Abstract
Most mitochondrial proteins are synthesized in the cytosol and imported into mitochondria. The N-terminal presequences of mitochondrial-precursor proteins contain a diverse consensus motif (φχχφφ, φ is hydrophobic and χ is any amino acid), which is recognized by the Tom20 protein on the mitochondrial surface. To reveal the structural basis of the broad selectivity of Tom20, the Tom20–presequence complex was crystallized. Tethering a presequence peptide to Tom20 through a disulfide bond was essential for crystallization. Unexpectedly, the two crystals with different linker designs provided unique relative orientations of the presequence with respect to Tom20, and neither configuration could fully account for the hydrophobic preference at the three hydrophobic positions of the consensus motif. We propose the existence of a dynamic equilibrium in solution among multiple states including the two bound states. In accordance, NMR 15N relaxation analyses suggested motion on a sub-millisecond timescale at the Tom20–presequence interface. We suggest that the dynamic, multiple-mode interaction is the molecular mechanism facilitating the broadly selective specificity of the Tom20 receptor toward diverse mitochondrial presequences.
Keywords: crystallography, mitochondrial protein import, NMR relaxation analysis, presequence, Tom20
Introduction
Amino-acid sequences that cannot be defined as a simple consensus motif are often used as recognition motifs in protein transport systems (Schatz and Dobberstein, 1996). Protein import into mitochondria represents a good example. Tom20, a 20-kDa subunit of the translocase of outer mitochondrial membrane complex, interacts with the N-terminal sequences of the precursor proteins destined for the mitochondrial matrix and inner membrane, and thereby functions as a receptor for the mitochondrial preproteins synthesized in the cytosol (Sollner et al, 1989; Ramage et al, 1993; Pfanner, 2000; Pfanner and Geissler, 2001; Endo and Kohda, 2002; Endo et al, 2003; Rapaport, 2003). These N-terminal-cleavable segments are collectively referred to as mitochondrial presequences. Statistical analyses revealed that a presequence typically consists of 15–40 amino-acid residues, with an abundance of positively charged residues, and tends to form an amphiphilic helical conformation (von Heijne, 1986). The Tom20 protein is anchored to the mitochondrial outer membrane by the N-terminal hydrophobic segment, and exposes its C-terminal domain to the cytosol. Two types of Tom20 protein exist in animal genomes, including human, mouse, and rat (Likic et al, 2005). The type II Tom20 gene is expressed ubiquitously in various mouse tissues, but the type I gene is specifically expressed in testis. In contrast, only one type of Tom20 (type I) exists in fungal genomes, including Saccharomyces cerevisiae and Neurospora crassa. Thus, in the vast majority of cells, a single Tom20 protein distinguishes ca 1000 mitochondrial proteins from other non-mitochondrial proteins and sorts them into mitochondria. In other words, Tom20 recognizes the targeting signal encrypted in the diverse amino-acid sequences of mitochondrial presequences, and at the same time does not accept other amino-acid sequences in the N-terminal portion of many non-mitochondrial proteins.
The consensus motif for the Tom20 recognition has not been fully defined, but our previous NMR and peptide-library analyses revealed that the consensus motif was represented as φχχφφ (where φ is a hydrophobic amino acid, such as leucine, isoleucine, phenylalanine, tryptophan, valine, and tyrosine, and χ is any amino acid) for a subset of presequences (Muto et al, 2001; Obita et al, 2003). Recently, some presequences containing alanine mutations at the hydrophobic positions within the consensus motif have been reported to mediate the import into mitochondria, despite their reduced Tom20 binding (Mukhopadhyay et al, 2006). Since the deletion of Tom20 is not lethal in yeast (Lithgow et al, 1994), it is possible that the mutated preproteins bypass Tom20 and use an alternative import pathway. However, a more likely explanation is that the mutated presequences actually bind to Tom20 very weakly, at a level lower than the detection limit of the bacterial two-hybrid system, but still enough to support the import.
We previously determined the solution structure of the cytosolic domain (residues 51–145) of rat Tom20 (ubiquitous form, type II) complexed with a presequence peptide derived from rat mitochondrial aldehyde dehydrogenase (ALDH) by NMR spectroscopy (Abe et al, 2000). The ALDH presequence contains LSRLL as the Tom20 recognition motif. The core structure of Tom20 consists of four α-helices, with a fifth α-helix loosely attached to the core (Figure 1). The bound portion of the presequence peptide adopts an amphiphilic helical conformation. Since presequences alone exhibit very little secondary structure in aqueous solutions (Roise et al, 1988; Pak and Weiner, 1990), the helical formation is essential for keeping the three leucine residues close together, so they can interact with the hydrophobic groove of Tom20.
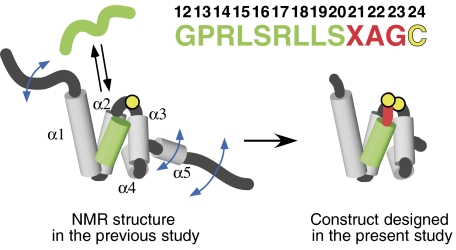
Figure 1.
Design of the Tom20–presequence complex. The solution structure of the cytosolic domain of Tom20, previously determined by NMR (Abe et al, 2000), consists of five α-helices, α1–α5 (gray), with flexible N- and C-terminal segments. The presequence associates with and dissociates from Tom20 rapidly in solution. The presequence from aldehyde dehydrogenase (ALDH) is shown in green. The presequence peptide has no tertiary structure in the free state, but assumes an α-helical conformation in the bound state. Note that the numbering starts at 12, since this sequence corresponds to the C-terminal half of the ALDH presequence (Farres et al, 1988). A cysteine residue (yellow) was attached to the C terminus after the three-residue linker (red). The intermolecular disulfide bond was formed with the single cysteine residue of Tom20 (Cys100, yellow circle), and the mobile α5 and two flexible termini were removed, to prepare the Tom20–presequence complexes for crystallization and the NMR relaxation study.
The NMR structure, however, did not provide more detailed information on the bound presequence peptide due to the insufficient number of distance constraints within the peptide and between the peptide and Tom20 even at saturated peptide concentrations (Abe et al, 2000). This implied that the peptide still had some residual mobility in the Tom20 groove. Consequently, although the structure of Tom20 itself was well determined, the position and structure of the peptide were not sufficiently defined for an in-depth discussion about the recognition mechanism of Tom20.
In the present study, we designed a fragment corresponding to the core structure of Tom20 for X-ray crystallography. Consideration of the NMR structure prompted us to remove the flexible N- and C-terminal segments, which may interfere with crystallization (Figure 1). Our first attempt to cocrystallize the Tom20 fragment with the ALDH presequence failed, probably due to the weak affinity of the presequence peptide for Tom20. Therefore, we tethered the presequence peptide to Tom20 via an intermolecular disulfide bond. This tethering method was successfully used to analyze the presequence recognition of Tom20 by a novel peptide-library approach (Obita et al, 2003); the reasonable consensus pattern thus obtained indicates that this strategy works well to fix the presequence peptide in the binding groove of Tom20 in an unbiased manner. We successfully obtained two forms of crystals suitable for data collection. The three-dimensional structures of the complex of Tom20 and the ALDH presequence peptide were determined at 1.9- and 2.1-Å resolutions. The comparison of the two crystal structures implied that a dynamic equilibrium exists among two or more bound states of the presequence peptide on Tom20 in solution. In accord with this model, an NMR relaxation study revealed motion on the sub-millisecond timescale at the interface between Tom20 and the presequence peptide. We propose a dynamic, multiple mode of recognition that explains the structural basis of the broadly selective specificity of Tom20 toward diverse mitochondrial presequences.
