Abstract
Recent advances in the field of molecular clonotype analysis have enabled detailed repertoire characterization of viably isolated antigen-specific T cell populations directly ex vivo. However, in the absence of a biologically contained FACS facility, peripheral blood mononuclear cell (PBMC) preparations derived from patients infected with agents such as HIV must be formaldehyde fixed to inactivate the pathogen; this procedure adversely affects nucleic acid template quality. Here, we developed and validated a method to amplify and sequence mRNA species derived from formaldehyde fixed PBMC specimens. Antigen-specific CD8+ cytotoxic T-lymphocyte populations were identified with standard fluorochrome-conjugated peptide-major histocompatibility complex class I tetramers refolded around synthetic peptides representing immunodominant epitopes from HIV p24 Gag (KRWII[M/L]GLNK/HLA B*2705) and CMV pp65 (NLVPMVATV/HLA A*0201 and TPRVTGGGAM/HLA B*0702), and acquired in separate laboratories with or without fixation. In the presence of proteinase K pre-treatment, the observed antigen-specific CD8+ T-cell repertoire determined by molecular clonotype analysis was statistically no different whether derived from fixed or unfixed PBMC. However, oligo-dT recovery methods were not suitable for use with fixed tissue as significant skewing of clonotypic representation was observed. Thus, we have developed a reliable RNA-based method for molecular clonotype analysis that is compatible with formaldehyde fixation and therefore suitable for use with primary human samples isolated by FACS outside the context of a biological safety level 3 containment facility.
Keywords: clonotype, repertoire, mRNA, T-cell, formaldehyde
Introduction
Observations in both primate and murine models indicate that quality, as well as magnitude, can impact upon the effectiveness of antiviral CD8+ cytotoxic T-lymphocyte (CTL) responses (Price, 2004; Cornberg et al., 2006). One determinant of this quality is the nature of the cognate T-cell receptor (TCR) repertoire within the responding population. Virus-specific CTL populations can easily be identified directly ex vivo by flow cytometry after labelling with peptide-major histocompatibility complex (MHC) class 1 tetramers (Altman et al., 1996; Appay and Rowland-Jones, 2002). To characterise the diversity of these specific populations, investigators have employed phenotypic identification of antigen-specific T-cells using tetramers in combination with TCR ß-chain variable region-specific monoclonal antibodies. These techniques have since been superseded by PCR-based molecular methods such as ‘immunoscope’ and CDR3 junction sequence analysis of expressed TCR genes from purified populations isolated using fluorescence-activated cell-sorting (FACS) (Douek et al., 2002; Lee et al., 2002; Rufer, 2005). These technical advances allow high resolution decryption of the nature and breath of the clonal response to individual antigens and highlight that clonality can no longer simply be equated to a detectable expansion of cells as defined by a single Vß-specific monoclonal antibody (Maryanski et al., 1999; Lim et al., 2002; Price et al., 2004).
The isolation and purification of primary tissue by FACS poses a risk of operator exposure to pathogens due to the generation of aerosolised particles (Perfetto et al., 2003). Samples of interest may be derived from individuals infected with pathogens such as the human immunodeficiency virus (HIV), hepatitis C virus (HCV) or pathogenic Mycobacteria. In such cases, the only option for high purity isolation of these cells is to use a dedicated biologically contained cell sorter within a level-3 biological containment facility. This is an expensive solution available to only a small proportion of research institutes. For certain applications, contained sorting of antigen-specific cells can be achieved using alternative methods such as peptide-MHC class 1 proteins conjugated to magnetic beads. This method allows for the safe manipulation of samples within a class-2 bio-safety cabinet. However, in the context of TCR analysis, the relatively poor purity of such preparations leads to spurious results with respect to the breadth and complexity of the repertoire (Kelleher AD, Price DA, unpublished observations). Hence, our primary technical aim in this study was the effective purification by FACS and subsequent reliable molecular clonotypic analysis of antigen-specific T-cells isolated from primary human material without using biological containment.
The risk associated with the sorting of infected material is removed through formaldehyde fixation in conjunction with universal safety precautions (Schmid et al., 1999). This chemical fixative has been used as standard practice in analytical flow cytometry, allowing safety of the operator without the requirements for physical/biological containment (Lifson et al., 1986; Martin et al., 1987; Nicholson et al., 1993; Nicholson and Browning, 1994). The potent electrophilic reactivity of this compound fixes intra- and extra-cellular proteins and nucleic acids without altering the fluorescent properties of most standard fluorochromes. Furthermore, the small size of this compound allows for rapid diffusion and preservation of treated samples for several days. To achieve our research aim, we explored a methodology incorporating a formaldehyde fixation step to inactivate potential pathogens combined with FACS isolation of cells. We first determined the conditions required for effective fixation and inactivation of high-titre HIV-1 using formaldehyde. The limitation to this methodology is that formaldehyde profoundly interferes with the recovery nucleic acids, particularly RNA, using standardized methods (Diez et al., 1999; Masuda et al., 1999; Byers et al., 2004). To overcome these limitations, we have developed a method of recovering full-length mRNA template of high purity and quality from small numbers of formaldehyde-fixed antigen-specific CD8+ T-cells. Comparisons with paired samples isolated without fixation demonstrated that this methodology can be used for accurate and precise characterisation of the TCR repertoire, as determined by TRB CDR3 sequencing, from primary human samples without the need for a contained flow cytometric cell-sorting facility.
Materials and methods
Informed consent
Donors of peripheral blood samples provided informed consent according to local ethical guidelines with appropriate Institutional Review Board approval.
Peripheral blood mononuclear cell (PBMC) preparation
PBMC were isolated from acid-citrate dextrose anti-coagulated blood by standard density gradient centrifugation using Ficoll-Hypaque (Amersham Pharmacia). Cells were washed twice in RPMI-1640 medium supplemented with penicillin (100U/ml), streptomycin (100μg/ml) and glutamine (2mM), counted and then cryopreserved in 90% fetal calf serum (FCS)/10% dimethyl sulfoxide (JRH Biosciences) using a controlled rate freezer prior to storage in liquid nitrogen.
Assessment of RNA quality
RNA isolated from PBMC was assessed for quality according to the manufacturer's instructions using the Agilent 2100 Bioanalyzer and accompanying software (Agilent Technologies, Palo Alto, CA) with an RNA 6000 reference ladder (Ambion, Austin, TX).
HeLa/CD4/CCR5/CXCR4 (MAGIC-5) cell line infection with HIV-1 (NL4.3)
HeLa cells expressing human CD4, CCR5 and CXCR4 co-receptors (MAGIC-5) were maintained and infected with NL4.3 at an MOI of 25 as previously described (Hachiya et al., 2001). At day 3, once infection was established, aliquots of 1×105 cells were fixed by incubation for 60 minutes in varying concentrations of formaldehyde (0.0, 0.05, 0.1, 0.25 and 0.5% v/v) in PBS. Cells were then washed in PBS and transferred into a flask of uninfected MAGIC-5 cells for co-culture. Aliquots of supernatant were periodically tested over 22 days for the presence of the HIV-1 reverse transcriptase (RT) enzyme as previously described (Suzuki et al., 1995).
