Abstract
The nucleotide excision repair (NER) pathway corrects DNA damage caused by sunlight, environmental mutagens and certain antitumor agents. This multistep DNA repair reaction operates by the sequential assembly of protein factors at sites of DNA damage. The efficient recognition of DNA damage and its repair are orchestrated by specific protein–protein and protein–DNA interactions within NER complexes. We have investigated an essential protein–protein interaction of the NER pathway, the binding of the XPA protein to the ERCC1 subunit of the repair endonuclease ERCC1-XPF. The structure of ERCC1 in complex with an XPA peptide shows that only a small region of XPA interacts with ERCC1 to form a stable complex exhibiting submicromolar binding affinity. However, this XPA peptide is a potent inhibitor of NER activity in a cell-free assay, blocking the excision of a cisplatin adduct from DNA. The structure of the peptide inhibitor bound to its target site reveals a binding interface that is amenable to the development of small molecule peptidomimetics that could be used to modulate NER repair activities in vivo.
Keywords: DNA repair, ERCC1, NMR, nucleotide excision repair, XPA
Introduction
The repair of chemical insults to DNA caused by UV light and other mutagens is essential for coping successfully with the intrinsic reactivity of DNA and preserving genetic information. Inherited diseases resulting in the failure to correct spontaneous or environmentally induced damage are typically associated with genomic instability and a predisposition to various cancers (Friedberg et al, 2005). Contrarily, DNA repair is undesirable when DNA-damaging agents are used for chemotherapy of cancer and other diseases. In these settings, the ability to modulate the DNA repair activities of cells targeted for destruction is a desirable goal (Ding et al, 2006). The nucleotide excision repair (NER) pathway is essential for normal genomic maintenance, removing bulky chemical adducts from DNA that are otherwise mutagenic or would pose lethal blocks to replication.
NER involves over 30 proteins that recognize damaged sites in DNA and excise an oligonucleotide containing the damage (de Laat et al, 1999; Gillet and Schärer, 2006). Following excision of the damaged DNA segment, the resulting gap is filled by templated DNA synthesis and ligase seals the nick to complete the repair. Cell biological and biochemical studies have shown that NER operates by the sequential assembly of protein factors at the sites of DNA damage, rather than through the action of a preformed repairosome (Houtsmuller et al, 1999; Volker et al, 2001). The recruitment of NER factors into protein–DNA ensembles is guided at each step by numerous protein–protein interactions (Araujo et al, 2001), imparting specificity to the recognition and verification of damaged sites. Damage recognition in NER culminates in the incision of DNA 5′ and 3′ to the lesion site by ERCC1-XPF and XPG, respectively, to release a 24–32 nucleotide segment containing the damage (de Laat et al, 1999; Gillet and Schärer, 2006).
For the NER pathway, DNA cleavage by ERCC1-XPF requires physical interaction with XPA, a scaffold protein that interacts with DNA and several repair proteins, including RPA, TFIIH and the ERCC1 subunit of ERCC1-XPF (Li et al, 1994, 1995; Park and Sancar, 1994; Saijo et al, 1996). Although XPA was originally described as a DNA damage-specific sensor or verification protein, recent work suggests that XPA instead recognizes the DNA structural intermediates arising during processing by NER (Jones and Wood, 1993; Camenisch et al, 2006). XPA recruits ERCC1-XPF to NER complexes (Volker et al, 2001), positioning the XPF nuclease domain at the 5′ side of the damage site (Enzlin and Schärer, 2002). ERCC1-XPF has other roles in DNA metabolism outside of NER, notably in interstrand crosslink repair and homologous recombination (Hoy et al, 1985; Niedernhofer et al, 2001). The importance of these additional, NER-independent functions of ERCC1-XPF is underscored by the pronounced sensitivity to crosslinking agents caused by mutations of ERCC1 or XPF in mice and humans (McWhir et al, 1993; Weeda et al, 1997; Niedernhofer et al, 2006). However, the exact biochemical role(s) of ERCC1-XPF in crosslink repair remain to be discovered.
Li et al (1994, 1995) identified residues 59–114 in XPA as the site of interaction with ERCC1, and showed that deletion of three conserved glycines (Gly72, Gly73, Gly74) abrogates the XPA–ERCC1 interaction as well as the ability of the XPA protein to confer UV resistance to XP-A cells. Furthermore, the expression of a truncated protein comprising residues 59–114 of XPA renders cells sensitive to UV light and cisplatin (Rosenberg et al, 2001), suggesting that this region is sufficient to disrupt the interaction of the native XPA protein with ERCC1-XPF. Conversely, it can be inferred from several previous studies that residues 92–119 of ERCC1 are necessary for the interaction with XPA (Li et al, 1994; Sijbers et al, 1996; Gaillard and Wood, 2001).
Following these seminal studies, understanding of the biochemical and structural basis for XPA's interaction with ERCC1 has not advanced, although more is known about the individual proteins. XPA contains a well-defined central domain (residues 98–219; Figure 1A), although the remainder of the protein including the ERCC1 interaction domain appears to be poorly structured (Buchko et al, 1998, 2001; Ikegami et al, 1998; Iakoucheva et al, 2001). Residues 92–119 of ERCC1 fall within the central domain of ERCC1 (Tsodikov et al, 2005) that is structurally homologous to the nuclease domain of the archaeal XPF-like proteins Aeropyrum pernix Mus81/XPF (Newman et al, 2005) and Pyrococcus furiosus Hef (Nishino et al, 2003). A V-shaped groove on the surface of ERCC1 corresponds to the nuclease active site of XPF (Enzlin and Schärer, 2002; Nishino et al, 2003; Newman et al, 2005), except that ERCC1's groove lacks the catalytic residues of a nuclease active site and is instead populated with basic and aromatic residues (Gaillard and Wood, 2001). We previously showed that the central domain of ERCC1 binds to single-stranded DNA in vitro (Tsodikov et al, 2005), and proposed the V-shaped groove as the DNA-binding site.
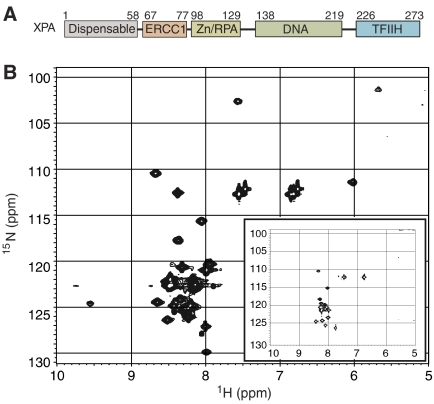
Figure 1.