Results
Crystal structure determination
We prepared a 68-residue fragment that corresponds to the core structure (residues 59–126) of Tom20. This fragment contains a single cysteine residue (Figure 1). The presequence peptide (GPRLSRLLSXAGC) consists of a sequence derived from mitochondrial pALDH (bold), a three-residue linker (italic), and a C-terminal cysteine residue. We used two different linker designs: an alanine residue at the X position (A linker) and a tyrosine residue at the same position (Y linker). The length of the linker was previously optimized by a novel peptide-library approach (Obita et al, 2003). The presequence peptide was incubated with Tom20 to prepare the disulfide-bond-linked complex by air oxidation (Figure 1). The results from a gel filtration analysis were consistent with a monomeric form of the Tom20–presequence complex in solution. The complex containing the Y-linker peptide eluted at the same volume as the complex containing the A-linker peptide, which corresponds to the monomeric molecular weight (data not shown). The resulting complex was purified by reverse-phase chromatography, concentrated, and crystallized (Igura et al, 2005). The three-dimensional structures of the two complexes with the two different linker designs were solved at resolutions of 1.9 and 2.1 Å (Table I).
Table 1.
Data collection and refinement statisticsa
| Tom20–SS-pALDH(A) (A-linker crystal) | Tom20–SS-pALDH(Y) (Y-linker crystal) | |
|---|---|---|
| Data collection | ||
| Space group | P21 | C2 |
| Cell dimensions, a, b, c (Å), β (deg) | 33.6, 27.6, 71.0, 103.0 | 151.8, 64.1, 68.0, 94.7 |
| No. of molecules in AU | 2 | 7 |
| Resolution (Å) | 50–1.90 (1.98–1.90) | 20–2.05 (2.12–2.05) |
| Rmergeb | 0.057 (0.193) | 0.067 (0.302) |
| Mean I/σ(I) | 24.4 (8.4) | 16.7 (2.5) |
| Completeness (%) | 98.9 (93.2) | 95.8 (77.7) |
| Redundancy | 3.1 (2.6) | 5.2 (3.4) |
| Refinement | ||
| Resolution (Å) | 50–1.90 (1.98–1.90) | 20–2.05 (2.11–2.05) |
| No. of unique reflections | 8906 (639) | 34 455 (1696) |
| Rworkc/Rfreed | 0.1781/0.2396 | 0.2481/0.3084 |
| No. of atoms | ||
| Protein | 1091 | 3271 |
| Peptide | 177 | 584 |
| Water | 168 | 378 |
| B-factors (average) (Å2) | ||
| Protein | 19.1 | 53.6 |
| Peptide | 21.5 | 57.1 |
| Water | 35.1 | 61.9 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.012 | 0.010 |
| Bond angles (deg) | 1.42 | 1.226 |
| PDB entry | 2V1T | 2V1S |
| Values in the parentheses are for the highest resolution shell. | ||
| aStatistics for data collection of the SeMet derivative of Tom20–SS-pALDH(Y) was summarized in the previous report (Igura et al, 2005). | ||
| bRmerge=∑h∑j∣〈I〉h−Ih,j∣/∑h∑jIh,j, where 〈I〉h is the mean intensity of symmetry-equivalent reflections. | ||
| cRwork=∑h∣∣Fo∣−∣Fc∣∣/∑h∣Fo∣, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively, for reflection h. | ||
| dRfree is equivalent to Rwork for a 9% test set of reflections. | ||
There are two complex molecules in the asymmetric unit of the crystal containing the A-linker peptide, and seven complex molecules in that of the crystal containing the Y-linker peptide (Supplementary Figure 1). The presequence peptide binds to the Tom20 molecule as a monomeric form in the crystal containing the A-linker peptide. In contrast, all of complex molecules in the crystal containing the Y-linker peptide exist as intertwined dimers, due to the exchange of the presequence peptides between adjacent Tom20 molecules. There are many examples of intertwined dimer formation in protein crystal structures. Although such intertwining phenomenon is proposed to be a mechanism for the emergence of oligomeric proteins and for protein misfolding and aggregation (Rousseau et al, 2003), in many cases, the proteins exist as a monomer in solution, as well as the disulfide-tethered Tom20–presequence complex. There are no contacts between the two Tom20 molecules in the intertwined dimers, whereas the two presequence peptides contact each other. Because non-essential serine residues are mainly involved in the peptide–peptide interaction, we consider that the intertwined dimer formation in the crystal is not biologically relevant. We reconstituted the monomeric forms by swapping the coordinates of the presequence peptides in the intertwined pairs. No special treatment was performed for the atoms near the disulfide bond, thus the two sulfur atoms are not in the bond distance. The reconstituted monomeric forms were used in the following discussion. The superposition of the two crystal structures is shown in Figures 2A and B.
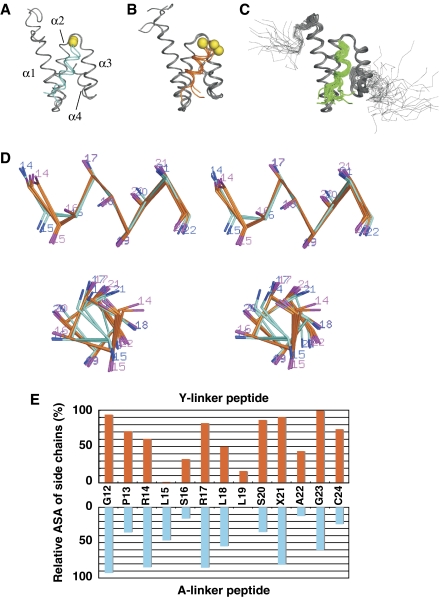
Figure 2.
Comparison of the crystal and solution structures of the Tom20–presequence complex. Superposition of (A) two A-linker crystal structures (disulfide-linked Tom20–pALDH complex containing the A linker), (B) five Y-linker crystal structures (disulfide-linked Tom20–pALDH complex containing the Y linker), and (C) 20 solution structures determined by NMR (PDB entry, 1OM2) (Abe et al, 2000). The Cα atoms of residues 65–121 of Tom20 were used for the superposition. The backbone atoms of the Tom20 protein are shown in gray, and those of the presequence peptide are cyan, orange, or green. The C-terminal cysteine residues of the peptide are depicted as yellow spheres. (D) Stereo views of the α-helical conformation of the presequence peptide in the A-linker structure (two structures, cyan) and in the Y-linker structure (five structures, orange). Residues 12, 13, 23, and 24 are omitted for clarity. (E) Relative accessible surface area (ASA) of the side chains of the presequence peptide in the complex. The relative ASA of the total of the side-chain atoms for each residue was calculated by the program NACESS, and the average values are plotted as a function of the residue number of the peptide.
Overall structures of the Tom20–presequence complexes
The core structure of the cytosolic domain of Tom20 consists of four α-helices, and the presequence peptide is accommodated in the groove formed between α1 and α3 of Tom20 (Figures 2A and B). Overall, the crystal structures fully agree with the NMR structure, which was determined in solution without the disulfide-bond tether (Figure 2C). Thus, neither the introduction of the intermolecular disulfide bond nor the crystal contacts had any deleterious effects.
Conformations of the presequence peptides in the bound states
The electron density maps demonstrated that the structures of the presequence peptides, including the linker regions, were well defined in both crystals (Supplementary Figure 2). Our previous NMR structure determination suggested that the bound peptide assumes an α-helical conformation, but a detailed analysis was not possible. In the present study, the crystal structures confirmed that the presequence peptide adopts an α-helical conformation (Figure 2D). The α-helix comprises not only the peptide region corresponding to the consensus motif (Leu15′–Leu19′, presequence numbering is marked by prime) but also the region close to the C terminus (Ser20′–Ala22′).