FACS analysis of antigen-specific CD8+ T-cells
Isolation of CD8+ antigen-specific T-cell populations by flow cytometry was performed in two separate facilities. Populations from healthy donors were isolated using a Becton Dickinson (BD) FACS Vantage-SE cytometer located at the St Vincent's Hospital Research Campus Core Flow Cytometry Suite. Cells specific for the CMVpp65 HLA B*0702-restricted epitope TPRVTGGGAM (TPR) from a healthy HIV uninfected donor (CPD) were sorted locally with and without formaldehyde fixation. Cells specific for the HIV p24 Gag HLA B*2705-restricted epitope KRWIILGLNK or its major variant KRWIIMGLNK (KRL/M), and cells specific for the CMVpp65 HLA A*0201-restricted epitope NLVPMVATV (NLV), were isolated with or without fixation from two HIV infected patients (057, 012) using a BD FACS Aria located in a biological safety level III (BSL-3) facility at the Vaccine Research Centre (VRC, NIH).
Monoclonal antibodies with the following specificities [clone] were used for flow cytometry at the St Vincent's Hospital Centre for Immunology (CFI): CD3-PerCP [SK7], CD8-FITC [SK1] (BD Biosciences) or CD8-ECD [SFCI21Thy2D3] (Beckman Coulter). Subsets of CD8+ T-cells were enumerated for TCR expression using FITC-conjugated antibodies specific for TCR Vß chains 1 [BL37.2], 2 [MPB2D5], 3 [CH92], 5.1 [IMMU157], 7 [ZOE], 8 [56C5.2], 11 [C21], 12 [VER2.32.1], 13.1 [IMMU222], 13.6 [JU74.3], 14 [CAS1.1.3], 16 [TAMAYA1.2], 17 [E17.5F3.15.13], 20 [ELL 1.4], 21.3 [IG125] or 22 [IMMU546] (Immunotech, Beckman Coulter). Monoclonal antibodies with the following specificities [clone] were used for flow cytometry at the VRC: CD8 [RPA-T8] (Invitrogen) conjugated to Q-dot 705 (Quantum Dot Corporation); CD14 [M5E5] (Invitrogen) and CD19 [internal VRC clone] conjugated to Pacific Blue together with violet amine reactive viability dye (ViViD; Invitrogen) were used to gate out irrelevant cells.
PBMC were thawed and washed in warm RPMI-1640 medium supplemented with penicillin (100U/ml), streptomycin (100μg/ml), glutamine (2mM) and 10% FCS (R10); viability was assessed with trypan blue staining. Sample preparation at the VRC comprised: incubation of PBMC with tetramers at 37°C for 20 minutes, followed by a wash with PBS and labelling with CD8 Q-dot 705, CD14-Pacific Blue and CD19-Pacific Blue monoclonal antibodies for 15 minutes at 4°C, followed by a second wash in PBS and staining with viability dye for 15 minutes before a final PBS wash and re-suspension in sort buffer. Sample preparation at the CFI comprised: incubation of PBMC with tetramers at 37°C for 20 minutes, followed by staining with CD3-PerCP and CD8-FITC/ECD at 4°C for 20 minutes, then a single wash with PBS prior to resuspension in sort buffer. Sort buffer consisted of PBS containing 1mM EDTA, 25mM HEPES (Gibco) and, where required, 1% (v/v) paraformaldehyde (Electron Microscopy Science, PA).
Cells collected at the VRC BSL-3 containment facility were gated via the small lymphocyte subset (forward scatter [FSC] versus side scatter [SSC]), followed by positive selection of CD8+ cells separated against negative selection of CD14, CD19 and viability dye positive cells (dump channel). Tetramer-labelled CD8+ cells (PE or APC) were collected directly into RNAlater as described previously (Price et al., 2004). The gating strategy used at the CFI differed from the above by firstly gating via FSC-A vs. FSC-H to remove doublets, then on the small lymphocyte subset, followed by positive selection of CD3+CD8+ T-cells. Tetramer-labelled CD3+CD8+ cells (PE) were gated and collected directly into an empty RNAse-free micro tube.
Preparation of peptide-MHC class I tetrameric complexes
Tetramers were prepared as described previously (Altman et al., 1996; Ogg et al., 1998). Briefly, tetramers presenting CMVpp65 epitopes were generated using either the HLA B*0702-restricted peptide TPRVTGGGAM (TPR) or the HLA A*0201-restricted peptide NLVPMVATV (NLV). HIVp24 Gag tetramers were generated using both variants of the HLA B*2705-restricted epitope KRWII(L/M)GLNK (KRL/M). Monomeric complexes were conjugated to streptavidin-R-phycoerythrin (R-PE) or streptavidin-allophycocyanin (APC) from ProZyme (Oxford GlykoSciences Ltd or Molecular Probes, Invitrogen). Aliquots of tetramers from the same monomer stock conjugated to either R-PE or APC were titrated so that similar results were obtained from either preparation.
Recovery of total RNA from sorted samples using Trizol reagent (CFI)
Fixed specimens were sorted directly into an RNAse-free 1.5mL micro tube and then centrifuged at 500g for 30 seconds. Recovery of total RNA from fixed specimens involved re-suspension in 200μL ice-cold lysis buffer consisting of 50mM Tris, 100mM NaCl, 5mM MgCl2, 0.5% Nonidet P-40 (USB), 40U Protector RNAse Inhibitor (Roche) and 1mM dithiothreitol (DTT) (Invitrogen). Sodium dodecyl-sulfate (Sigma Aldrich) was added to a final concentration of 0.5%, followed by 100μg Proteinase K (Roche). This was mixed, centrifuged at 12000g for 30 seconds and then incubated on a dry heating block at 55°C for 60 minutes. Samples were then cooled and centrifuged at 500g for 30 seconds at 4°C; monophasic phenol/guanidine isothiocyanate (Trizol-LS reagent; Invitrogen), referred to as Trizol from hereon, was then added at a 3:1 ratio to lysis buffer (v/v). Samples were mixed gently and incubated at room temperature for 5 minutes. To separate the phases, 200μL of chloroform (Sigma) was added and mixed thoroughly for 15 seconds, settled for 3 minutes at room temperature, then centrifuged at 12000g for 15 minutes. To precipitate RNA, 10μg of glycogen (Roche) and an equal original volume of 2-propanol was added and incubated on ice for 10 minutes; RNA was pelleted by centrifugation at 12000g for 10 minutes, washed with 1mL cold 70% EtOH, centrifuged at 7600g for 5 minutes and then dried for 10 minutes at 37°C. RNA was then resuspended in 20μL DEPC water pre-heated to 60°C and incubated at 60°C for a further 5 minutes. Samples were cooled on ice prior to freezing at −70°C.