XPA domain organization and structure of the ERCC1-binding peptide. (A) The ERCC1-binding region of XPA (residues 67–77) is located between the central domain (Zn2+-binding and DNA-binding subdomains; residues 98–219) and an N-terminal region (residues 1–58) that is dispensable for functional complementation of NER in whole-cell extracts from XP-A mutant cells (Miyamoto et al, 1992) and a TFIIH-binding region (Park et al, 1995). (B) 15N HSQC spectrum of 15N-labeled XPA59−93 in complex with unlabeled ERCC1, and in the unbound state (inset). The spectrum of the unbound XPA59−93 (inset) is characteristic of an unfolded peptide. The appearance of new well-dispersed NMR peaks in the XPA spectrum upon addition of ERCC192−214 (shown in the larger spectrum) indicates that a portion of the XPA peptide adopts a defined conformation in complex with ERCC1.
These observations prompted us to investigate the structural and functional basis for the interaction of XPA and ERCC1, and its role in recruiting the XPF-ERCC1 endonuclease to sites of NER. Here, we describe the structure of a short peptide motif from XPA in complex with the central domain of ERCC1. We show that this XPA peptide specifically inhibits the NER reaction in vitro, creating an opportunity for structure-based design of NER inhibitors targeting this protein–protein interaction. Point mutations in the corresponding region in XPA abolish NER activity in vitro, underscoring the importance of the XPA–ERCC1 interactions for NER. In addition to providing insights into how protein–protein interactions mediate progression through the NER pathways, our studies will provide a blueprint to develop small molecules that intercept the interaction between XPA and ERCC1. Such molecules should be valuable for studying the biochemical functions of ERCC1-XPF in NER and other repair pathways including DNA interstrand crosslink repair and homologous recombination by selectively inhibiting NER.
Results
Induced fit of the XPA peptide upon interaction with ERCC1
Previous reports have suggested that the ERCC1-interacting region of XPA (Figure 1A) is unfolded in solution, based on NMR studies and its sensitivity to proteolytic cleavage (Buchko et al, 2001; Iakoucheva et al, 2001). To investigate the structure of the XPA ligand, we collected HSQC NMR spectra of an 15N-labeled XPA59−93 peptide alone and in complex with unlabeled central domain of ERCC1 (the ERCC192−214 protein; Figure 1B). In the absence of ERCC1, the resonance signals for XPA cluster in a narrow range of chemical shifts (Figure 1B, inset) that is characteristic of an unstructured polypeptide with poor spectral dispersion. In the complex with ERCC1, a subset of XPA backbone amides become well dispersed and the peaks are broader. These changes are indicative of a well-structured region within the bound XPA peptide. Only a few resonance peaks are markedly perturbed when XPA59−93 is bound to ERCC1, and among these, three glycine residues (assigned as Gly72, Gly73 and Gly74) are strongly perturbed in the complex. In order to overcome the peak broadening observed in NMR spectra of the XPA59−93 peptide at concentrations above 0.1 mM, we sought to identify shorter XPA peptide ligands for ERCC1. A minimal, 14-residue XPA67−80 peptide (described below) was identified by examining a series of overlapping XPA fragments from the region previously shown to interact with ERCC1 (Li et al, 1994).
The structure of XPA in complex with ERCC1
A synthetic XPA67−80 peptide with amino-acid sequence KIIDTGGGFILEEE forms a stable complex with ERCC196−214 that can be purified by gel filtration chromatography. Like full-length XPA protein, the XPA59−93 and the XPA67−80 peptides behave similarly and efficiently co-purify with ERCC1, suggesting that XPA67−80 contains all significant binding determinants. We confirmed that XPA and ERCC1 form a stoichiometric 1:1 complex by estimating the amount of each subunit in the purified complex using an Edman degradation reaction, and by analytical centrifugation of the complex. Equilibrium sedimentation data for the complex (Supplementary Figure 1C) were best fit to the expected masses for a 1:1 complex of XPA59−93 and ERCC192−214 (Mw=(19.4±1.2) kDa) and unbound ERCC192−214 (Mw=(15.0±1.0) kDa). We confirmed that ERCC196−214 binds stoichiometrically to the XPA67−80 peptide with a KD of 0.78 μM (Supplementary Figure 2). A structure of the XPA67−80–ERCC196−214 complex (Figure 2A) was determined by a combination of NMR-derived distance restraints and X-ray diffraction data extending to 4 Å resolution (Table 1 in the Supplementary data and Materials and methods) as described below.
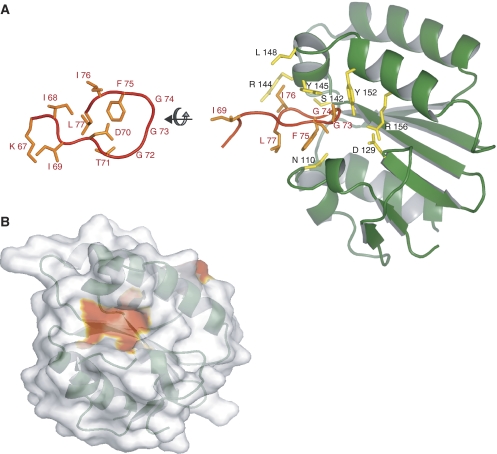
Figure 2.
Structure of the XPA–ERCC1 complex. (A) The XPA67−80 peptide (orange) is bound to a V-shaped groove of the central domain of ERCC196−214 (green). An orthogonal view of the bound XPA peptide (left side) is shown in comparison to the peptide in complex with ERCC1 (right-hand side). (B) The XPA-binding site on the surface of ERCC1 (colored red) was identified by resonance perturbations larger than 0.2 ppm that are indicative of direct interactions with XPA.