The covalent trapping could change the position of the presequence peptide and strain its conformation. Despite the concern, the helical regions of the peptides are well superimposed on each other, whereas the N-terminal region (Gly12′–Pro13′) and the C-terminal linker region (Gly23′–Cys24′), which are not in contact with Tom20, have no preferred positions, as compared to the region in contact with Tom20 (Figures 2A and B). Furthermore, we attempted to reduce the tethering disulfide bond in situ by soaking the crystals in a DTT-containing mother liquor. The crystal structure determination revealed that the disulfide bond was successfully cleaved in the A-linker crystal, but the overall structure was not changed except that the C-terminal cysteine residue of the peptide became disordered and invisible (to be published elsewhere). The attempts using the Y-linker crystal are now in progress. These results argue against the position and its conformation being forced by its covalent attachment.
Comparison of the two structures
The differences between the two crystal structures were analyzed with the program DynDom (Hayward and Lee, 2002), which detects domain movement by comparing two conformations of the same protein. The Tom20 structure consists of two helix–turn–helix domains, α1–α2 and α3–α4. Cys100, which was used as a tethering point, actually acts as the hinge of the domain rearrangement. In the Y-linker structure, the α1–α2 domain moves upward relative to α3–α4, with a 10 deg rotation around an axis as compared to the A-linker structure (Figure 3). In a coordinated manner, the presequence helix tilts its C terminus upward in the Y-linker structure as compared to the A-linker structure.
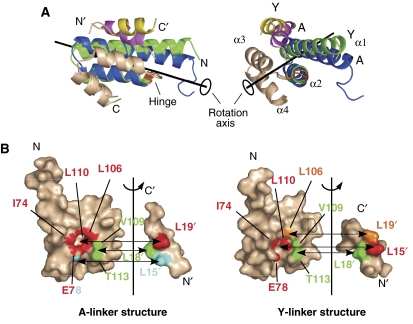
Figure 3.
Detailed comparison of the two crystal structures. (A) Conformational differences between the two Tom20 structures. The two crystal structures were compared using the program DynDom (Hayward and Lee, 2002). The Tom20 structure consists of two domains, α1–α2 (residues 59–99) and α3–α4 (residues 101–126). The α3–α4 domain (white) is superimposed. The α1–α2 domain of the Y-linker structure (green) rotates 10 deg around the axis and moves 0.2 Å along the axis relative to the A-linker structure (blue). The hinge residue was identified as Cys100 (orange). The bound presequence peptide of the A-linker structure is magenta, and that of the Y-linker structure is yellow. (B) Open-book representations of the contact residues at the Tom20–presequence interface. The surface of Tom20 that contacts one of the three leucine residues of the presequence peptide is displayed in the same color. Two atoms are defined as being in contact if the distance is less than 4.0 Å. The buried surface areas are similar: 305±2 Å2 for the A-linker structure versus 338±32 Å2 for the Y-linker structure.
The nearly parallel arrangement of the α1 and α3 helices forms a shallow groove, which serves as the binding site for the presequence. The contacting residues at the Tom20–peptide interface were examined by calculations of the inter-atom distances (Figure 3B). The three leucine residues of the presequence peptide are involved in the contacts. The side chain of Leu18′ in the presequence contacts the side chains of Val109 and Thr113 of Tom20 in both crystal structures. The side chain of Leu18′ is partially buried and rests against the rim formed by the methyl groups of Val109 and Thr113. The side chain of Leu19′ in the presequence is accommodated in a hydrophobic pocket created by Ile74, Glu78, Leu106, and Leu110 of Tom20 in the A-linker structure. To our surprise, the pocket formed by the same residues holds the side chain of Leu15′ in the presequence in the Y-linker structure. That is, the leucine residues at different positions in the presequence are recognized by the same hydrophobic pocket. The interaction is tight, since the relative accessible surface area (ASA) of Leu19′ in the A-linker structure and that of Leu15′ in the Y-linker structure are virtually 0% (Figure 2E). Moreover, the side chain of the third leucine residue, Leu15′ of the A-linker structure and Leu19′ of the Y-linker structure, has few contacts with the Tom20 surface (Figure 3B). In summary, Tom20 is equipped with only two hydrophobic subsites (subsite 1: the rim formed by residues 109 and 113; subsite 2: the pocket formed by residues 74, 78, 106, and 110) for the recognition of the three hydrophobic side chains in the φχχφφ motif.
The amino-acid residues of Tom20 involved in the two hydrophobic subsites are evolutionally conserved (Supplementary Figure 3). It is notable that the residue at position 78 is Glu or Asp, and that at position 113 is Ser or Thr. Thus, the side chains of these conserved non-hydrophobic residues seem to be functionally important. The functional groups, carboxylate and hydroxyl, are involved in hydrogen bond networks including bound water molecules (Supplementary Figure 4). The replacement of Glu78 by Ala, however, only slightly reduced the preprotein binding (Abe et al, 2000).
Mutational analysis of the amino-acid residues involved in the hydrophobic subsites
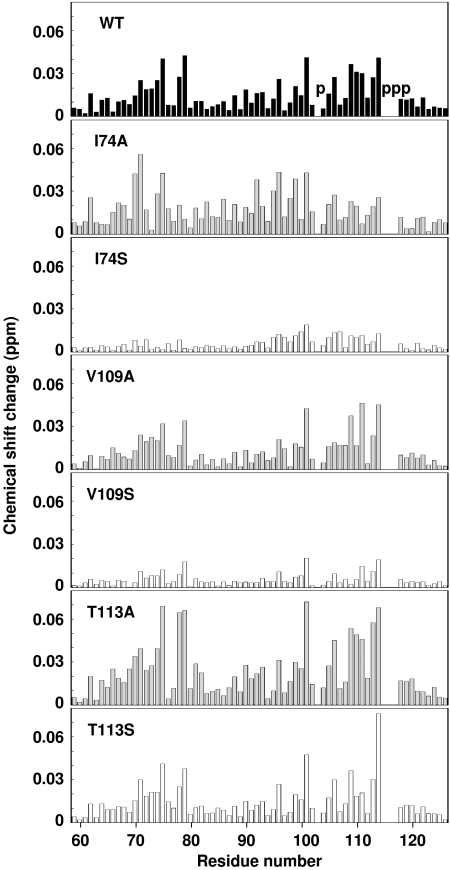
We examined the effects of mutations of the two hydrophobic subsites. Val109 and Thr113 from the subsite 1 and Ile74 from the subsite 2 were selected and changed to Ala or Ser. Figure 4 summarizes the results of the NMR titration experiments with the pALDH presequence peptide. The wild-type Tom20 used in this study (residues 59–126) bound to the pALDH presequence (residues 12–22) with the dissociation constant of 0.4 mM. Our previous NMR titration experiment using a longer version of Tom20 (residues 51–145) with a longer pALDH (residues 1–22) reported a dissociation constant of 0.02 mM (Abe et al, 2000). The present higher dissociation constant may be explained by the different lengths of Tom20 and pALDH used in this study. The substitutions of Ile74 and Val109 to an Ala residue did not change the extent and pattern of the chemical shift changes, as compared to the wild-type Tom20, suggesting that the Ala residues at positions 74 and 109 maintained the hydrophobic interactions with the Leu side chains of the presequence peptide. In contrast, the substitutions of Ile74 and Val109 to a Ser residue resulted in the substantial reduction of the chemical shift changes. This indicates that the hydrophobicity contributes to the presequence binding at both the subsites 1 and 2. The substitution of Thr113 to Ser did not change the interaction, which is consistent with the group conservation of Thr and Ser at this position (Supplementary Figure 3). Finally, the substitution of Thr113 to Ala increased the chemical shift changes. The mutation apparently strengthened the interaction, but the underlying explanation is unclear. A detailed study, for example a systematic mutation at position 113, is necessary in future.