Recovery of messenger RNA from sorted samples using oligo-dT beads (VRC)
Direct mRNA recovery from formaldehyde-fixed specimens using oligo-dT beads (Oligotex kit, Qiagen) involved re-suspension in a urea/SDS-based lysis buffer consisting of 700mM urea (Sigma), 500mM NaCl, 50mM HEPES (pH 7.3), 1% SDS, 10mM DTT, 40U Protector RNAse inhibitor and 100μg Proteinase K. Samples were treated in urea/SDS lysis buffer at 55°C for 60 minutes and centrifuged at 14000g for 3 minutes. To isolate mRNA, the sample was mixed with 20μL oligo-dT beads and mRNA recovered according to manufacturer's instructions.
Specimens without formaldehyde treatment were sorted into 100μL RNAlater (Ambion) prior to RNA recovery using the CFI or VRC methods. Samples were processed in accordance with the manufacturers' instructions.
Amplification of expressed TRB genes
First-strand cDNA template generated from random primers used a reaction mix containing 1.2μg random hexamer primers and 2.5mM dNTP (both Invitrogen), 40U Protector RNAse Inhibitor, Expand RTase buffer, 10mM DTT, and 1μL Expand reverse transcriptase (all Roche). Template was incubated for 60 minutes at 42°C before storage at −20°C.
TRBV3-specific amplification was performed using BV3 (5' – TCTAGAGAGAAGAAGGAGCGC – 3') and BC (5' – CTTCTGATGGCTCAAACAC – 3') primers at 500nM, Expand Hi-Fidelity PCR buffer, 250nM dNTPs and 3U Expand Hi-Fidelity DNA Polymerase (Roche). PCR conditions were 94°C for 2 minutes, 35 cycles at 94°C for 15 seconds, 54°C for 30 seconds and 72°C for 1 minute with a final extension period of 72°C for 10 minutes.
Unbiased TRB first-strand cDNA template was generated using a non-nested anchored template-switch RT-PCR with a 3' TRB constant region primer (5' – TTCTGATGGCTCAAACACAGCGAC – 3') as described previously (Douek et al., 2002). Conditions for thermal cycling were 94°C for 30 seconds, 5 rounds at 94°C for 5 seconds, 72°C for 2 minutes, 5 rounds at 94°C for 5 seconds, 70°C for 10 seconds, 72°C for 2 minutes and 35 rounds at 94°C for 5 seconds, 68°C for 10 seconds, 72°C for 2 minutes. Product amplified from sorted cells was visualised on a 2% agarose gel and purified. Purified product was ligated into either a TOPO-TA cloning vector (Invitrogen) and transformed into chemically competent TOP10 E.coli (CFI), or ligated into a pGEMT easy vector (Promega) and transformed into E.coli DH5α (VRC). Colonies were selected by blue/white screening and carriage of inserts was confirmed by PCR amplification of inserts using generic M13 primers. A minimum of 50 clones was sequenced per sample using a BigDye v3.1 sequencing reaction on an ABI 3730xl capillary sequencing machine (AB systems).
Clonotype identification
Subsets of CD8+ T-cells identified by flow cytometry for TCR expression using antibodies and primers specific for TCR Vß chains were designated according to the nomenclature of Arden (Arden et al., 1995). Sequences derived from unbiased TCR amplification were aligned using Sequencher (Gene Codes Corporation) and clonotype identity confirmed using the Immunogenetics online sequence analysis algorithm (IMGT, the international ImMunoGeneTics information system® http://imgt.cines.fr (Initiator and coordinator: Marie-Paule Lefranc, Montpellier, France)). Sequence analysis and presented clonotype data are based on amino acid alignment of the expressed TRB gene products. There were no or only rare incidences of multiple nucleotide sequences encoding the same product.
Statistical analysis
The similarity between paired sequence data sets was quantified using the Morisita-Horn similarity index (C-MH) (reviewed in (Magurran, 2003)), which accounts for both the variation of clonotypes and the number of copies of each clonotype. The statistical significance of the similarity measure between each pair of sequence data sets was determined using a randomisation test (Venturi et al., 2007). The p-value associated with each similarity measure represents the probability that a similarity less than or equal to the observed similarity could have been obtained by chance (i.e. the p-value indicates wether the two samples are significantly different).
Results
Inactivation of high titre HIV-1 in MAGIC-5 cell culture by formaldehyde fixation
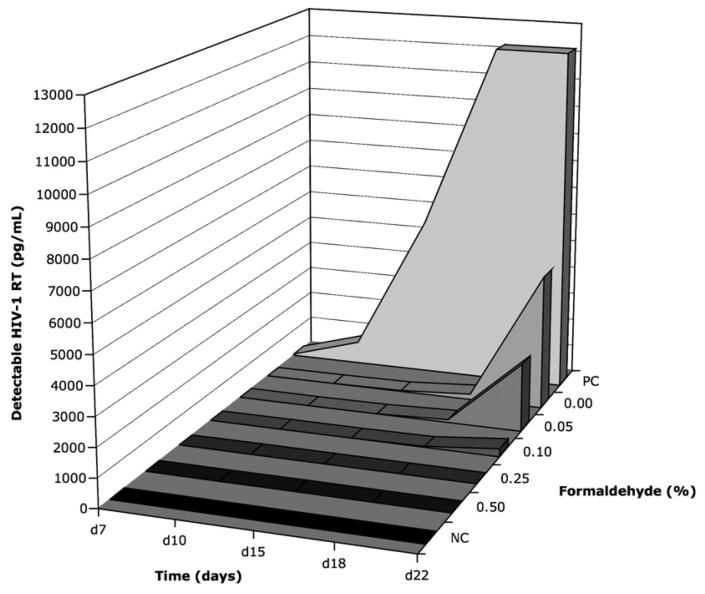
To assess the efficacy of viral inactivation with formaldehyde treatment at various concentrations, measurements of viral replication in MAGIC-5 cell co-culture supernatant were collected up to day 22 after seeding with treated cells or controls, after which time the experiment was terminated (Figure 1). RT enzyme was detectable in the positive control culture (PC) at all time points, increasing from 58pg/mL at day 7 to >12000pg/mL at day 15. Undetectable activity levels were observed in supernatants at all time points from the negative control (NC) at no greater 0.41pg/mL. The 0.25% and 0.5% formaldehyde treated cultures were below this cut-off at 0.21pg/mL for all time points measured. When formaldehyde concentrations were reduced further, cultures treated with 0.10% formaldehyde displayed no detectable RT enzyme activity before day 18 (29pg/mL); cultures treated with 0.0% and 0.05% formaldehyde exhibited undetectable RT activity before day 15 (10 and 30pg/mL, respectively). At day 22, the presence of HIV-1 virus replicating within these cultures (2401pg/mL for 0.05% and 4783pg/mL for 0.0% formaldehyde fixation) was coincident with syncytia formation (data not shown). Thus, from these data, we did not detect HIV-1 RT in MAGIC-5 cell cultures fixed at concentrations above 0.25% formaldehyde (v/v). Since a minimum concentration of 0.25% is therefore required for effective HIV-1 inactivation, the use of 1.0% (v/v) formaldehyde should introduce a robust safety margin for cell sorting by flow cytometry. A standardized operating procedure was developed for use in conjunction with universal safety precautions (Schmid et al., 1997; Schmid et al., 1999; Perfetto et al., 2004). This procedure was approved by the local Institutional Biosafety committee prior to commencement of sorting experiments.