Identification of the ERCC1-binding site in complex with XPA
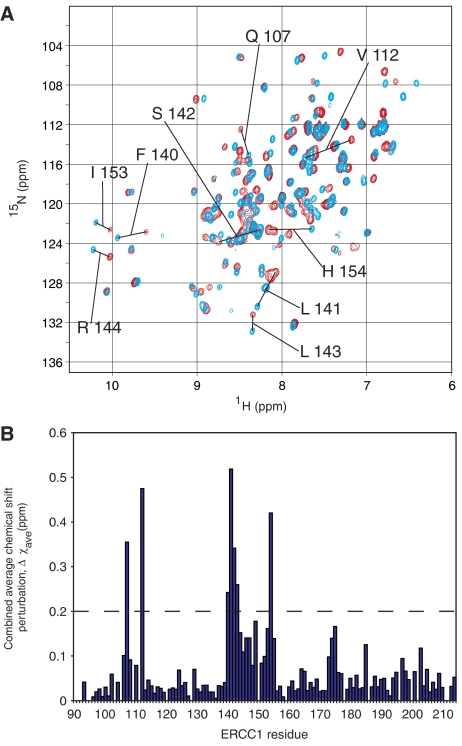
The binding site for XPA on the surface of ERCC1 (Figure 2B) was identified using two-dimensional HSQC experiments. The spectrum of unliganded 15N-labeled ERCC192−214 (blue, Figure 3) showed significant differences from that of the complex with unlabeled XPA67−80 (red, Figure 3). However, complexes of ERCC196−214 with either XPA67−80 or XPA59−93 were identical, suggesting that the shorter XPA peptide makes all of the significant binding contacts. The 15N HSQC spectrum of the ERCC1–XPA complex is consistent with a slow-exchange regime, implying a dissociation equilibrium constant below 1 μM for the complex. The XPA-binding site on the ERCC1 central domain was identified using the backbone assignments for ERCC196−214 alone and in complex with XPA (see Materials and methods). A comparison of the 15N HSQC spectra for ERCC1 in the presence and absence of XPA reveals that, with only one exception, the most prominent changes in chemical shifts involve a cluster of residues within a V-shaped groove of the ERCC1 central domain (Figures 2B and 3).
Figure 3.
XPA67−80 binds in a shallow groove of ERCC1. (A) A comparison of the two-dimensional HSQC spectra for 15N-labeled ERCC192−214 in the presence and absence of an unlabeled XPA67−80 peptide. The 15N HSQC spectra reveal significant chemical shift changes for some ERCC1 residues in the absence (blue) or presence (red) of unlabeled XPA67−80. (B) Combined average chemical shift perturbations are calculated as Δχave=(((Δχ1H)2+(Δχ15N/5)2)/2)1/2 for each backbone amide of ERCC1 and shown as a histogram.
The bound XPA peptide fits snugly into the V-shaped groove of ERCC1 (Figure 2) that we previously speculated could be a binding site for single-stranded DNA (Tsodikov et al, 2005). Three consecutive glycines (Gly72, Gly 73, Gly74) of the XPA peptide insert into the groove, making a U-turn with close steric complementary to the binding site. These are the same three conserved glycines previously reported to be essential for the interaction of XPA with ERCC1 and required for the functional complementation of XP-A cells (Li et al, 1994, 1995). A total of 1039 Å2 of accessible surface area from XPA peptide is buried in the complex with ERCC1, accounting for 61% of the solvent accessible surface area of XPA residues 67–77, which are in close proximity to the binding site. The XPA ligand derives many interactions from the core sequence motif (shown in boldface; KIIDTGGGFILEEE) of the XPA67−80 peptide. The side chains of Phe75, Leu77 and Thr71 of XPA are clustered together at the mouth of the V-shaped groove (Figure 2A) where Phe75 stacks against Asn110 of ERCC1, and the Ile76 side chain packs against the aliphatic portion of ERCC1 side chains Arg144 and Leu148. The binding groove in ERCC1 is capped by XPA Leu77.
The glycine-rich loop of XPA67−80 extends far into the groove of ERCC1 where main chain atoms of these XPA residues stack against the side chains of Tyr145 and Tyr152 from ERCC1 (Figure 2A). The main chain amides of these glycines could participate in hydrogen-bonding interactions with the ERCC1-binding site, although these interactions cannot be directly observed from our NMR experiments nor can they be reliably confirmed by low-resolution (4 Å) X-ray diffraction data (Table 1 in the Supplementary data). Based on the proximity of atoms modeled in the complex, we infer that the carbonyl oxygen of Gly74 may bond with the main chain amide of Ser142 from ERCC1. The orientations of the Tyr145 and Tyr152 side chains from ERCC1 would permit their hydroxyl groups to make hydrogen-bonding interactions with the backbone carbonyls of Thr71 and Gly73, respectively. The side chain of XPA Asp70 could participate in electrostatic interactions with the side chain His149 of ERCC1. It is notable that a solvent-exposed salt bridge between the side chains of Asp129 and Arg156 of ERCC1 (PDB code 2A1I; Tsodikov et al, 2005) becomes almost completely buried when XPA is bound.
Phe75 of XPA is completely buried within the ERCC1-binding site (Figure 2A). We tested whether an alanine substitution at this position interferes with binding to 15N-labeled ERCC1 by measuring chemical shifts in the 15N HSQC spectra in the presence of the mutant peptide designated XPA67−80 F75A. Addition of the mutant peptide failed to perturb the chemical shifts of ERCC1 seen upon addition of wild-type XPA67−80 (data not shown), indicating that the mutant peptide does not bind to ERCC1. The TGGGFI-binding motif of the XPA ligand and the corresponding residues of the ERCC1-binding site are strictly conserved in higher eukaryotes. In lower eukaryotes, the corresponding sequences of both proteins have diverged from this consensus, perhaps indicating the coevolution of these two proteins and their functions.
The XPA peptide inhibits NER in mammalian cell extracts
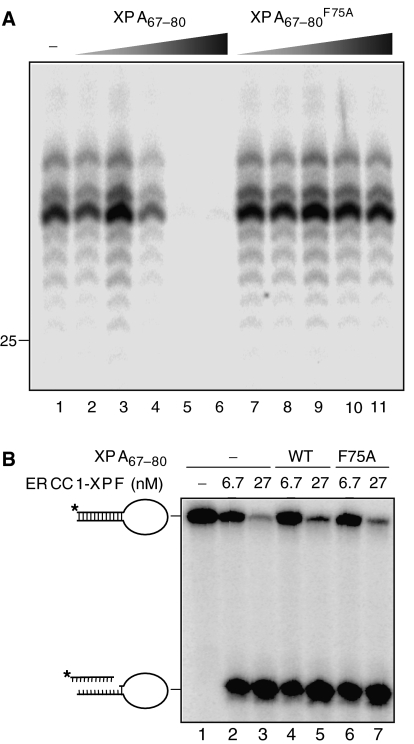
The direct interaction of XPA67−80 peptide with the ERCC1-binding pocket raised the possibility that this peptide might specifically interfere with the recruitment of the ERCC1-XPF nuclease into the NER reaction pathway. We investigated the effect of XPA67−80 and the mutant XPA67−80 F75A peptide on the dual incision of a DNA lesion during NER in cell-free extracts. A plasmid containing a single site-specific 1,3-cisplatin intrastrand crosslink was incubated with HeLa cell-free extract in the presence of increasing concentrations of XPA peptide (Shivji et al, 1999). In the absence of XPA peptide, the characteristic NER excision products of 28–33 nucleotides containing the lesion were evident (Figure 4A, lane 1). Increasing concentrations of XPA67−80 interfered with excision of the oligonucleotide, and complete inhibition was achieved at a concentration of XPA peptide in the low micromolar range (Figure 4A, lanes 2–6). In contrast, the addition of XPA67−80 F75A did not affect NER activity at concentrations up to 92 μM, the maximum concentration tested (Figure 4A, lanes 7–11).