Figure 4.
Chemical shift changes of backbone amides of the wild-type and mutated Tom20s upon binding to the pALDH presequence peptide at the molar ratio of 1:1. The chemical shift change of each backbone amide in [1H, 15N]HSQC spectra was calculated according to the equation, [Δδ(1H)2+(Δδ(15N)/5)2]1/2. The positions of proline residues are indicated with the letter ‘p' in the top panel.
NMR relaxation analyses
We carried out NMR 15N relaxation experiments using the disulfide-tethered Tom20–presequence complex to analyze the protein dynamics in solution. We used the A-linker for the detailed relaxation study, but the choice of the linker was not critical, because a preliminary study on the complex with the Y-linker provided consistent results. Two labeling schemes were used to minimize the spectral overlaps. In one preparation, the presequence peptide was 15N-labeled, and in the other preparation, Tom20 was labeled (Supplementary Figure 5).
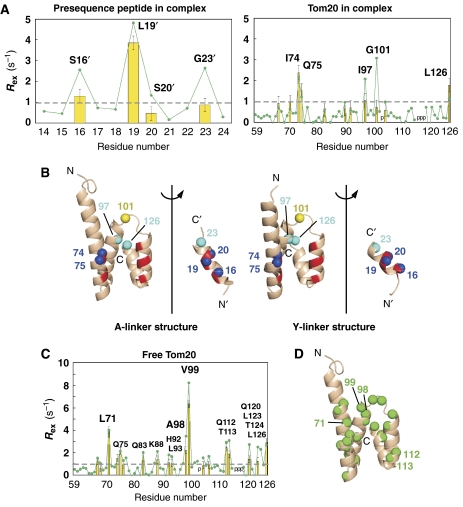
We used two methods to estimate the exchange-induced spin-relaxation rate, Rex. First, we used the model-free analysis under the isotropic rotational diffusion model, using the measured R1, R2, and {1H} 15N steady-state heteronuclear NOE values for 1H–15N (Lipari and Szabo, 1982). The overall rotational diffusion time, τm, was calculated independently for the [15N]peptide and [15N]Tom20 in the complex. The good agreement (5.4 versus 5.6 ns) shows the validity of the model-free analysis (Supplementary Figure 6). As a second approach, we measured the transverse CSA/dipolar cross-relaxation rate (ηxy) for 1H–15N, which is independent of chemical exchange processes (Tjandra et al, 1996; Kroenke et al, 1998). The Rex values were estimated from a simple approximate equation, Rex=R2−aηxy (see Materials and methods). The Rex values obtained by the two methods agree well, which demonstrates the reliability of the determined Rex values (Figure 5A). This cautious approach was taken because each method has its own drawbacks in approximation and simplification. We also measured the relaxation dispersion curves by a Carr–Purcell–Meiboom–Gill experiment (Supplementary Figure 7). No obvious changes were observed in the τcp range from 1 to 65 ms, indicating that the motion is not occurring in this time range. We estimate that the time range of the motion is on the order of the sub-millisecond timescale (see details in Materials and methods). For comparison, we estimated the Rex rates of the [15N]Tom20 in the free form (Figures 5C and Supplementary Figure 6).
Figure 5.
Distribution of slow motions. (A) Plot of Rex as a function of residue number of the disulfide-bond-tethered complex of Tom20 and the presequence. The yellow bar represents the Rex values estimated from the model-free analysis, and the green line with the closed circles is Rex from the measurement of transverse CSA/dipolar cross-relaxation rates (ηxy). The broken line indicates the threshold for the Rex mapping on the three-dimensional structures. The positions of proline residues are indicated with the letter ‘p'. (B) Open-book representations of the spatial distributions of amide 15N spins with an Rex value larger than 1 s−1 from the ηxy measurement in the two crystal structures. The amide nitrogen atoms, shown as blue spheres, of Ser16′, Leu19′, and Ser20′ of the presequence peptide and of Ile74 and Gln75 of Tom20 are located at the hydrophobic contact surface (red patches on the ribbons). The amide nitrogen atoms, shown as cyan spheres, of Gly23′, Ile97, and Leu126 probably correspond to the conformational changes accompanying the motion of the presequence peptide. Finally, the yellow sphere represents the amide nitrogen atom of Gly101, which shows exceptional discrepancy between the two Rex values. This could arise from the variation in the Rex value calculated from ηxy due to the variation in magnitude and orientation of the CSA tensor. Note that the large values of the three 15N spins at positions 16, 19, and 20 of the presequence suggest a very interesting rule: an amide 15N spin in an α-helical conformation monitors the side-chain motion of the preceding residue, in the present case, Leu15′, Leu18′, and Leu19′. (C) Plot of Rex as a function of residue number of the free form of Tom20. The yellow bar represents the Rex values estimated from the model-free analysis, and the green line with the closed circles represents Rex from the ηxy measurement. The broken line indicates the threshold for the Rex mapping on the three-dimensional structure. The positions of proline residues are indicated with the letter ‘p'. (D) The spatial distribution of amide 15N spins with an Rex value larger than 1 s−1 from the ηxy measurement. The Tom20 structure in the A-linker crystal structure was used instead of the free structure. The amide nitrogen atoms with large Rex values, shown as green spheres, are widely distributed in the Tom20 structure.
Discussion
We successfully obtained the high-resolution crystal structures of a Tom20–presequence complex. Our success was a consequence of the sophisticated design of the complex, with reference to our previous NMR structure: (i) The flexible N-terminal 8 residues and C-terminal 19 residues were omitted, to produce a shorter fragment of Tom20 that corresponds to the core structure. (ii) The single cysteine residue in the Tom20 fragment was used as an anchoring point. This cysteine residue is located at the optimal position for anchoring. (iii) A cysteine residue was added to the C terminus of the presequence peptide after the linker sequence. The optimal length of the linker (three residues) was previously determined in our peptide-library experiment (Obita et al, 2003). (iv) An intermolecular disulfide bond was formed to tether the presequence peptide onto the Tom20 protein.
Necessity of the intermolecular disulfide tethering
The fixation of the presequence peptide to Tom20 and the reduction of the residual mobility of the peptide in the bound state, by the introduction of the disulfide tether, were essential for the successful crystallization. In fact, no crystals have ever been obtained by simply mixing Tom20 with the presequence peptide. Previously, the local concentration of the peptide ligand was increased by attaching it as an N- or C-terminal extension, with a flexible polypeptide linker sequence, in crystallization and NMR studies (Fremont et al, 1996; Hennecke et al, 2000; Pellegrini et al, 2002; Freund et al, 2003). We also attempted this strategy: the ALDH presequence was fused to the N terminus of the Tom20 fragment (residues 64–126) via a 9- or 14-residue Gly-rich linker sequence. However, these constructs did not produce any crystals. We consider that the increase in the local concentration was not sufficient for the crystallization of the Tom20–presequence complex. As the previous NMR study suggested (Abe et al, 2000), the peptide has substantial mobility in the bound state. The long linker allows the multiple conformations to exist, thus disturbing the crystallization. In contrast, the short linker effectively traps one of the possible conformations in a crystal lattice, i.e. a state conformationally locked by the crystal contacts (Supplementary Figure 1). A linker as short as three residues is only possible by using disulfide bond tethering, in the case of Tom20.
