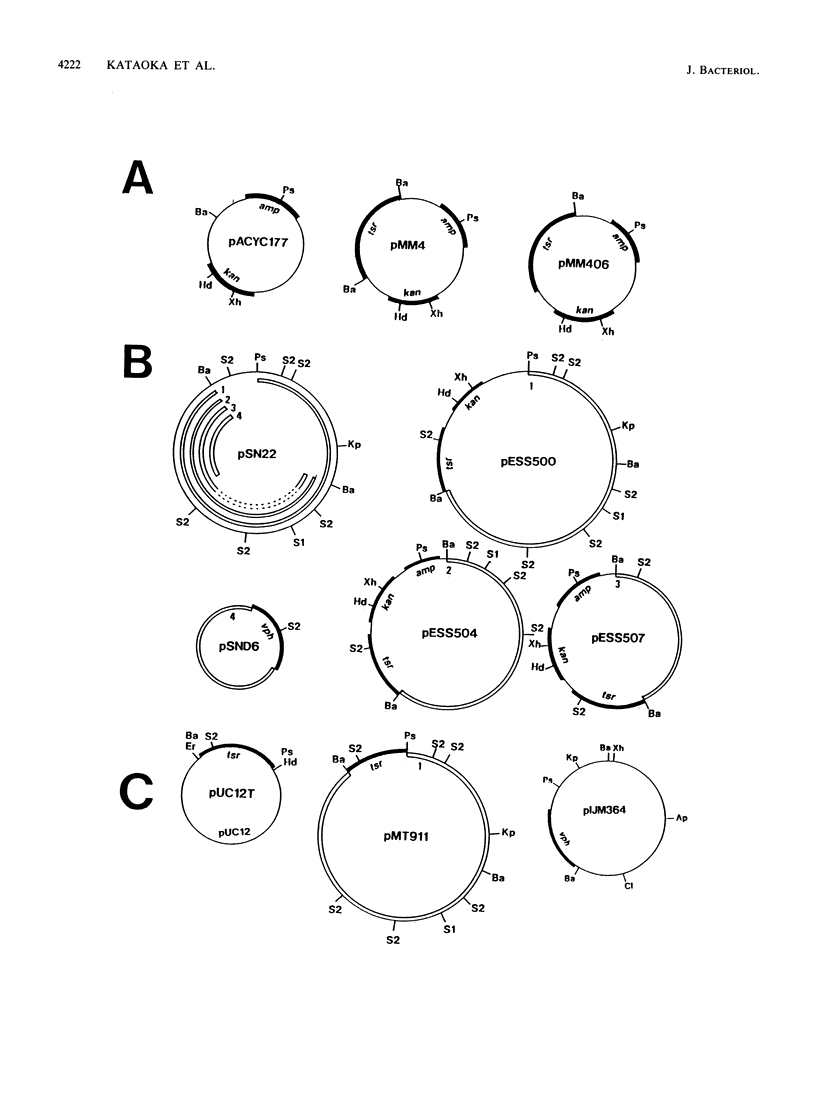
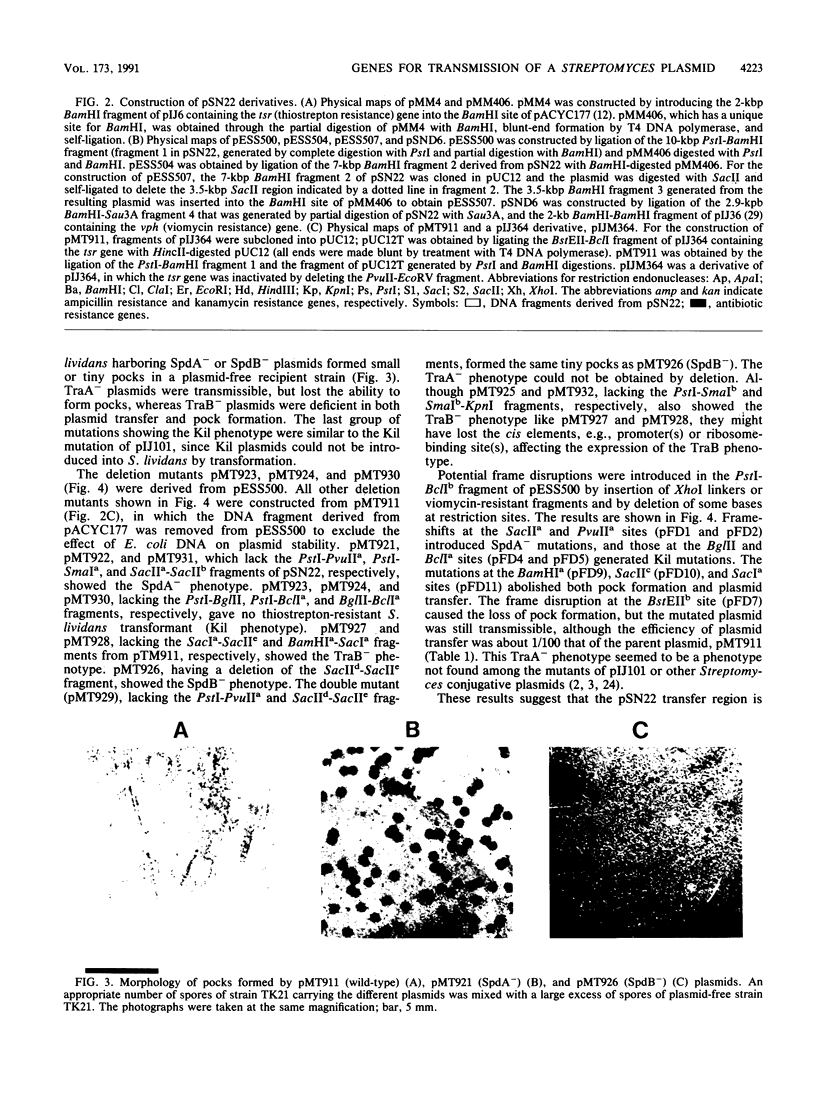
An 11-kbp multicopy plasmid, pSN22, was isolated from Streptomyces nigrifaciens SN22. pSN22 is self-transmissible (conjugative), is maintained stably in S. lividans, and forms pocks in a wide range of Streptomyces strains. Mutational analyses showed that a fragment of pSN22 contained five genes involved in plasmid transfer and pock formation. traB was essential for plasmid transfer. traA was required for pock formation, but not for plasmid transfer. spdA or spdB were concerned with pock size; mutations in these genes decreased pock size. The fifth gene, traR, could be deleted together with other genes to give nontransmissible plasmids, but plasmids with insertions or deletions only within traR became nonviable. traR is probably needed to counterbalance the lethal effects of another plasmid gene. Transfer of pSN22 promoted the cotransfer of nontransmissible plasmids and enhanced chromosome recombination between the host and recipient strains, suggesting that plasmid transfer accompanies cytoplasmic mixing.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akagawa H., Kawaguchi K., Ichihara M. Plasmids of Streptomyces kasugaensis MB273: their pock formation, their dispensable endonuclease cleavage sites for pock formation, and transformation of S. kasugaensis MB273 by them. J Antibiot (Tokyo) 1984 Sep;37(9):1016–1025. doi: 10.7164/antibiotics.37.1016. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccard F., Smokvina T., Pernodet J. L., Friedmann A., Guérineau M. The integrated conjugative plasmid pSAM2 of Streptomyces ambofaciens is related to temperate bacteriophages. EMBO J. 1989 Mar;8(3):973–980. doi: 10.1002/j.1460-2075.1989.tb03460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAVALLI L. L., LEDERBERG J., LEDERBERG E. M. An infective factor controlling sex compatibility in Bacterium coli. J Gen Microbiol. 1953 Feb;8(1):89–103. doi: 10.1099/00221287-8-1-89. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J. S., Carlton B. C. Conjugal plasmid transfer in Bacillus thuringiensis. Basic Life Sci. 1985;30:453–467. doi: 10.1007/978-1-4613-2447-8_33. [DOI] [PubMed] [Google Scholar]
- Clewell D. B. Plasmids, drug resistance, and gene transfer in the genus Streptococcus. Microbiol Rev. 1981 Sep;45(3):409–436. doi: 10.1128/mr.45.3.409-436.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Boyer H. W., Helling R. B. Construction of biologically functional bacterial plasmids in vitro. Proc Natl Acad Sci U S A. 1973 Nov;70(11):3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski D. H., Pohlman R. F., Bechhofer D. H., Prince A. S., Kelton C. A. Broad host range plasmid RK2 encodes multiple kil genes potentially lethal to Escherichia coli host cells. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1935–1939. doi: 10.1073/pnas.79.6.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant S. R., Lee S. C., Kendall K., Cohen S. N. Identification and characterization of a locus inhibiting extrachromosomal maintenance of the Streptomyces plasmid SLP1. Mol Gen Genet. 1989 Jun;217(2-3):324–331. doi: 10.1007/BF02464900. [DOI] [PubMed] [Google Scholar]
- HAYES W. [Observations on a transmissible agent determining sexual differentiation in Bacterium coli]. J Gen Microbiol. 1953 Feb;8(1):72–88. doi: 10.1099/00221287-8-1-72. [DOI] [PubMed] [Google Scholar]
- Hopwood D. A., Chater K. F., Dowding J. E., Vivian A. Advances in Streptomyces coelicolor genetics. Bacteriol Rev. 1973 Sep;37(3):371–405. doi: 10.1128/br.37.3.371-405.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood D. A., Kieser T., Wright H. M., Bibb M. J. Plasmids, recombination and chromosome mapping in Streptomyces lividans 66. J Gen Microbiol. 1983 Jul;129(7):2257–2269. doi: 10.1099/00221287-129-7-2257. [DOI] [PubMed] [Google Scholar]
- Hopwood D. A., Lydiate D. J., Malpartida F., Wright H. M. Conjugative sex plasmids of Streptomyces. Basic Life Sci. 1985;30:615–634. doi: 10.1007/978-1-4613-2447-8_43. [DOI] [PubMed] [Google Scholar]
- Ippen-Ihler K. A., Minkley E. G., Jr The conjugation system of F, the fertility factor of Escherichia coli. Annu Rev Genet. 1986;20:593–624. doi: 10.1146/annurev.ge.20.120186.003113. [DOI] [PubMed] [Google Scholar]
- Kendall K. J., Cohen S. N. Complete nucleotide sequence of the Streptomyces lividans plasmid pIJ101 and correlation of the sequence with genetic properties. J Bacteriol. 1988 Oct;170(10):4634–4651. doi: 10.1128/jb.170.10.4634-4651.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall K. J., Cohen S. N. Plasmid transfer in Streptomyces lividans: identification of a kil-kor system associated with the transfer region of pIJ101. J Bacteriol. 1987 Sep;169(9):4177–4183. doi: 10.1128/jb.169.9.4177-4183.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid. 1984 Jul;12(1):19–36. doi: 10.1016/0147-619x(84)90063-5. [DOI] [PubMed] [Google Scholar]
- Kinashi H., Shimaji M., Sakai A. Giant linear plasmids in Streptomyces which code for antibiotic biosynthesis genes. 1987 Jul 30-Aug 5Nature. 328(6129):454–456. doi: 10.1038/328454a0. [DOI] [PubMed] [Google Scholar]
- Lomovskaya N. D., Mkrtumian N. M., Gostimskaya N. L., Danilenko V. N. Characterization of temperate actinophage phi C31 isolated from Streptomyces coelicolor A3(2). J Virol. 1972 Feb;9(2):258–262. doi: 10.1128/jvi.9.2.258-262.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydiate D. J., Malpartida F., Hopwood D. A. The Streptomyces plasmid SCP2*: its functional analysis and development into useful cloning vectors. Gene. 1985;35(3):223–235. doi: 10.1016/0378-1119(85)90001-0. [DOI] [PubMed] [Google Scholar]