Figure 4.
The XPA67−80 peptide is an effective inhibitor of NER activity. (A) XPA67−80 inhibits the in vitro NER reaction, whereas the mutant XPA67−80 F75A peptide has no effect. HeLa cell extracts were incubated with a plasmid containing a 1,3-intrastrand cisplatin adduct in the presence of increasing concentrations of either XPA67−80 or XPA67−80 F75A (lane 1, no XPA; lanes 2 and 7, 46 nM XPA peptide; lanes 3 and 8, 460 nM; lanes 4 and 9, 4.6 μM; lanes 5 and 10, 46 μM; lanes 6 and 11, 92 μM). Products were visualized by a fill-in reaction following annealing to an oligonucleotide complementary to the excision product with a 4-nt overhang (Shivji et al, 1999). The marker DNA ladder is labeled LMW DNA ladder. (B) XPA67−80 and XPA67−80 F75A do not affect the intrinsic nuclease activity of ERCC1-XPF. The stem12-loop22 substrate (6.6 nM) was incubated with different concentrations of ERCC1-XPF (lanes 2, 4 and 6: 6.7 nM ERCC1-XPF; lanes 3, 5 and 7: 26.8 nM) and 0.4 mM MnCl2 in the presence of no peptide (lanes 1–3), 92 μM XPA67−80 (lanes 4 and 5), and 92 μM XPA67−80 F75A (lanes 6 and 7). The DNA substrate and the cleavage products are indicated.
The XPA peptide might inhibit NER activity in vitro by directly interfering with the endonuclease activity of ERCC1-XPF, instead of blocking its interaction with XPA. To account for the former possibility, we tested the effects of XPA peptides on the DNA incision reaction catalyzed by purified ERCC1-XPF using stem-loop DNA substrate (de Laat et al, 1998). ERCC1-XPF efficiently cleaves on the 5′ side of the loop and the XPA peptide has no effect on this activity (Figure 4B) even at a concentration (92 μM) that completely abolishes NER activity (Figure 4A). We conclude that the inhibitory effect of XPA67−80 on the NER reaction results from disrupting the interaction of ERCC1 with XPA, an essential protein–protein interaction for the dual incision of DNA by the NER pathway.
Mutations in the ERCC1-binding epitope of XPA abolish NER
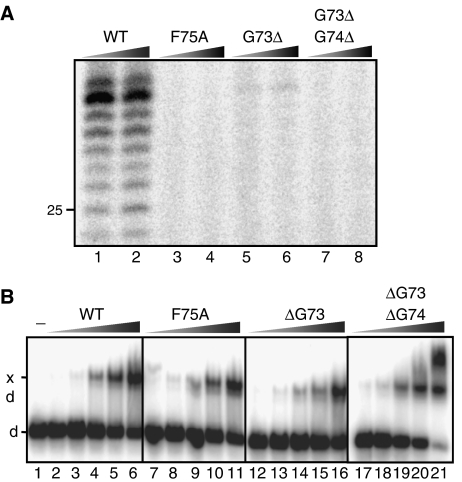
The specificity of inhibition of NER by XPA67−80 suggested that mutations of single residues such as F75 might diminish the NER activity of the XPA protein. We generated mutant XPA proteins containing an F75A mutation and ΔG73 single and ΔG73/ΔG74 double deletion, and compared their activities to that of the wild-type XPA protein. The ability of the XPA proteins to mediate NER activity was tested by incubating a plasmid containing a 1,3-cisplatin intrastrand crosslink with a cell-free extract generated from XPA-deficient cells supplemented with purified wild-type or mutant XPA protein (Shivji et al, 1999). Addition of wild-type XPA protein to this mixture resulted in robust NER activity, as evidenced by formation of the characteristic excision products of 24–32 nts in length (Figure 5A, lanes 1–2). By contrast, no NER activity was observed following addition of the F75A or G73Δ/G74Δ mutants, while the G73Δ single deletion mutant displayed marginal NER activity.
Figure 5.
Mutation of the ERCC1-binding epitope of XPA abolishes NER but not DNA-binding activity. (A) XP-A (XP2OS) cell extracts were incubated with a plasmid containing a 1,3-intrastrand cisplatin adduct in the presence of wild-type XPA (XPA-WT) or mutant XPA proteins (XPA-F75A, XPA-G73Δ or XPA-G73Δ/G74Δ). The reaction products were visualized by a fill-in reaction after annealing the excision product to an complementary oligonucleotide with a 4-nt overhang (Shivji et al, 1999). Different XPA concentrations of 200 nM (lanes 1, 3, 5 and 7) and 800 nM (lanes 2, 4, 6 and 8) were tested. The position of a 25 mer of the LMW DNA ladder is indicated. (B) A 5′-labeled DNA three-way junction (1 nM) was incubated with wild-type and mutant XPA proteins for 30 min at room temperature, then the XPA-bound (xd) and free DNA (d) oligonucleotides were separated on an 8% native polyacrylamide gel. The reaction products generated with different concentrations of XPA are shown: 0 (lane 1), 4 nM (lanes 2, 7, 13, 17), 10 nM (lanes 3, 8, 13, 18), 25 nM (lanes 4, 9, 14, 19), 60 nM (lanes 5, 10, 15, 20) and 150 nM (lanes 6, 11, 16, 21).