Note that the short linker itself has sufficient flexibility in solution, since the peptide-library experiment using the three-residue linker produced a reasonable consensus motif (Obita et al, 2003). This should be accompanied by the (partial) melting of the α-helical conformation of the linker region (Ser20′–Ala22′), to increase the conformational degrees of freedom in solution. No steric hindrance imposed by the short linker assures the use of the disulfide-tethered Tom20–presequence complex for the NMR relaxation studies. It is possible to use the 15N-labeled peptide without tethering, at a saturated concentration in the presence of Tom20, but the fast exchange involving the free state would obscure the effects of the exchange among the multiple bound states on the relaxation.
A multiple-mode interaction
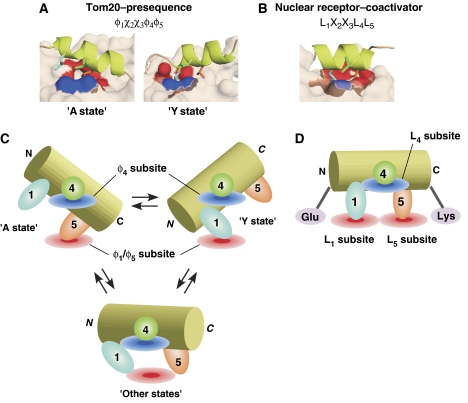
Two crystal structures with different linker designs were determined. It is striking that in each structure, one of the three leucine residues of the presequence has few contacts with Tom20 (Figure 3B). In fact, the mutagenesis study revealed that these three leucine residues are all necessary for binding to Tom20 (Abe et al, 2000). In contrast, Tom20 is equipped with only two hydrophobic subsites. The mutagenesis study showed that both the two hydrophobic subsites are important for the interactions of Tom20 with the presequence (Figure 4). Thus, neither of the crystal structures alone can explain the need for the three hydrophobic leucine side chains of the ALDH presequence. We consider both crystal structures to represent a snapshot of the solution states of the Tom20–presequence complex. The sequence, L1S2R3L4L5, in the ALDH presequence is recognized as a rigid α-helix by the two states of Tom20 (Figure 6A).
Figure 6.
Comparison of the recognition mode of Tom20 with that of the nuclear receptor. (A) Close-up view of the binding interface of Tom20 with the presequence peptide, and (B) that of the nuclear receptor with a coactivator peptide containing the LXXLL motif (estrogen receptor α complex (Shiau et al, 1998); PDB entry 3ERD). The three leucine side chains of the peptide ligands, at positions 1, 4, and 5, are colored cyan, green, and orange, respectively. The molecular surface contacting the leucine residue at position 4 is blue, and that contacting the leucine residue at position 1 or 5 is red. Schematic representation of the binding modes (C) of Tom20 and (D) of the nuclear receptor. The peptide adopts an α-helical conformation in both the bound states. The helices are drawn as cylinders to imply the firmness of the helical structure. The side chain at position 4 lies along the edge of the interaction surface (blue disks). In the case of Tom20, the two states exist in a rapid equilibrium and the side chain at position 4 functions as a fulcrum. In each state, one of the side chains at positions 1 and 5 is recognized by the single hydrophobic subsite of Tom20 (red disk). It is likely that the third or more bound states exist, and they are in equilibrium with the A and Y states. In the nuclear receptor, both the side chains at positions 1 and 5 are buried in the two hydrophobic subsites of the nuclear receptor (red disks). The charge clamp residues, Glu and Lys, position the helix and determine its length.
We propose that the presequence peptide has mobility within the binding groove of Tom20 without releasing the peptide. There are at least two different modes of interaction, represented by the A- and Y-linker structures (Figure 6C). L1 and L4 are recognized in the ‘Y-state' configuration in a given moment, and L4 and L5 are recognized in the ‘A-state' configuration in another moment. In other words, L4 is a molecular fulcrum held by two supporting residues (φ4 subsite: Val109 and Thr113) of Tom20. L1 or L5 of the presequence is accommodated in the same hydrophobic pocket (φ1/φ5 subsite: Ile74, Glu78, Leu106, and Leu110) in each state of Tom20 (Figure 3B). This is concertedly accomplished by the combination of the tilting of the presequence helix, the rotation of the side chains of L1 and L5, and the conformational change of Tom20 (Figure 3A).
To prove the hypothetical motion of the presequence peptide on the Tom20 molecule, we measured the 15N relaxation of the disulfide-tethered Tom20–presequence complex. The tethering of the presequence peptide to Tom20 suppresses the rapid exchange between the free and bound states of the peptide, by increasing the local concentration. The exchange-induced spin-relaxation rate, Rex, was estimated for each amide 15N spin (Figure 5A). The two independent methods provided consistent Rex values, which demonstrate the reliability of our Rex determination. The amide 15N spins with a large Rex value (>1 s−1, from the ηxy measurement) are mapped onto the two crystal structures (Figure 5B). The model-free Rex data (yellow bars) are not used in the following analysis, because the inclusion of the Rex parameter for a given residue is dependent on the model selection from the five possible sets of model-free parameters (Mandel et al, 1995). The amide nitrogen atoms of Ser16′, Leu19′, and Ser20′ of the presequence peptide, and those of Ile74 and Gln75 of Tom20 are located at the hydrophobic contact surface. This suggests molecular motion of the presequence peptide in the binding groove of Tom20. In this respect, our previous NMR structure (Abe et al, 2000) should be considered as the average of the structures corresponding to the two or more modes of interaction. The three other amide nitrogen atoms, located in Tom20 and the linker of the presequence peptide, probably correspond to the conformational change accompanying with the motion of the presequence peptide (Figure 3A).
We also estimated the Rex values using the [15N]Tom20 in the absence of the presequence peptide (Figure 5C). Unexpectedly, relatively large Rex values were obtained over the entire structure, indicating pervasive distribution of conformational fluctuations on sub-millisecond timescales (Figure 5D). This suggests that, in the free form, the core structure of Tom20 is regarded as a loosely packed four-helix bundle. Upon binding to a presequence, the motions converge to a small number of states that are necessary for the recognition of presequences. The motions seen in the free form are unrelated to the collective motions in the bound form, since the amide nitrogen spins with a large Rex value are totally different between the complex and the free forms (Figures 5B and D).
LXXLL-motif recognition by nuclear receptors
Nuclear receptors are transcription factors that are activated by the binding of small hydrophobic ligands, such as steroid hormones (Savkur and Burris, 2004; Plevin et al, 2005). The hormone-bound nuclear receptors undergo a conformational change and gain the ability to bind to a variety of proteins called coactivators. The LXXLL motifs (L is leucine and X is any amino acid, also designated as the NR box), which are commonly found in the coactivators, mediate the interaction between the coactivators and their cognate nuclear receptors. The requirement of the leucine side chains is strict: the replacement with a valine residue was not tolerated, for example, in the case of the RIP-140 coactivator (Heery et al, 1997). The structural basis of the strict specificity toward the three leucine residues was established by several crystal structures (Darimont et al, 1998; Nolte et al, 1998; Shiau et al, 1998). Figure 6B shows a close-up view of the interface of the estrogen receptor α–LXXLL peptide complex (Shiau et al, 1998). The LXXLL portion of the 13-residue peptide (KHKILHRLLQDSS) adopts an α-helical conformation, and the side chains of the three leucine residues are recognized simultaneously by three hydrophobic subsites. The position and the length (two turns) of the short α-helix are specified by two helix-capping interactions (Savkur and Burris, 2004). The ‘charge clamp' comprises two highly conserved, charged residues of the nuclear receptors: the γ-carboxylate group of glutamate and the ɛ-amino group of lysine form hydrogen bonds with the N-terminal backbone amide group and the C-terminal backbone carbonyl group of the LXXLL α-helix, respectively. The complementary interface and the helix-capping interaction facilitate the strict recognition of the LXXLL motifs by a conventional key-and-lock mechanism.