Figure 1. Inactivation of HIV-1 using formaldehyde (0.00 - 0.50%) as measured by RT enzyme (pg/mL) production in MAGIC-5 co-culture supernatant.
Background for the reverse transcriptase (RT) assay was negative control uninfected MAGIC-5 culture (NC). Positive control (PC) cultures were established from untreated samples. The lag observed in viral replication for 0.00% treated culture is associated with removal of free virus through necessary wash steps after formaldehyde fixation.
Qualitative assessment of RNA recovery from fixed samples
Lysis and extraction of RNA from cells was based on a previously described protocol (Church and Buckler, 1999). This is a gentle detergent-based lysis protocol, rather than the more common mechanical homogenisation techniques. Post-extraction RNA quality was examined in paired samples of 1 ×106 unsorted PBMC. Cells were treated for 60 minutes at 4°C using PBS containing 1% formaldehyde or PBS only. Quality was assessed by measuring the ratio of 28S:18S ribosomal RNA peaks, using an Agilent Bioanalyzer (Figure 2).
Figure 2. RNA electropherograms to assess RNA quality according to treatment conditions.
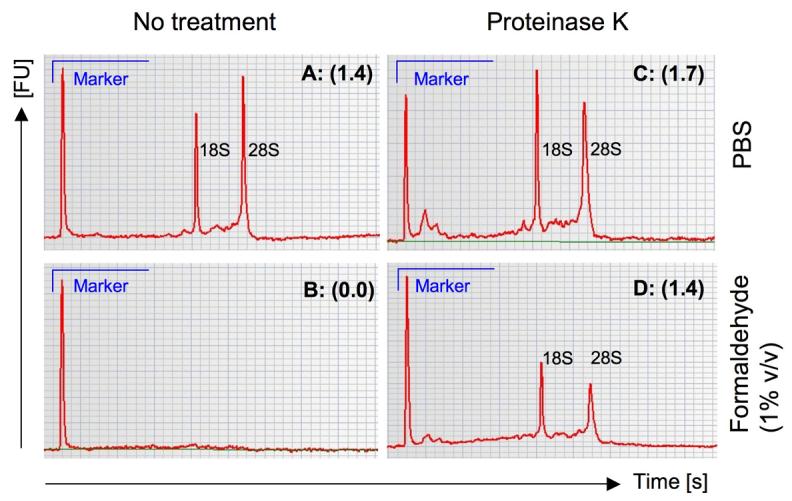
Measurements of 28S:18S ratio (brackets) were calculated based on the area under the curve, detected as fluorescence units [FU] over time in seconds [s] for individual peaks. Whole PBMC fractions without pre-treatment are shown for both unfixed (A) and fixed (B) conditions. Identical sample treatments using proteinase K pre-treatment were compared for PBS in (C) and buffered formaldehyde in (D).
Several methods were tested for recovery of mRNA template from fixed PBMC; these included silica-based based spin columns and whole liquid techniques used in combination with heat and/or enzymatic digestion buffers with or without Proteinase K, according to both in-house and commercial protocols (data not shown). In brief, quantitative recovery was generally poorer when extracting total RNA using spin-columns as compared to the CFI or VRC methods. In all circumstances, there was no recovery of RNA after formaldehyde fixation without ancillary pre-treatment. If higher temperature incubations were used to reverse fixation, the quality of the RNA suffered substantially especially at low cell numbers.
Mock fixation (PBS alone) of a PBMC sample yielded a 28S:18S rRNA ratio of 1.4 following Trizol based extraction and was used as our comparator (Figure 2A). As expected, no measurable RNA was recovered from formaldehyde-fixed PBMC using Trizol only (Figure 2B). Mock fixation using PBS only with proteinase K pre-treatment for 30 minutes at 37°C improved the quality of recovered material, yielding a ratio of 1.7 (Figure 2C). Importantly, proteinase K pre-treatment improved the quality and quantity of RNA recovered using Trizol from PBMC fixed using 1% formaldehyde, with the 28S:18S ratio improving from 0.0 to 1.4 (Figure 2D). We established from this data that the quality of template recovered from formaldehyde-fixed PBMC using proteinase K pre-treatment is suitable to attempt cDNA synthesis.
In order to demonstrate that this template could be reverse transcribed for amplification of rearranged TCR mRNA by RT-PCR, a CD8+ T-cell population with known Vß usage was identified. PBMC from a healthy CMV seropositive donor (CPD from hereon) were stained with a panel of specific anti-TCR Vβ antibodies. Expanded sub-populations were defined as greater than two standard deviations above the population mean for detected Vß pools. In CPD, Vß3 was over-represented (23.2%) within the total population and suitable for use as a positive control for amplification of transcribed TCR mRNA.
Template recovered using the CFI method from 5 × 104 PBMC and reverse transcribed using random hexamers was amplified via PCR using a BV3 gene-specific and BV constant region primers. The quantity of the PCR amplicon generated was indirectly measured by band density and used to judge the optimal conditions for proteinase K pre-treatment. All PBMC samples were fixed in buffered 1% formaldehyde and subsequently incubated with proteinase K for various times (5, 15, 30 or 60 minutes) at a range of temperatures (0 – 4°C, 23°C, 37°C and 55°C), prior to extraction using the CFI method (Figure 3A).
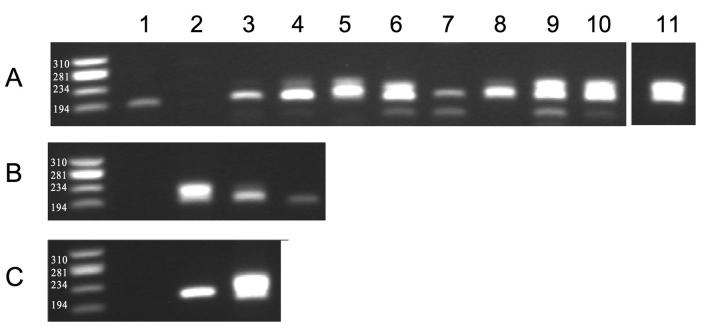
Figure 3. Optimisation of template recovery from formaldehyde-fixed PBMC as assessed by amplification of rearranged T-cell receptor.