To test if these XPA mutations only affected binding to ERCC1, we also compared the DNA-binding activities of wild-type and mutant XPA proteins. We investigated the binding of wild-type and mutant XPA to a DNA three-way junction, representing a high-affinity target for XPA in band-shift assays (Missura et al, 2001). The wild-type, F75A, G73Δ and G73Δ/G74Δ XPA proteins all bound with similar affinity to a three-way junction (Figure 5B), indicating that the mutant proteins are fully proficient in DNA binding and unlikely to be misfolded or otherwise inactive. These results show that single point mutations in XPA can result in a defect in NER activity by weakening the interaction between ERCC1 and XPA. Due to the highly cooperative nature of NER (Moggs et al, 1996), other NER functions and interactions may be disrupted as a result of blocking the recruitment of XPF–ERCC1.
XPA competes with single-stranded DNA for binding to ERCC1
Because XPA binds in the groove on ERCC1 (Figure 2) that was previously implicated in DNA-binding activity (Tsodikov et al, 2005), we directly tested whether or not XPA competes with single-stranded DNA for binding to ERCC1. DNA-binding activity was measured by monitoring fluorescence anisotropy, using single-stranded 40-mer oligonucleotide labeled on the 5′ end with 6-carboxyfluorescein. The XPA67−80 peptide does not detectably bind to DNA (not shown), although it does compete with DNA for binding to ERCC1 (Supplementary Figure 3). This result confirms that the DNA-binding site on ERCC1's central domain overlaps with the XPA-binding site. The EC50 for binding of XPA67−80 is in the micromolar range, but quenching of the fluorescent probe by high concentrations of XPA precluded an accurate measurement of the binding constant. We previously reported an equilibrium-binding constant of 1.5 μM for DNA binding to the central domain of ERCC1 (Tsodikov et al, 2005). By fitting the XPA competition titration data to a competitive binding model (Equation 9 in the Supplementary data), we obtain the estimated binding constant of Kd=(540±280) nM for the XPA–ERCC1 complex. This result agrees well with the affinity determined directly for this interaction (Supplementary Figure 2). Thus, XPA binds to the central domain of ERCC1 with approximately three-fold higher affinity than single-stranded DNA.
Discussion
The removal of bulky and helix-distorting DNA lesions by the NER pathway requires the coordinated assembly of a large multiprotein complex (Houtsmuller et al, 1999; Volker et al, 2001) that exposes the damaged DNA strand and excises an oligonucleotide containing the lesion (Gillet and Schärer, 2006). We have investigated one of the essential protein–protein interactions in this pathway. The specific interaction of XPA with ERCC1 is responsible for recruitment of the ERCC1-XPF nuclease to the DNA repair complex (Li et al, 1994). Our structural studies have defined the XPA ligand as a TGGGFI sequence motif that inserts into a pocket of the central domain of ERCC1 (Figure 2). It was previously shown that deletion of the GGG triplet within this motif abolishes the interaction of XPA with ERCC1 (Li et al, 1995). These glycines insert deep into the ERCC1-binding site and are likely to make hydrogen-bonding interactions using main chain atoms (Figure 2). The binding site of ERCC1 is mainly a nonpolar surface that is punctuated by several large aromatic side chains (Phe145, Phe152) and a buried salt bridge between Arg156 and Asp129. We show that single point mutations in XPA (F75A or G73Δ) effectively abolish NER activity in vitro, underscoring the high specificity of the binding interaction between ERCC1 and XPA.
The XPA peptide ligand is unstructured in solution (Figure 1B). It is therefore remarkable that a short peptide segment binds to ERCC1 with submicromolar affinity, given the associated entropic penalty for binding. This peptide–protein interaction is sufficient to block NER activity in cell-free extracts (Figure 4A), raising the possibility that peptidomimetic ligands could be developed to specifically block NER activity in vivo. Although the competing XPA peptide prevents the double incision of lesioned DNA during NER, the peptide does not interfere with the cleavage of a model DNA substrate by purified ERCC1–XPF (Figure 4B). These results show that the XPA peptide does not block the nuclease activity of XPF-ERCC1 and is instead likely to interfere with the recruitment of the nuclease into the NER protein complex. Mutations in the ERCC1-binding domain of XPA similarly abolish NER activity without affecting the intrinsic DNA-binding activity of XPA. Intriguingly, the central domain of ERCC1 binds to single-stranded DNA in vitro (Tsodikov et al, 2005) and this DNA-binding activity is blocked by XPA (Supplementary Figure 3). Although these competing activities at first appear contradictory, our cleavage assays employed full-length ERCC1 protein in complex with XPF, whereas the in vitro binding studies used the central domain only of ERCC1. Since ERCC1-XPF has multiple DNA-binding sites (Newman et al, 2005; Tripsianes et al, 2005; Tsodikov et al, 2005; Nishino et al, 2005a, 2005b), it is likely that some of the other DNA-binding surfaces of the XPF–ERCC1 heterodimer can compensate for the interference by the XPA peptide. With respect to the overall NER reaction, it is conceivable that DNA and the XPA protein alternatively bind to the same site on ERCC1 during different steps of the repair process. In this regard, a molecular handoff of ERCC1 from XPA to one strand of the unwound DNA substrate could be envisioned as one of multiple, individually weak interactions that drive the progression through the NER pathway in a concerted fashion (Stauffer and Chazin, 2004; Gillet and Schärer, 2006).
Homologs of the XPA and ERCC1 proteins are found only in eukaryotic organisms, despite the presence of homodimeric XPF-like endonucleases in the Archaea (Nishino et al, 2003). The ERCC1 residues constituting the XPA-binding site are poorly conserved in Saccharomyces cerivisiae (Rad10 protein) and S. pombe (Swi10 protein), and the XPA homologs (S. saccharomyces Rad14; S. pombe Rhp14) are highly divergent from mammalian XPA. Indeed, a different interaction site has recently been reported for Rad14 and Rad10/Rad1, the respective yeast homologs of XPA and ERCC1–XPF (Guzder et al, 2006). These observations suggest that XPA and ERCC1 may have coevolved to interact specifically with each other in higher eukaryotes, perhaps in response to the added complexity and distinct functional organization of the eukaryotic NER pathway. Correspondingly, a BLAST search does not identify the TGGGFI motif of the XPA ligand in any other mammalian protein.