The paxillin LD motif, LDXLLXXL (where L is leucine, D is aspartate, and X is any amino acid), is another example of a short motif containing leucine residues (Hurley, 2003). It should be emphasized that the structural basis of the interaction of the LD motif and the focal adhesion targeting domain shares many common features with that of the LXXLL motif and the nuclear receptors (Hoellerer et al, 2003).
A dynamic equilibrium among multiple bound states
A structural comparison with the nuclear receptor highlights the dynamic features of the presequence recognition by Tom20 (Figure 6). Even though the peptide sequences are very similar to each other (LSRLL for Tom20 versus LHRLL for estrogen receptor α), the recognition modes of the two systems differ considerably. Our previous peptide-library experiment revealed that Tom20 accepted Leu, Phe, Trp, and Ile at position 1, Leu, Ile, Trp, Phe, and Met at position 4, and Leu, Phe, Val, Trp, and Ile at position 5 of the ALDH presequence (Obita et al, 2003). In fact, Tom20 must recognize more diverse five-residue sequences in other protein presequences. The experimentally deduced examples include WKRCM of the yeast F1β (F1-ATPase β-subunit) presequence and LRRAY of the yeast mitochondrial heat shock protein 60 presequence (Muto et al, 2001). To achieve such broadly selective specificity, a static mode of recognition, like that of nuclear receptors, seems to be inadequate even if an induced fit mechanism is considered, because Tom20 needs to adapt simultaneously to the two side chains at positions 1 and 5, in addition to the third side chain at position 4. In contrast, dynamic recognition is an attractive mechanism. One state just recognizes two of the three hydrophobic residues, which increases the chance of adapting to various hydrophobic side chains by an induced fit mechanism. The change in the relative orientation of the two helix–turn–helix units (Figure 3A) is likely to be the induced fit mechanism of Tom20.
The dynamic equilibrium between the two states revealed by the two crystal structures are apparently enough to explain the five-residue consensus motif of Tom20. The fact that a preliminary S-linker structure (complex containing the pALDH presequence peptide with the Ser-linker) is almost identical to that of the A-linker structure supports this notion. However, it is quite likely that the real motion of the presequence peptide in the binding groove is far more complicated dynamic equilibrium among multiple bound states than simple exchange between the two states. A series of crystal structures with different linker designs are desired to prove or reject the existence of the third or more bound states. Finally, we suggest that the multiple-mode dynamic recognition of presequences by Tom20 could be necessary for the transfer of presequences to the next receptor, Tom22, as it is likely that Tom20 and Tom22 recognize opposite sides of the same presequence; Tom20 recognizing the hydrophobic surface and Tom22 the hydrophilic surface (Pfanner, 2000).
Not all presequences contain a consensus motif that exactly matches the pattern of φχχφφ; e.g. the yeast cytochrome c oxidase subunit IV) presequence and the rat ornithine transcarbamylase presequence do not (Muto et al, 2001). Thus, Tom20 may recognize similar, but different, patterns from the φχχφφ motif. The pervasive conformational fluctuations seen in the free form of Tom20 (Figure 5D), in combination with the multiple-mode recognition, might explain the outstanding specificity of Tom20.
Concluding remarks
We designed the Tom20–presequence complex for crystallography with reference to the NMR structure. This is a good example of NMR-assisted crystallography. Unexpectedly, two different structures were obtained, and neither could fully account for the hydrophobic preference at the three hydrophobic positions of the presequence. NMR relaxation analyses suggested motion on a sub-millisecond timescale at the Tom20–presequence interface. In view of the high-resolution crystal structures supplemented with the NMR dynamics data in solution, we propose a multiple-mode recognition model: two or more modes of interaction exist in dynamic equilibrium, and each mode recognizes a different feature of the short, but diverse, consensus motif. In this context, it is interesting to ask whether the plant Tom20, which convergently evolved from a distinct ancestral gene, shares a similar dynamic mechanism for the recognition of plant mitochondrial presequences (Perry et al, 2006).
Materials and methods
Sample preparation
The core structure of the cytosolic domain of Tom20, encompassing Asp59–Leu126 from Rattus norvegicus (accession no. Q62760), was produced and purified, as described (Igura et al, 2005). The 15N-labeled protein was prepared as described (Abe et al, 2000). The peptide sequence is Gly12–Pro13–Arg14–Leu15–Ser16–Arg17–Leu18–Leu19–Ser20–X21–Ala22–Gly23–Cys24. The numbering starts at 12, because the sequence in bold is derived from the C-terminal half of the presequence of mitochondrial ALDH from R. norvegicus (pALDH, Accession no. P11884) (Farres et al, 1988). The linker region contains an alanine residue (A-linker peptide) or a tyrosine residue (Y-linker peptide) at the position of X. The peptides used for crystallization were custom-made with an amidated C terminus by Operon Biotechnologies (Tokyo, Japan). The 15N-labeled peptides were produced as a GST–SUMO–fusion protein in E. coli BL21(DE3)pLysS cells. The transformed E. coli cells were cultured in M9 minimum medium containing 15NH4Cl. After purification by glutathione Sepharose 4B resin (Amersham), each fusion protein was digested with SENP2 protease (100:1, w/w). The resulting mixture was fractionated on a reverse-phase HPLC column (Cosmosil 5C18-AR-II) to purify the peptide. The disulfide-bond-tethered complex with a presequence peptide was prepared by air oxidation, as described (Igura et al, 2005).
Gel filtration analysis
The gel filtration experiment was performed on a TSK-GEL super SW2000 column (Tosoh, Tokyo, Japan), preequilibrated with 0.1 M sodium phosphate buffer, pH 7.0, containing 0.5 M NaCl, at a flow rate of 0.5 ml min−1. The following protein standards were used: bovine serum albumin (67 kDa), ovalbumin (43 kDa), and lysozyme (14 kDa). The injected protein concentration was 0.33 mg ml−1 and the eluted protein concentration was estimated to be about 0.05 mg ml−1.
Data collection and structure determination
The disulfide-bond-tethered complexes were crystallized as described (Igura et al, 2005). Native and SeMet (selenomethionyl) derivative data sets were collected at 100 K at beamlines BL40B2 and BL41XU, SPring-8, Harima, Japan, as described (Igura et al, 2005). All diffraction data sets were processed and scaled with the DENZO/Scalepack package (Otwinowski and Minor, 1997). A starting model was obtained from the data set of the SeMet Y-linker complex; the initial phase calculations were performed by the MAD method using SOLVE (Terwilliger and Berendzen, 1999), and, subsequently, a solvent-flattening procedure was carried out using RESOLVE (Terwilliger, 2000). The model building was carried out manually with O (Jones et al, 1991) with reference to the NMR structure (PDB entry, 1OM2). The subsequent molecular dynamics refinement was performed using CNS (Brünger et al, 1998) and Refmac (CCP4, 1994) with translation libration screw parameters, up to 2.05-Å resolution, with the native data set of the Y-linker complex. The final model of the Y-linker structure comprises six complex molecules and 355 water molecules. In the later stages of the refinement, parts of the seventh complex molecule (chains G and N) could be built, but their electron densities were not clear enough to build all of the residues of both the protein chain and peptide chain. The final model of the Y-linker structure, comprising seven complex molecules and 378 water molecules, was refined to an R factor of 24.8% and an Rfree factor of 30.8%. This model was used to solve the structure of the A-linker complex by the molecular replacement procedure, using MOLREP in the CCP4 package (CCP4, 1994). The final structure of the A-linker complex, comprising two complex molecules and 168 water molecules, was refined to an R factor of 17.8% and an Rfree factor of 24.0% with the translation libration screw parameters. Statistics for data collection and refinement is summarized in Table I.