A. Effect of proteinase K pre-treatment of fixed material across a range of temperatures and time as measured by amplification of TCR BV3 mRNA template. Treatment conditions were as follows: room temperature for 60 minutes (lane 1); ice for 60 minutes (lane 2); samples digested at 37°C for 5, 15, 30 and 60 minutes (lanes 3, 4, 5 and 6 respectively); samples digested at 55°C for 5, 15, 30 and 60 minutes (lanes 7, 8, 9 and 10 respectively). Lane 11 shows the positive control for unfixed material digested at 37°C for 30 minutes. B. Detectable limit of template as measured by TRBV3 amplification after digestion at 55°C for 60 minutes. Negative control (lane 1), and dilution of template material from 5000 (lane 2) to 500 (lane 3) and 50 (lane 4) cells. C. Detection of TRBV3 amplified template from antigen-specific CD8+ T-cells sorted using the CMVpp65 HLA B*0702/TPR tetramer. Negative control (lane 1), HLA B*0702/TPR-labelled (tetramer +) cells (lane 2) and non-specific (tetramer −) cells (lane 3).
Digestion in lysis buffer at 37°C over a period of 60 minutes incrementally increased the recovery of RNA (at 5, 15, 30 and 60 minutes; lanes 3 to 6, respectively) as detected by the strength of amplified product. The same pattern of incremental RNA recovery from fixed PBMC was noted at 55°C. Incubation at 55°C appeared to improve upon the corresponding yield from cells incubated at 37°C. By observation of the gel, the yield from fixed PBMC pre-treated with proteinase K at 55°C of 60 minutes was no different to the unfixed control (lane 11).
Next, we determined the minimum cell number from which we could amplify rearranged BV3 template. This was assessed by using three serial 10-fold reductions in the number of input cells using the recovery conditions described above (Figure 3B). Amplification of product equivalent in size (200 – 240bp) was observed down to an input number of 50 cells. We were confident at this stage to proceed with collection of fixed tetramer-labelled antigen-specific CD8+ T-cells by FACS for subsequent amplification of TCR mRNA, as the process worked at a cell number substantially below the minimum anticipated yield of sorted populations.
Donor CPD has a known antigen-specific response to the HLA-B*0702 restricted CMV epitope TPRVTGGGAM. Surface staining of PBMC using HLA B*0702/TPR-PE tetramer and anti-BV3 monoclonal antibody indicated that the magnitude of response using antigen-specific Vß3+ CD8+ T-cells was 5.91%. PBMC were separately labelled with the HLA B*0702/TPR-PE tetramer, fixed with 1% formaldehyde and isolated by FACS; total RNA was then extracted using the CFI method. BV3-specific cDNA template was amplified from a total of 1200 tetramer-labelled cells (Figure 3C, lane 2). The amplified PCR product was ligated into a TA cloning vector and sequenced. Generated sequences were confirmed as rearranged BV28 using the IMGT online sequence algorithm (data not shown).
Equivalence of fixed and unfixed templates for molecular clonotype analysis
We now had a method to amplify from first-strand cDNA template the post-transcriptional TCR mRNA expressed in FACS-separated, formaldehyde-fixed, tetramer-labelled cells. However, to validate our approach formally, we needed to demonstrate that neither fixation using formaldehyde nor subsequent processing steps introduced a bias into the sequences obtained from these procedures.
To achieve this, we compared identical antigen-specific CD8+ T cell populations isolated from both fixed and unfixed samples. First-strand cDNA synthesis and unbiased amplification of expressed TRB gene products using a TRBC primer were conducted as described previously (Douek et al., 2002). A minimum of 50 TA sub cloned amplicons was sequenced for each sorted population. Clonotypes were identified firstly according to TRBV family (as identified by comparison with the IMGT database), but were specifically compared by alignment of CDR3 sequences. The frequencies of aligned clonotypes for data sets from fixed and unfixed samples were paired statistically using randomisation analysis and the extent of similarity of the two populations was determined.
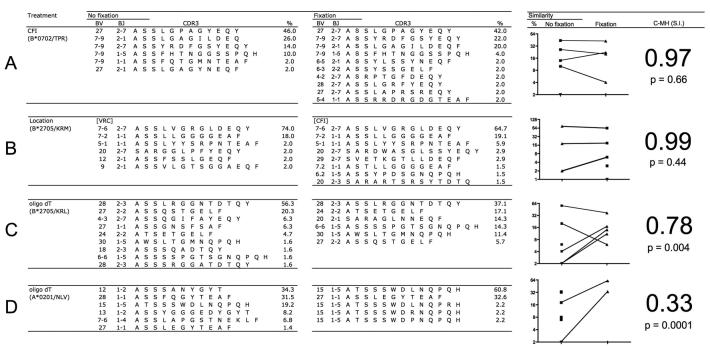
The antigen-specific repertoire of CMVpp65 HLA B*0702/TPR-specific CD8+ T-cells in donor CPD without fixation contained 6 individual clonotypes that varied in frequency; one immunodominant BV 27 clonotype used BJ 2-7 (46% of the population), four BV 7-9 clonotypes used BJ 2-7, 2-1, 1-5 and 1-1 (2 – 26%), and one minor BV 27 clonotype used BJ 2-1 (2% of the population) (Figure 4A). For the paired formaldehyde-fixed sample, ten clonotypes were observed in the repertoire; the same immunodominant BV27 clonotype using BJ 2-7 (42%) and three of the same four subdominant BV7-9 clonotypes were present (4 – 22%). Below this frequency (2%), the apparent difference in the populations appeared to be a product of the limitations of the screening process because statistical analysis indicated that the populations were not significantly different (C-MH = 0.97, p = 0.66).
Figure 4. Comparison of treatments and recovery methods for sorted antigen-specific CD8+ T-cells.
Groups are separated by treatment without (no fixation) and with (fixation) buffered formaldehyde. The Morisita-Horn similarity index (C-MH: S.I.) was used to assess the similarity and the associated p-value indicates whether two samples are significantly different. The frequencies (%) and complementarity-determining region 3 (CDR3) amino acid sequence identities of recovered TRB gene rearrangements are shown for each condition. Phenol/guanine isothiocyanate was used to isolate template in (A); location and FACS sorting strategies are compared in (B); and oligo-dT bead-based mRNA recovery data from sorted CD8+ T-cell populations specific for antigens derived from HIV and CMV are shown in (C) and (D) respectively.
Sampling strategy does not bias statistical outcome
Next we conducted a comparison of cells from HIV-1 infected patients. This necessitated using FACS isolation protocols in a biosafety level-3 containment facility for the unfixed samples and the methodologies detailed above for the formaldehyde-fixed PBMC. These comparisons were made across two separate research facilities. An identical aliquot of cryopreserved PBMC was used to isolate HIV-specific CD8+ T-cells from the same time point in an HIV-infected donor (057) at both locations (CFI and VRC) using the HIV p24 Gag HLA B*2705/KRM tetramer. This aliquot was fixed according to the containment requirements of the facility in which the sample was sorted; that is, PBMC were fixed in formaldehyde for cell sorting and recovery using the CFI method. PBMC were unfixed when cell sorting in a BSL-3 containment facility for recovery using the VRC method.