In conclusion, we have established that only a short peptide segment of XPA is sufficient to form a stable and specific 1:1 complex with ERCC1. The interactions of three consecutive glycines (Gly72, Gly73, Gly74) and several flanking residues of XPA complement a V-shaped, hydrophobic groove in the central domain of ERCC1. This protein–protein interaction is essential for NER activity, and the XPA peptide is an effective inhibitor of NER activity in a cell-free reaction. This work paves the way for development of specific NER inhibitors targeting the surface of ERCC1 involved in XPA binding. ERCC1 has served as a molecular marker for clinical resistance to cisplatin-based chemotherapy (Reed, 2006), raising the possibility of using ERCC1 antagonists as sensitizing agents for tumors resistant to this and other DNA-damaging agents in the treatment of cancer.
Materials and methods
Peptide and DNA
The XPA67−80 peptide corresponding to residues 67–80 of the XPA protein and the mutant peptide XPA67−80 F75A were synthesized by solid phase methods, then HPLC-purified by the Molecular Biology Core Facility at Tufts University (Boston, MA). A 40-mer DNA oligomer 5′-CCGGTGGCCAGCGCTCGGCGT20–3′ with a 5′ 6FAM label (Integrated DNA Technologies) was gel-purified by conventional techniques.
Protein expression and purification
The central domain of ERCC1 (constructs ERCC192−214 or ERCC196−214) with an N-terminal His6 tag was expressed and purified as previously described (Tsodikov et al, 2005). Fragments of the XPA protein (XPA1−273, XPA59−273, XPA59−219, XPA59−93) were cloned into pET19bpps, in which an N-terminal (His)10 tag is separated from the XPA sequences by a Prescission protease cleavage site (gift of Dr Tapan Biswas), between NdeI and XhoI sites. Full-length XPA protein was expressed in bacteria from pET15b-XPA and purified by Ni2+-NTA, gel filtration, and heparin chromatography. Protein expression and purification are described in detail in the Supplementary data.
Analytical ultracentrifugation
Sedimentation equilibrium experiments with ERCC192−214 and the complex ERCC192−214–XPA59−93 were performed using Beckman XLA Analytical Centrifuge. In both cases, proteins were at concentrations of 0.3–0.5 mg/ml in NMR Buffer (20 mM Tris buffer pH 7.2, 50 mM NaCl, 2 mM β-mercaptoethanol and 0.1 mM EDTA). Sedimentation equilibrium data were analyzed as described in the Supplementary data.
Crystallization of XPA–ERCC1 complex, data collection and analysis
Protein crystallization and X-ray data collection are described in the Supplementary data. A complete and redundant X-ray data set was collected and processed using HKL2000 (Otwinowski and Minor, 1997). The crystals belong to space group I4132 with one ERCC1–XPA complex in the asymmetric unit. The structure was determined by molecular replacement (MR) methods using the program PHASER (McCoy et al, 2005) and the crystallographic model of the ERCC1 central domain (Tsodikov et al, 2005; PDB code 2A1I) in which the residues C-terminal to residue 214 were deleted. A difference (Fo−Fc) electron density map calculated with phases from the MR solution revealed the bound XPA peptide. The XPA peptide was built into the difference density using distance restraint information from NMR experiments and the structure of the complex was then refined as described below, with strong geometric restraints imposed on the ERCC1 subunit due to the low-resolution diffraction data and the absence of intramolecular distance information for ERCC1. All experimental XPA–ERCC1 distance restraints were accommodated without violations using the structure of unbound ERCC1, suggesting that ERCC1 does not undergo significant conformational changes upon binding to XPA.
NMR experiments and determination of the structure of XPA–ERCC1 complex
All NMR data were acquired in the NMR Buffer described above. The protein concentrations were 0.25 mM for free ERCC196−214 or ERCC192−214 (which behaved similarly in all experiments), 0.25 mM for ERCC192−214 in complex with a synthetic XPA67−80 peptide and 0.1 mM for ERCC192−214 complex with XPA59−93 fragment. Higher protein concentrations resulted in line broadening and lower quality NMR spectra. Backbone assignments of the free ERCC192−214 and ERCC1–XPA67−80 complex were performed using a standard set of triple-resonance experiments: HNCA/HN(CO)CA, HN(CA)CB/HN(COCA)CB and HNCO/HN(CA)CO.
Structural information for the ERCC1–XPA complex was obtained with a differentially labeled sample in which ERCC1 was 15N-labeled and perdeuterated and the synthetic XPA fragment was unlabeled (D,N-ERCC1/U-XPA) (Walters et al, 1997, 2001). The assignment of the XPA peptide in this sample was performed using homonuclear 2D NOESY and 2D TOCSY experiments acquired in both H2O and D2O buffers. The total of 92 intramolecular distance constraints for the XPA peptide were derived from the 2D NOESY experiment acquired in H2O with 100 ms mixing time. Intermolecular distance restraints were derived from a 15N-dispersed NOE-HSQC experiment acquired on the D,15N-ERCC1/U-XPA sample using 200 ms mixing time. A total of 23 intermolecular distance restraints between the amide protons of ERCC1 and the protons of XPA were derived from this experiment. The structure of the ERCC1–XPA complex was calculated using simulated annealing procedure in XPLOR-NIH (Schwieters et al, 2003). The total energy term used in the calculation incorporated all of the NMR-derived distance restraints as well as the 4 Å X-ray data. Ten lowest energy structures out of 100 calculated were deposited in the PDB with accession code 2JNW. The solvent accessible surface areas were calculated for the lowest energy structure using Surface Racer 4.0 (Tsodikov et al, 2002) with the solvent probe radius of 1.4 Å.
Competitive binding equilibrium titrations
Fluorescence anisotropy measurements were performed as previously reported (Tsodikov et al, 2005). The equilibrium titrations and their analysis are described in detail in the supplementary data.
Construction and expression of mutant XPA proteins
Site-directed mutagenesis using the QuikChange kit (Stratagene) introduced point mutations in the expression vector pET15b-XPA was performed. pET15b-XPA served as template and oligonucleotide primers used to generate the mutations contained the desired mutation, and a marker restriction site for selection. The following primers were used (restriction site are underlined and indicated, modified nucleotides are shown in italics):
XPA-F75A: GACACAGGAGGAGGCGCCATTCTAGAAGAGGAAGAAG (XbaI)
XPA-ΔG74: GACACAGGAGGATTCATTCTAGAAGAGGAAGAAG (XbaI)
XPA-ΔG73/74: GATAATTGACACAGGATTCATTCTAGAAGAGGAAGAAG (XbaI).
Positive clones were fully sequenced to rule out the introduction of additional mutations. Mutant XPA proteins were expressed in Escherichia coli BL21(DE3)pLyS cells and purified by chromatography on nickel-NTA, gel filtration and heparin columns.