Structural analysis
The stereochemical quality analysis and the secondary structure assignment were carried out with PROCHECK (Laskowski et al, 1993). The Ramachandran plot of the backbone angles of the non-glycine and non-proline residues of the A-linker structure gave 95.6% in the most favored region and 4.4% in the additionally allowed region. The Ramachandran plot of the Y-linker structure gave 92.6% in the most favored region, 5.9% in the additionally allowed region, 1.2% in the generously allowed region, and 0.2% in the disallowed region. The distribution of these five residues in the generously allowed region and one residue in the disallowed region was not directly correlated with the disulfide bond formation, intertwined dimer formation, or Tom20–peptide interaction. The ASA was calculated with NACESS (http://wolf.bi.umist.ac.uk/naccess/) with a probe radius of 1.4 Å. The contribution of the linker region was omitted in the calculations of ASA. Domain rearrangement and hinge-bending motion were analyzed with DynDom (Hayward and Lee, 2002). The average structures of the A-linker structures and that of the Y-linker structures were used as input with the parameters of window length=5 (default), minimum domain size=15, and ratio of inter- to intradomain displacement=1.0 (default). The figures were prepared with PyMOL (DeLano Scientific; http://pymol.sourceforge.net/).
NMR spectroscopy
NMR spectra were recorded at 298 K on Avance600 spectrometer equipped with a TXI cryoprobe. The NMR samples contained 0.4 mM (spectral assignment and relaxation study) or 0.1 mM (titration study) 15N- and/or 13C-labeled proteins in 90% 1H2O/10% 2H2O, containing 20 mM MOPS-NaOH, pH 7.0, and 50 mM NaCl. The spectral assignments of the wild-type and mutated Tom20s, and the disulfide-tethered complexes were made as described (Abe et al, 2000).
Single amino-acid-substituted Tom20 proteins were 15N labeled, and titrated with the pALDH peptide, Ac-GPRLSRLLSYA-NH2, to monitor the binding, at the molar ratio of 0:1, 0.5:1, 1:1, 1.5:1, 2:1, 2.5:1, and 3:1. DTT was added to the sample solutions at the final concentration of 5 mM to prevent the formation of Tom20 dimers during the titration. The titration curves were analyzed with the program xcrvfit ver 4.0.9, developed by R Boyko and BD Sykes (University of Alberta; http://www.bionmr.ualberta.ca/bds/software/xcrvfit).
Spectra for 15N longitudinal relaxation rates, R1, 15N transverse relaxation rates, R2, and {1H} 15N steady-state heteronuclear NOE values were acquired at 298 K using [15N]Tom20–SS-presequence(A-linker), Tom20–SS-[15N]presequence(A-linker), and [15N]Tom20C100S samples at 600 MHz 1H frequency. The cysteine residue was replaced by serine for the prevention of dimer formation during the prolonged period of experimental time. The pulse sequences used were the gradient sensitivity enhancement version (Farrow et al, 1994). R1 was determined from the data with relaxation delays of 30, 80, 150, 300, 500, and 800 ms, and R2 from the data with relaxation delays of 57.6, 86.4, 115.2, 144.0, 172.8, and 201.6 ms. In the heteronuclear NOE experiment, 1H saturation for 3.0 s during the relaxation delay was applied for NOE enhancement. The model-free analysis was performed (Lipari and Szabo, 1982) with the programs MODELFREE-4.10 (Mandel et al, 1995) and TENSOR2 (Dosset et al, 2000). The model-free parameters for each amide 15N spin were selected, as described in the MODELFREE manual. We used the isotropic diffusion model for the model-free analysis, but the axially symmetric diffusion model and the fully anisotropic diffusion model, combined with the two crystal coordinates, provided essentially the same results. The overall shapes of the Tom20–SS-presequence complexes and the free form of Tom20 are both regarded as a prolate ellipsoid with the ratio D∣∣/D⊥=1.2, and thus a sphere shape is a good approximation. The transverse CSA/dipolar cross-relaxation rate (ηxy) for 1H–15N was measured as described (Tjandra et al, 1996; Kroenke et al, 1998). The Rex values were estimated from a simple approximate equation Rex=R2−aηxy (a=1.3 at 600 MHz 1H frequency) (Fushman et al, 1998). The relaxation dispersion curves were obtained using the relaxation-compensated Carr–Purcell–Meiboom–Gill experiment, as described (Millet et al, 2000). The R2 value was determined from the data with τcp of 1.0, 2.0, 4.0, 6.6, 10.8, 21.6, and 64.8 ms.
We estimate the rate constant of the motion of the peptide in the binding groove. We assume exchange between two states for calculation. The relaxation dispersion experiment suggested that the motion occurs in the fast-exchange regime (i.e. kex≫Δω, Supplementary Figure 7), and so the following expression approximates the effect of two-site chemical exchange: Rex=pApB(Δω)2/kex, where pA and pB are the populations of the two bound states (pA+pB=1), Δω is the difference in chemical shift of the 15N spins in the two bound states, and kex is the chemical exchange rate constant (kex=k1+k−1, k1 is the forward exchange rate and k−1 is the reverse exchange rate). It is assumed that the two states are equally represented in solution, i.e. pA=pB=0.5. The Rex values are found in the range of 1–4 s−1 (Figure 5A). Assuming a reasonable value of 1–4 ppm (60–240 Hz at 60 MHz 15N frequency) for Δω, we estimate the rate constant of the motion to be on the order of 10 000s−1.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7
Acknowledgments
We thank the staff of beamlines BL40B2 and BL41XU at the SPring-8 (Harima, Japan). The experiments at the SPring-8 were approved by the Japan Synchrotron Radiation Research Institute, as proposals nos. 2004A0690 and 2004B0790. KM and DK were supported by Grants-in-Aid for Scientific Research in Priority Areas and the National Project on Target Protein Analyses from the Ministry of Education, Culture, Sports, Science and Technology of Japan. T Obita and T Ose were supported by research fellowships from the Japan Society for the Promotion of Science. Atomic coordinates and structure factors for the Tom20–presequence complexes have been deposited in the Protein Data Bank (accession codes 2V1S and 2V1T).