The HLA B*2705/KRM antigen-specific CD8+ T-cells derived from the unfixed sample contained six clonotypes: an immunodominant BV 7-6 clonotype that used BJ 2-7 (74%) and individual subdominant clonotypes ranging in frequency from 2 to 18% (Figure 4B). The fixed sample contained eight clonotypes, with the same immunodominant BV 7-6/BJ 2-7 clonotype (64%) and a range of subdominant clonotypes in the same hierarchical order from BV 7-2 using BJ 1-1 (19.1% fixed, versus 18% unfixed) to BV 5-1 using BJ 1-1 (5.9% fixed, versus 2.0% unfixed). The remaining clonotypes in the unfixed and fixed populations were unrelated (three and five clonotypes, respectively). Measured similarity (C-MH: 0.99) indicated that the paired samples were not significantly different (p = 0.44). It is also important to note that since extraction was performed separately using the CFI method for fixed total RNA and the VRC method for recovery of unfixed mRNA template, this data serves as evidence that there is no difference between sampling at separate institutes for outcome of repertoire analysis.
Limitation to recovery methods for fixed samples
There are theoretical advantages to the use of oligo-dT-based methods of mRNA isolation in conjunction with molecular clonotype analysis over a Trizol-based methodology, which isolates total cellular RNA. However, the chemistry of the interaction between formaldehyde and primary amine groups of the poly-adenylated (poly-A) tail has the potential to interfere with its subsequent hybridisation to an oligo-dT primer. To address this, we compared recovery of fixed and unfixed template using the VRC method from a second HIV-infected HLA-B*2705 positive donor (012). A single aliquot of PBMC was split and resuspended in sort buffer with or without 1% formaldehyde. Antigen-specific CD8+ T-cells were isolated by FACS after staining with the HLA-B*2705/KRL tetramer. Nine clonotypes were detected in the unfixed sample, with frequencies ranging from 1.6% to 56.3% (Figure 4C). The immunodominant clonotype representing 56.3% of the population used BV 28 and BJ 2.3. This clonotype was also detected as dominant in the fixed sample, albeit at a lower frequency (37.1%). Discrepancies in the repertoire between the two samples emerge in the hierarchy of subdominant clonotypes when fixation was used. There were six clonotypes in the fixed sample, of which the subdominant clonotypes BV 24 and BV 6-6 significantly increased in frequency from 4.7 and 1.6 % respectively in the unfixed sample to 17.1 and 14.3% respectively in the fixed sample. Furthermore, a BV 27 clonotype using BJ 2.2, which represented 20.3% of the unfixed sample was detected at a substantially lower frequency in the fixed sample (5.7%). The data indicate that use of the VRC method results in significant differences in repertoire distribution for the fixed and the unfixed samples (C-MH = 0.78, p = 0.004).
To confirm that this result was not related to cytometry-based error (eg. dead cells or non-specific labelling with insoluble fluorochrome), a second sample from the same donor was analysed using the HLA A*0201/NLV tetramer to sort CMV-specific CD8+ T-cells. The sample was again split and treated with or without formaldehyde. There were six clonotypes in the unfixed sample with the immunodominant BV 12 clonotype representing 34.3% of the repertoire for unfixed cells. This clonotype was not detected in the fixed sample (Figure 4D). The subdominant clonotype BV 15 with the CDR3 sequence ATSSSWDLNQPQH (19.2%) and the clonotype BV 27 (1.4%) in the unfixed sample were detected far more frequently in the fixed sample representing >93% of the observed repertoire. Overall, the distribution of clonotypes in these paired samples was again significantly different (C-MH = 0.33, p = 0.0001).
Discussion
Molecular analysis of RNA transcripts from infectious human peripheral blood samples sorted by flow cytometry has to date only been possible within biosafety level-3 containment facilities. In the present study, we have developed a reproducible method that enables high quality RNA recovery from formaldehyde-fixed PBMC samples collected directly ex vivo by FACS. Our in vitro formaldehyde inactivation data support previous publications, which outline the requirements for safe inactivation of human pathogens in tissues to be used for flow cytometric sorting (Lifson et al., 1986; Martin et al., 1987; Nicholson et al., 1993). Used in combination with universal safety precautions, this technique allows sorting of primary human samples for molecular analysis in a general laboratory setting. The concentration of formaldehyde used for fixation of pathogens and subsequent recovery of RNA was four times greater than the minimum inactivation of high-titre HIV-1 infection of MAGIC-5 cells.
We found that the integrity of the recovered RNA template could be maintained by using an optimised proteinase K treatment. Clonal sequencing of rearranged TCR cDNA from virus-specific CD8+ T-cells isolated by FACS using tetramers yielded comparable data to that derived from unfixed samples. Furthermore, our observations indicate that, irrespective of the exact gating strategy used, the statistical similarity of the TCR repertoire between paired fixed and unfixed samples was robust provided that stringent definition of the antigen-specific population was maintained.
A critical component of the methodology described herein is the addition of a proteinase K digestion step prior to the extraction of RNA. This appeared to work best if moderate temperatures were used over a short period. The rationale for this step is similar to that governing the use of proteinase K in longer genomic DNA extractions (Gross-Bellard, 1973). Genomic DNA is bound to a range of proteins, predominantly histones. One of the critical steps in a nucleic acid purification protocol is dissociation of proteins from nucleic acids. Proteinase K is a serine-specific endopeptidase, which, by digesting proteins, allows separation of DNA from the histone backbone. This step is often not necessary in standard mRNA extraction protocols. In formaldehyde-fixed tissue, the cross-linking of protein results in non-specific high avidity associations between mRNA and their associated cellular proteins for differential RNA processing at the time of formaldehyde infiltration. This association is not disrupted by the traditional RNA extraction protocols and transcripts are lost in the purification process, discarded with associated proteins in the phase separation step. The use of proteinase K with heat disrupts the association between RNA and protein by digesting the cross-linked protein, allowing subsequent purification of total RNA by essentially standard methodology.
A second critical step in the methodology is the need to extract total RNA rather than preferentially purifying mRNA by the use of reagents such as oligo-dT beads. The clonotype profile of fixed template recovered using oligo-dT beads is significantly different to unfixed samples. While experimental data explaining this observation are not presented here, existing data in the published literature allow a coherent explanation. The poly-A tail of mRNA template is especially susceptible to electrophilic attack from formaldehyde (Masuda et al., 1999). Adenine has a primary amine group that is accessible to addition of a mono-methylol group (N-CH2-OH) per nucleotide. Such a covalent modification to the poly-A tail would be prohibitive to thymine bond formation and thus hybridisation to oligo-dT.