Nuclease assay
ERCC1-XPF was purified and nuclease assays using a stem-loop substrate were carried out as described previously (Enzlin and Schärer, 2002) (see Supplementary data for details).
DNA-binding assays
The three-way junction DNA substrate described previously (substrate 7 in Table 1 of Hohl et al, 2003) was 5′-32P-end labeled and incubated at 1 nM concentration with various amounts of XPA in EMSA buffer (25 mM HepesKOH pH 8.0, 30 mM KCl, 10% glycerol, 1 mM DTT, 1 mM EDTA, 0.1 mg/ml BSA) at a reaction volume of 15 μl. After equilibration at room temperature for 30 min, the samples were loaded on a 5% (37.5:1) native polyacrylamide gel containing 0.5 × TBE and electrophoresed at 90 V for 2 h. Gels were dried and the radioactive bands visualized by autoradiography.
In vitro NER assay
This assay was performed using an established protocol (Shivji et al, 1999), which is described in detail in the Supplementary data.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Dr Tapan Biswas for comments and assistance with the figures. This work was funded by grants from the National Institutes of Health (GM52504) to TE, the Human Frontier Science Program (RGP0298/2001-M) to TE and ODS, and the New York State Office of Science and Technology and Academic Research (NYSTAR) grant. no. C040069 to ODS. DI is a recipient of a Scholar Award from the Harvard Center for AIDS Research.
References
- Araujo SJ, Nigg EA, Wood RD (2001) Strong functional interactions of TFIIH with XPC and XPG in human DNA nucleotide excision repair, without a preassembled repairosome. Mol Cell Biol 21: 2281–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchko GW, Isern NG, Spicer LD, Kennedy MA (2001) Human nucleotide excision repair protein XPA: NMR spectroscopic studies of an XPA fragment containing the ERCC1-binding region and the minimal DNA-binding domain (M59-F219). Mutat Res 486: 1–10 [DOI] [PubMed] [Google Scholar]
- Buchko GW, Ni S, Thrall BD, Kennedy MA (1998) Structural features of the minimal DNA binding domain (M98-F219) of human nucleotide excision repair protein XPA. Nucleic Acids Res 26: 2779–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camenisch U, Dip R, Schumacher SB, Schuler B, Naegeli H (2006) Recognition of helical kinks by xeroderma pigmentosum group A protein triggers DNA excision repair. Nat Struct Mol Biol 13: 278–284 [DOI] [PubMed] [Google Scholar]
- de Laat WL, Appeldoorn E, Jaspers NG, Hoeijmakers JH (1998) DNA structural elements required for ERCC1-XPF endonuclease activity. J Biol Chem 273: 7835–7842 [DOI] [PubMed] [Google Scholar]
- de Laat WL, Jaspers NG, Hoeijmakers JH (1999) Molecular mechanism of nucleotide excision repair. Genes Dev 13: 768–785 [DOI] [PubMed] [Google Scholar]
- Ding J, Miao ZH, Meng LH, Geng MY (2006) Emerging cancer therapeutic opportunities target DNA-repair systems. Trends Pharmacol Sci 27: 338–344 [DOI] [PubMed] [Google Scholar]
- Enzlin JH, Schärer OD (2002) The active site of the DNA repair endonuclease XPF-ERCC1 forms a highly conserved nuclease motif. EMBO J 21: 2045–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E, Walker G, Siede W, Wood R, Schultz R, Ellenberger T (2005) DNA Repair and Mutagenesis. Washington, DC: ASM Press [Google Scholar]
- Gaillard PH, Wood RD (2001) Activity of individual ERCC1 and XPF subunits in DNA nucleotide excision repair. Nucleic Acids Res 29: 872–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet LC, Schärer OD (2006) Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev 106: 253–276 [DOI] [PubMed] [Google Scholar]
- Guzder SN, Sommers CH, Prakash L, Prakash S (2006) Complex formation with damage recognition protein Rad14 is essential for Saccharomyces cerevisiae Rad1–Rad10 nuclease to perform its function in nucleotide excision repair in vivo. Mol Cell Biol 26: 1135–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl M, Thorel F, Clarkson SG, Schärer OD (2003) Structural determinants for substrate binding and catalysis by the structure-specific endonuclease XPG. J Biol Chem 278: 19500–19508 [DOI] [PubMed] [Google Scholar]
- Houtsmuller AB, Rademakers S, Nigg AL, Hoogstraten D, Hoeijmakers JH, Vermeulen W (1999) Action of DNA repair endonuclease ERCC1/XPF in living cells. Science 284: 958–961 [DOI] [PubMed] [Google Scholar]
- Hoy CA, Thompson LH, Salazar EP, Stewart SA (1985) Different genetic alterations underlie dual hypersensitivity of CHO mutant UV-1 to DNA methylating and cross-linking agents. Somat Cell Mol Genet 11: 523–532 [DOI] [PubMed] [Google Scholar]
- Iakoucheva LM, Kimzey AL, Masselon CD, Bruce JE, Garner EC, Brown CJ, Dunker AK, Smith RD, Ackerman EJ (2001) Identification of intrinsic order and disorder in the DNA repair protein XPA. Protein Sci 10: 560–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami T, Kuraoka I, Saijo M, Kodo N, Kyogoku Y, Morikawa K, Tanaka K, Shirakawa M (1998) Solution structure of the DNA- and RPA-binding domain of the human repair factor XPA. Nat Struct Biol 5: 701–706 [DOI] [PubMed] [Google Scholar]
- Jones CJ, Wood RD (1993) Preferential binding of the xeroderma pigmentosum group A complementing protein to damaged DNA. Biochemistry 32: 12096–12104 [DOI] [PubMed] [Google Scholar]
- Li L, Elledge SJ, Peterson CA, Bales ES, Legerski RJ (1994) Specific association between the human DNA repair proteins XPA and ERCC1. Proc Natl Acad Sci USA 91: 5012–5016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Peterson CA, Lu X, Legerski RJ (1995) Mutations in XPA that prevent association with ERCC1 are defective in nucleotide excision repair. Mol Cell Biol 15: 1993–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr 61: 458–464 [DOI] [PubMed] [Google Scholar]
- McWhir J, Selfridge J, Harrison DJ, Squires S, Melton DW (1993) Mice with DNA repair gene (ERCC-1) deficiency have elevated levels of p53, liver nuclear abnormalities and die before weaning. Nat Genet 5: 217–224 [DOI] [PubMed] [Google Scholar]