- Marinus M. G. Location of DNA methylation genes on the Escherichia coli K-12 genetic map. Mol Gen Genet. 1973 Dec 14;127(1):47–55. doi: 10.1007/BF00267782. [DOI] [PubMed] [Google Scholar]
- Omer C. A., Cohen S. N. Plasmid formation in Streptomyces: excision and integration of the SLP1 replicon at a specific chromosomal site. Mol Gen Genet. 1984;196(3):429–438. doi: 10.1007/BF00436190. [DOI] [PubMed] [Google Scholar]
- Pernodet J. L., Simonet J. M., Guérineau M. Plasmids in different strains of Streptomyces ambofaciens: free and integrated form of plasmid pSAM2. Mol Gen Genet. 1984;198(2):35–41. doi: 10.1007/BF00328697. [DOI] [PubMed] [Google Scholar]
- Stein D. S., Kendall K. J., Cohen S. N. Identification and analysis of transcriptional regulatory signals for the kil and kor loci of Streptomyces plasmid pIJ101. J Bacteriol. 1989 Nov;171(11):5768–5775. doi: 10.1128/jb.171.11.5768-5775.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas W. D., Jr, Archer G. L. Identification and cloning of the conjugative transfer region of Staphylococcus aureus plasmid pGO1. J Bacteriol. 1989 Feb;171(2):684–691. doi: 10.1128/jb.171.2.684-691.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. J., Kieser T., Ward J. M., Hopwood D. A. Physical analysis of antibiotic-resistance genes from Streptomyces and their use in vector construction. Gene. 1982 Nov;20(1):51–62. doi: 10.1016/0378-1119(82)90086-5. [DOI] [PubMed] [Google Scholar]
- Thompson C. J., Ward J. M., Hopwood D. A. Cloning of antibiotic resistance and nutritional genes in streptomycetes. J Bacteriol. 1982 Aug;151(2):668–677. doi: 10.1128/jb.151.2.668-677.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. J., Ward J. M., Hopwood D. A. DNA cloning in Streptomyces: resistance genes from antibiotic-producing species. Nature. 1980 Jul 31;286(5772):525–527. doi: 10.1038/286525a0. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Weber J. M., Wierman C. K., Hutchinson C. R. Genetic analysis of erythromycin production in Streptomyces erythreus. J Bacteriol. 1985 Oct;164(1):425–433. doi: 10.1128/jb.164.1.425-433.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieslander L. A simple method to recover intact high molecular weight RNA and DNA after electrophoretic separation in low gelling temperature agarose gels. Anal Biochem. 1979 Oct 1;98(2):305–309. doi: 10.1016/0003-2697(79)90145-3. [DOI] [PubMed] [Google Scholar]
- Wright L. F., Hopwood D. A. Identification of the antibiotic determined by the SCP1 plasmid of Streptomyces coelicolor A3(2). J Gen Microbiol. 1976 Jul;95(1):96–106. doi: 10.1099/00221287-95-1-96. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]