While the methodology presented here is applicable to fixation with low percentages of formaldehyde over relatively short periods of time, it may not be applicable to tissues exposed to formaldehyde for much longer periods. Extended fixation beyond hours or days negatively impacts on RNA quality and subsequent amplification of transcripts because of truncation and /or fragmentation of transcripts (Masuda et al., 1999; Macabeo-Ong et al., 2002; Gloghini et al., 2004). We have not addressed amplification of full-length mRNA transcripts beyond the scope of what was required to obtain our sequences of interest, that is amplicons up to 600 base pairs in length.
One incongruity is why, when relying upon oligo-dT-based first-strand cDNA synthesis, no significant difference in repertoire is apparent. It is feasible that the heat provided by increasing the reaction temperature to 72°C for first-strand cDNA synthesis primer annealing impacts upon removal of covalent modifications from the poly-A tail. This is not the case when using oligo-dT beads for recovery, as this degree of heating has not occurred by this stage. This potentially helps explain the observed differences in repertoire obtained when using of oligo-dT beads for extraction of mRNA from fixed and unfixed samples.
Molecular analysis of the T-cell repertoire in combination with ex vivo isolation of antigen-specific CD8+ T-cells detected by peptide-MHC class I tetramers has become a powerful approach for examining the quality and in vivo dynamics of such immune responses to viral infection (Wilson et al., 1998; Kedzierska et al., 2004; Meyer-Olson et al., 2004; Price et al., 2004; Betts et al., 2005; Price et al., 2005; Leslie et al., 2006). From the work presented herein, these techniques can now be applied to fixed specimens, thereby enabling primary human sample analysis in a general flow cytometry suite.
Acknowledgements
We would like to thank the donors who provided blood samples for experimental use and the staff who performed the PBMC isolation. We are grateful to the Institutional Biosafety Committee of the St Vincent's Hospital Research Campus in their review and approval of the virus inactivation methodology. We thank Jerome Darakdjian (CFI) and David Ambrozak (VRC) for their assistance with FACS isolation of high purity antigen-specific CD8+ populations. This project was supported by program grant funding from the Australian National Health and Medical Research Council (NHMRC) and the National Institutes of Health (USA). The National Centre in HIV Epidemiology and Clinical Research (NCHECR) is supported by the Commonwealth Department of Health and Ageing through the Australian National Council on AIDS, Hepatitis C and Related Diseases. DvB is the recipient of a Dora Lush postgraduate scholarship from the NHMRC. ADK is supported by a practitioner fellowship. DAP is a Medical Research Council (UK) Senior Clinical Fellow.
Footnotes
Abbreviations: CTL, cytotoxic T-lymphocyte; MHC, major histocompatibility complex; TCR, T-cell receptor; TRB, T-cell receptor B; CDR3, complementarity determining region (3); HIV, human immunodeficiency virus; CMV, cytomegalovirus; FACS, fluorescence-activated cell sort; PBMC, peripheral blood mononuclear cell; PCR, polymerase chain reaction; RNA, ribonucleic acid; DNA, deoxyribonucleic acid; APC, Allophycocyanin; FITC, Fluorescein isothiocyanate; PerCP, Peridinin chlorophyll protein; PE, Phycoerythrin; ECD, Phycoerythrin-Texas Red
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altman JD, Moss PAH, Goulder PJR, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic Analysis of Antigen-Specific T Lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- Arden B, Clark SP, Kabelitz D, Mak TW. Human T-cell receptor variable gene segment families. Immunogenetics. 1995;42:455–500. doi: 10.1007/BF00172176. [DOI] [PubMed] [Google Scholar]
- Appay V, Rowland-Jones SL. The assessment of antigen-specific CD8+ T cells through the combination of MHC class I tetramer and intracellular staining. J Immunol Methods. 2002;268:9–19. doi: 10.1016/s0022-1759(02)00195-3. [DOI] [PubMed] [Google Scholar]
- Betts MR, Exley B, Price DA, Bansal A, Camacho ZT, Teaberry V, West SM, Ambrozak DR, Tomaras G, Roederer M, Kilby JM, Tartaglia J, Belshe R, Gao F, Douek DC, Weinhold KJ, Koup RA, Goepfert P, Ferrari G. Characterization of functional and phenotypic changes in anti-Gag vaccine-induced T cell responses and their role in protection after HIV-1 infection. Proc Natl Acad Sci U S A. 2005;102:4512–7. doi: 10.1073/pnas.0408773102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers R, Roebuck J, Sakhinia E, Hoyland J. PolyA PCR amplification of cDNA from RNA extracted from formalin-fixed paraffin-embedded tissue. Diagn Mol Pathol. 2004;13:144–50. doi: 10.1097/01.pdm.0000133154.73846.66. [DOI] [PubMed] [Google Scholar]
- Church DM, Buckler AJ. Gene identification by exon amplification. Methods Enzymol. 1999;303:83–99. doi: 10.1016/s0076-6879(99)03008-6. [DOI] [PubMed] [Google Scholar]
- Cornberg M, Chen AT, Wilkinson LA, Brehm MA, Kim SK, Calcagno C, Ghersi D, Puzone R, Celada F, Welsh RM, Selin LK. Narrowed TCR repertoire and viral escape as a consequence of heterologous immunity. J Clin Invest. 2006;116:1443–56. doi: 10.1172/JCI27804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez C, Bertsch G, Simm A. Isolation of full-size mRNA from cells sorted by flow cytometry. J Biochem Biophys Methods. 1999;40:69–80. doi: 10.1016/s0165-022x(99)00020-2. [DOI] [PubMed] [Google Scholar]
- Douek DC, Betts MR, Brenchley JM, Hill BJ, Ambrozak DR, Ngai KL, Karandikar NJ, Casazza JP, Koup RA. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J Immunol. 2002;168:3099–104. doi: 10.4049/jimmunol.168.6.3099. [DOI] [PubMed] [Google Scholar]