- Missura M, Buterin T, Hindges R, Hubscher U, Kasparkova J, Brabec V, Naegeli H (2001) Double-check probing of DNA bending and unwinding by XPA-RPA: an architectural function in DNA repair. EMBO J 20: 3554–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto I, Miura N, Niwa H, Miyazaki J, Tanaka K (1992) Mutational analysis of the structure and function of the xeroderma pigmentosum group A complementing protein. Identification of essential domains for nuclear localization and DNA excision repair. J Biol Chem 267: 12182–12187 [PubMed] [Google Scholar]
- Moggs JG, Yarema KJ, Essigmann JM, Wood RD (1996) Analysis of incision sites produced by human cell extracts and purified proteins during nucleotide excision repair of a 1,3-intrastrand d(GpTpG)-cisplatin adduct. J Biol Chem 271: 7177–7186 [DOI] [PubMed] [Google Scholar]
- Newman M, Murray-Rust J, Lally J, Rudolf J, Fadden A, Knowles PP, White MF, McDonald NQ (2005) Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition. EMBO J 24: 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Essers J, Weeda G, Beverloo B, de Wit J, Muijtjens M, Odijk H, Hoeijmakers JH, Kanaar R (2001) The structure-specific endonuclease Ercc1-Xpf is required for targeted gene replacement in embryonic stem cells. EMBO J 20: 6540–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, van Leeuwen W, Theil AF, Vermeulen W, van der Horst GT, Meinecke P, Kleijer WJ, Vijg J, Jaspers NG, Hoeijmakers JH (2006) A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature 444: 1038–1043 [DOI] [PubMed] [Google Scholar]
- Nishino T, Komori K, Ishino Y, Morikawa K (2003) X-ray and biochemical anatomy of an archaeal XPF/Rad1/Mus81 family nuclease: similarity between its endonuclease domain and restriction enzymes. Structure 11: 445–457 [DOI] [PubMed] [Google Scholar]
- Nishino T, Komori K, Ishino Y, Morikawa K (2005a) Structural and functional analyses of an archaeal XPF/Rad1/Mus81 nuclease: asymmetric DNA binding and cleavage mechanisms. Structure 13: 1183–1192 [DOI] [PubMed] [Google Scholar]
- Nishino T, Komori K, Tsuchiya D, Ishino Y, Morikawa K (2005b) Crystal structure and functional implications of Pyrococcus furiosus hef helicase domain involved in branched DNA processing. Structure 13: 143–153 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Park CH, Mu D, Reardon JT, Sancar A (1995) The general transcription-repair factor TFIIH is recruited to the excision repair complex by the XPA protein independent of the TFIIE transcription factor. J Biol Chem 270: 4896–4902 [DOI] [PubMed] [Google Scholar]
- Park CH, Sancar A (1994) Formation of a ternary complex by human XPA, ERCC1, and ERCC4(XPF) excision repair proteins. Proc Natl Acad Sci USA 91: 5017–5021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed E (2006) ERCC1 measurements in clinical oncology. N Engl J Med 355: 1054–1055 [DOI] [PubMed] [Google Scholar]
- Rosenberg E, Taher MM, Kuemmerle NB, Farnsworth J, Valerie K (2001) A truncated human xeroderma pigmentosum complementation group A protein expressed from an adenovirus sensitizes human tumor cells to ultraviolet light and cisplatin. Cancer Res 61: 764–770 [PubMed] [Google Scholar]
- Saijo M, Kuraoka I, Masutani C, Hanaoka F, Tanaka K (1996) Sequential binding of DNA repair proteins RPA and ERCC1 to XPA in vitro. Nucleic Acids Res 24: 4719–4724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieters CD, Kuszewski JJ, Tjandra N, Marius Clore G (2003) The Xplor-NIH NMR molecular structure determination package. J Magn Reson 160: 65–73 [DOI] [PubMed] [Google Scholar]
- Sijbers AM, van der Spek PJ, Odijk H, van den Berg J, van Duin M, Westerveld A, Jaspers NG, Bootsma D, Hoeijmakers JH (1996) Mutational analysis of the human nucleotide excision repair gene ERCC1. Nucleic Acids Res 24: 3370–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivji MK, Moggs JG, Kuraoka I, Wood RD (1999) Dual-incision assays for nucleotide excision repair using DNA with a lesion at a specific site. Methods Mol Biol 113: 373–392 [DOI] [PubMed] [Google Scholar]
- Stauffer ME, Chazin WJ (2004) Structural mechanisms of DNA replication, repair, and recombination. J Biol Chem 279: 30915–30918 [DOI] [PubMed] [Google Scholar]
- Tripsianes K, Folkers G, Ab E, Das D, Odijk H, Jaspers NG, Hoeijmakers JH, Kaptein R, Boelens R (2005) The structure of the human ERCC1/XPF interaction domains reveals a complementary role for the two proteins in nucleotide excision repair. Structure 13: 1849–1858 [DOI] [PubMed] [Google Scholar]
- Tsodikov OV, Enzlin JH, Scharer OD, Ellenberger T (2005) Crystal structure and DNA binding functions of ERCC1, a subunit of the DNA structure-specific endonuclease XPF-ERCC1. Proc Natl Acad Sci USA 102: 11236–11241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsodikov OV, Record MT Jr, Sergeev YV (2002) Novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J Comput Chem 23: 600–609 [DOI] [PubMed] [Google Scholar]
- Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH (2001) Sequential assembly of the nucleotide excision repair factors in vivo. Mol Cell 8: 213–224 [DOI] [PubMed] [Google Scholar]
- Walters KJ, Ferentz AE, Hare BJ, Hidalgo P, Jasanoff A, Matsuo H, Wagner G (2001) Characterizing protein–protein complexes and oligomers by nuclear magnetic resonance spectroscopy. Methods Enzymol 339: 238–258 [DOI] [PubMed] [Google Scholar]
- Walters KJ, Matsuo H, Wagner G (1997) A simple method to distinguish intermonomer nuclear overhauser effects in homodimeric proteins with C-2 symmetry. J Am Chem Soc 119: 5958–5959 [Google Scholar]
- Weeda G, Donker I, de Wit J, Morreau H, Janssens R, Vissers CJ, Nigg A, van Steeg H, Bootsma D, Hoeijmakers JH (1997) Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr Biol 7: 427–439 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information