References
- Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nishikawa S, Endo T, Kohda D (2000) Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell 100: 551–560 [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Darimont BD, Wagner RL, Apriletti JW, Stallcup MR, Kushner PJ, Baxter JD, Fletterick RJ, Yamamoto KR (1998) Structure and specificity of nuclear receptor–coactivator interactions. Genes Dev 12: 3343–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosset P, Hus JC, Blackledge M, Marion D (2000) Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J Biomol NMR 16: 23–28 [DOI] [PubMed] [Google Scholar]
- Endo T, Kohda D (2002) Functions of outer membrane receptors in mitochondrial protein import. Biochim Biophys Acta 1592: 3–14 [DOI] [PubMed] [Google Scholar]
- Endo T, Yamamoto H, Esaki M (2003) Functional cooperation and separation of translocators in protein import into mitochondria, the double-membrane bounded organelles. J Cell Sci 116: 3259–3267 [DOI] [PubMed] [Google Scholar]
- Farres J, Guan KL, Weiner H (1988) Sequence of the signal peptide for rat liver mitochondrial aldehyde dehydrogenase. Biochem Biophys Res Commun 150: 1083–1087 [DOI] [PubMed] [Google Scholar]
- Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G (1994) Backbone dynamics of a free and a phosphopeptide-complexed src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33: 5984–6003 [DOI] [PubMed] [Google Scholar]
- Fremont DH, Hendrickson WA, Marrack P, Kappler J (1996) Structures of an MHC class II molecule with covalently bound single peptides. Science 272: 1001–1004 [DOI] [PubMed] [Google Scholar]
- Freund C, Kuhne R, Park S, Thiemke K, Reinherz EL, Wagner G (2003) Structural investigations of a GYF domain covalently linked to a proline-rich peptide. J Biomol NMR 27: 143–149 [DOI] [PubMed] [Google Scholar]
- Fushman D, Tjandra N, Cowburn D (1998) Direct measurement of 15N chemical shift anisotropy in solution. J Am Chem Soc 120: 10947–10952 [Google Scholar]
- Hayward S, Lee RA (2002) Improvements in the analysis of domain motions in proteins from conformational change: DynDom version 1.50. J Mol Graph Model 21: 181–183 [DOI] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG (1997) A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387: 733–736 [DOI] [PubMed] [Google Scholar]
- Hennecke J, Carfi A, Wiley DC (2000) Structure of a covalently stabilized complex of a human alpha beta T-cell receptor, influenza HA peptide and MHC class II molecule, HLA-DR1. EMBO J 19: 5611–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoellerer MK, Noble ME, Labesse G, Campbell ID, Werner JM, Arold ST (2003) Molecular recognition of paxillin LD motifs by the focal adhesion targeting domain. Structure 11: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Hurley JH (2003) Leucine in the sky with diamonds. Structure 11: 1192–1193 [DOI] [PubMed] [Google Scholar]
- Igura M, Ose T, Obita T, Sato C, Maenaka K, Endo T, Kohda D (2005) Crystallization and preliminary X-ray analysis of mitochondrial presequence receptor Tom20 in complex with a presequence from aldehyde dehydrogenase. Acta Crystallogr F 61: 514–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kroenke CD, Loria JP, Lee LK, Rance M, Palmer AG III (1998) Longitudinal and transverse 1H–15N dipolar/15N chemical shift anisotropy relaxation interference: unambiguous determination of rotational diffusion tensors and chemical exchange effects in biological macromolecules. J Am Chem Soc 120: 7905–7915 [Google Scholar]
- Laskowski R, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Likic VA, Perry A, Hulett J, Derby M, Traven A, Waller RF, Keeling PJ, Koehler CM, Curran SP, Gooley PR, Lithgow T (2005) Patterns that define the four domains conserved in known and novel isoforms of the protein import receptor Tom20. J Mol Biol 347: 81–93 [DOI] [PubMed] [Google Scholar]
- Lipari G, Szabo A (1982) Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. J Am Chem Soc 104: 4546–4570 [Google Scholar]
- Lithgow T, Junne T, Wachter C, Schatz G (1994) Yeast mitochondria lacking the two import receptors Mas20p and Mas70p can efficiently and specifically import precursor proteins. J Biol Chem 269: 15325–15330 [PubMed] [Google Scholar]
- Mandel AM, Akke M, Palmer AG III (1995) Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J Mol Biol 246: 144–163 [DOI] [PubMed] [Google Scholar]
- Millet O, Loria JP, Kroenke CD, Pons M, Palmer AG III (2000) The static magnetic field dependence of chemical exchange line broadening defines the NMR chemical shift time scale. J Am Chem Soc 122: 2867–2877 [Google Scholar]
- Mukhopadhyay A, Yang C, Weiner H (2006) Binding of mitochondrial leader sequences to Tom20 assessed using a bacterial two-hybrid system shows that hydrophobic interactions are essential and that some mutated leaders that do not bind Tom20 can still be imported. Protein Sci 15: 2739–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto T, Obita T, Abe Y, Shodai T, Endo T, Kohda D (2001) NMR identification of the Tom20 binding segment in mitochondrial presequences. J Mol Biol 306: 137–143 [DOI] [PubMed] [Google Scholar]
- Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, Rosenfeld MG, Willson TM, Glass CK, Milburn MV (1998) Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature 395: 137–143 [DOI] [PubMed] [Google Scholar]
- Obita T, Muto T, Endo T, Kohda D (2003) Peptide library approach with a disulfide tether to refine the Tom20 recognition motif in mitochondrial presequences. J Mol Biol 328: 495–504 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pak YK, Weiner H (1990) Import of chemically synthesized signal peptides into rat liver mitochondria. J Biol Chem 265: 14298–14307 [PubMed] [Google Scholar]
- Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, Venkitaraman AR (2002) Insights into DNA recombination from the structure of a RAD51–BRCA2 complex. Nature 420: 287–293 [DOI] [PubMed] [Google Scholar]
- Perry AJ, Hulett JM, Likic VA, Lithgow T, Gooley PR (2006) Convergent evolution of receptors for protein import into mitochondria. Curr Biol 16: 221–229 [DOI] [PubMed] [Google Scholar]
- Pfanner N (2000) Protein sorting: recognizing mitochondrial presequences. Curr Biol 10: R412–R415 [DOI] [PubMed] [Google Scholar]
- Pfanner N, Geissler A (2001) Versatility of the mitochondrial protein import machinery. Nat Rev Mol Cell Biol 2: 339–349 [DOI] [PubMed] [Google Scholar]
- Plevin MJ, Mills MM, Ikura M (2005) The LxxLL motif: a multifunctional binding sequence in transcriptional regulation. Trends Biochem Sci 30: 66–69 [DOI] [PubMed] [Google Scholar]
- Ramage L, Junne T, Hahne K, Lithgow T, Schatz G (1993) Functional cooperation of mitochondrial protein import receptors in yeast. EMBO J 12: 4115–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D (2003) Finding the right organelle targeting signals in mitochondrial outer-membrane proteins. EMBO Rep 4: 948–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roise D, Theiler F, Horvath SJ, Tomich JM, Richards JH, Allison DS, Schatz G (1988) Amphiphilicity is essential for mitochondrial presequence function. EMBO J 7: 649–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau F, Schymkowitz JWH, Itzhaki LS (2003) The unfolding story of three-dimensional domain swapping. Structure 11: 243–251 [DOI] [PubMed] [Google Scholar]
- Savkur RS, Burris TP (2004) The coactivator LXXLL nuclear receptor recognition motif. J Pept Res 63: 207–212 [DOI] [PubMed] [Google Scholar]
- Schatz G, Dobberstein B (1996) Common principles of protein translocation across membranes. Science 271: 1519–1526 [DOI] [PubMed] [Google Scholar]
- Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL (1998) The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95: 927–937 [DOI] [PubMed] [Google Scholar]
- Sollner T, Griffiths G, Pfaller R, Pfanner N, Neupert W (1989) MOM19, an import receptor for mitochondrial precursor proteins. Cell 59: 1061–1070 [DOI] [PubMed] [Google Scholar]
- Terwilliger TC (2000) Maximum-likelihood density modification. Acta Crystallogr D 56: 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J (1999) Automated MAD and MIR structure solution. Acta Crystallogr D 55: 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjandra N, Szabo A, Bax A (1996) Protein backbone dynamics and 15N chemical shift anisotropy from quantitative measurement of relaxation interference effects. J Am Chem Soc 118: 6986–6991 [Google Scholar]
- von Heijne G (1986) Mitochondrial targeting sequences may form amphiphilic helices. EMBO J 5: 1335–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7