- Gloghini A, Canal B, Klein U, Dal Maso L, Perin T, Dalla-Favera R, Carbone A. RT-PCR analysis of RNA extracted from Bouin-fixed and paraffin-embedded lymphoid tissues. J Mol Diagn. 2004;6:290–6. doi: 10.1016/S1525-1578(10)60524-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Bellard MO,P, Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur. J. Biochem. 1973;36:32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- Hachiya A, Aizawa-Matsuoka S, Tanaka M, Takahashi Y, Ida S, Gatanaga H, Hirabayashi Y, Kojima A, Tatsumi M, Oka S. Rapid and simple phenotypic assay for drug susceptibility of human immunodeficiency virus type 1 using CCR5-expressing HeLa/CD4(+) cell clone 1-10 (MAGIC-5) Antimicrob Agents Chemother. 2001;45:495–501. doi: 10.1128/AAC.45.2.495-501.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedzierska K, Turner SJ, Doherty PC. Conserved T cell receptor usage in primary and recall responses to an immunodominant influenza virus nucleoprotein epitope. Proc Natl Acad Sci U S A. 2004;101:4942–4947. doi: 10.1073/pnas.0401279101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Xu Z, Lieberman J, Shankar P. The functional CD8 T cell response to HIV becomes type-specific in progressive disease. J Clin Invest. 2002;110:1339–47. doi: 10.1172/JCI16028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie A, Price DA, Mkhize P, Bishop K, Rathod A, Day C, Crawford H, Honeyborne I, Asher TE, Luzzi G, Edwards A, Rousseau CM, Mullins JI, Tudor-Williams G, Novelli V, Brander C, Douek DC, Kiepiela P, Walker BD, Goulder PJ. Differential selection pressure exerted on HIV by CTL targeting identical epitopes but restricted by distinct HLA alleles from the same HLA supertype. J Immunol. 2006;177:4699–708. doi: 10.4049/jimmunol.177.7.4699. [DOI] [PubMed] [Google Scholar]
- Lifson JD, Sasaki DT, Engleman EG. Utility of formaldehyde fixation for flow cytometry and inactivation of the AIDS associated retrovirus. J Immunol Methods. 1986;1:143–149. doi: 10.1016/0022-1759(86)90278-4. [DOI] [PubMed] [Google Scholar]
- Lim A, Baron V, Ferradini L, Bonneville M, Kourilsky P, Pannetier C. Combination of MHC-peptide multimer-based T cell sorting with the Immunoscope permits sensitive ex vivo quantitation and follow-up of human CD8+ T cell immune responses. J Immunol Methods. 2002;261:177–94. doi: 10.1016/s0022-1759(02)00004-2. [DOI] [PubMed] [Google Scholar]
- Macabeo-Ong M, Ginzinger DG, Dekker N, McMillan A, Regezi JA, Wong DT, Jordan RC. Effect of duration of fixation on quantitative reverse transcription polymerase chain reaction analyses. Mod Pathol. 2002;15:979–87. doi: 10.1097/01.MP.0000026054.62220.FC. [DOI] [PubMed] [Google Scholar]
- Magurran AE. Measuring Biological Diversity. Blackwell Publishing; Malden, MA: 2003. [Google Scholar]
- Martin LS, Loskoski SL, McDougal JS. Inactivation of human Tlymphotropic virus type III/lymphadenopathy-associated virus by formaldehyde-based reagents. Appl Environ Microbiol. 1987;4:708–709. doi: 10.1128/aem.53.4.708-709.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryanski JL, Attuil V, Bucher P, Walker PR. A quantitative, single-cell PCR analysis of an antigen-specific TCR repertoire selected during an in vivo CD8 response: direct evidence for a wide range of clone sizes with uniform tissue distribution. Mol Immunol. 1999;36:745–53. doi: 10.1016/s0161-5890(99)00088-7. [DOI] [PubMed] [Google Scholar]
- Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 1999;27:4436–43. doi: 10.1093/nar/27.22.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Olson D, Shoukry NH, Brady KW, Kim H, Olson DP, Hartman K, Shintani AK, Walker CM, Kalams SA. Limited T cell receptor diversity of HCV-specific T cell responses is associated with CTL escape. J Exp Med. 2004;200:307–19. doi: 10.1084/jem.20040638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Browning SW. Ability of 'OptiLyse' lysing/fixing reagents to inactivate HIV-infected H9 cells in whole blood. J Immunol Methods. 1994;168:283–4. doi: 10.1016/0022-1759(94)90066-3. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Browning SW, Orloff SL, McDougal JS. Inactivation of HIV-infected H9 cells in whole blood preparations by lysing/fixing reagents used in flow cytometry. J Immunol Methods. 1993;160:215–8. doi: 10.1016/0022-1759(93)90180-f. [DOI] [PubMed] [Google Scholar]
- Ogg GS, Jin X, Bonhoeffer S, Dunbar PR, Nowak MA, Monard S, Segal JP, Cao Y, Rowland-Jones SL, Cerundolo V, Hurley A, Markowitz M, Ho DD, Nixon DF, McMichael AJ. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–6. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- Perfetto S, Ambrozak D, Koup RA, Roederer M. Measuring Containment of Viable Infectious Cell Sorting in High-Velocity Cell Sorters. Cytometry. 2003;52A(part A):122–130. doi: 10.1002/cyto.a.10033. [DOI] [PubMed] [Google Scholar]
- Perfetto SP, Ambrozak DR, Roederer M, Koup RA. Viable infectious cell sorting in a BSL-3 facility. Methods Mol Biol. 2004;263:419–24. doi: 10.1385/1-59259-773-4:419. [DOI] [PubMed] [Google Scholar]
- Price DA, Brenchley JM, Ruff LE, Betts MR, Hill BJ, Roederer M, Koup RA, Migueles SA, Gostick E, Wooldridge L, Sewell AK, Connors M, Douek DC. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J Exp Med. 2005;202:1349–61. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DA, West SM, Betts MR, Ruff LE, Brenchley JM, Ambrozak DR, Edghill-Smith Y, Kuroda MJ, Bogdan D, Kunstman K, Letvin NL, Franchini G, Wolinsky SM, Koup RA, Douek DC. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity. 2004;21:793–803. doi: 10.1016/j.immuni.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Rufer N. Molecular tracking of antigen-specific T-cell clones during immune responses. Curr Opin Immunol. 2005;17:441–7. doi: 10.1016/j.coi.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Schmid I, Kunkl A, Nicholson JK. Biosafety considerations for flow cytometric analysis of human immunodeficiency virus-infected samples. Cytometry. 1999;38:195–200. doi: 10.1002/(sici)1097-0320(19991015)38:5<195::aid-cyto1>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Schmid I, Nicholson JK, Giorgi JV, Janossy G, Kunkl A, Lopez PA, Perfetto S, Seamer LC, Dean PN. Biosafety guidelines for sorting of unfixed cells. Cytometry. 1997;28:99–117. doi: 10.1002/(sici)1097-0320(19970601)28:2<99::aid-cyto2>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Saito T, Kondo M, Osanai M, Watanabe S, Kano T, Kano K, Imai M. Poly A-linked non-isotopic microtiter plate reverse transcriptase assay for sensitive detection of clinical human immunodeficiency virus isolates. J Virol Methods. 1995;55:347–56. doi: 10.1016/0166-0934(95)00073-5. [DOI] [PubMed] [Google Scholar]
- Venturi V, Kedzierska K, Turner SJ, Doherty PC, Davenport MP. Methods for comparing the diversity of samples of the T cell receptor repertoire. J Immunol Methods. 2007;321:182–95. doi: 10.1016/j.jim.2007.01.019. [DOI] [PubMed] [Google Scholar]
- Wilson JD, Ogg GS, Allen RL, Goulder PJ, Kelleher AD, Sewell AK, O'Callaghan CA, Rowland-Jones SL, Callan MF, McMichael AJ. Oligoclonal expansions of CD8(+) T cells in chronic HIV infection are antigen specific. J Exp Med. 1998;188:785–90. doi: 10.1084/jem.188.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]