Abstract
Trf4 is the poly(A) polymerase component of TRAMP4, which stimulates nuclear RNA degradation by the exosome. We report that in Saccharomyces cerevisiae strains lacking Trf4, cryptic transcripts are detected from regions of repressed chromatin at telomeres and the rDNA intergenic spacer region (IGS1-R), and at CEN3. Degradation of the IGS1-R transcript was reduced in strains lacking TRAMP components, the core exosome protein Mtr3 or the nuclear-specific exosome component Rrp6. IGS1-R has potential binding sites for the RNA-binding proteins Nrd1/Nab3, and was stabilized by mutation of Nrd1. IGS1-R passes through the replication fork barrier, a region required for rDNA copy number control. Strains lacking Trf4 showed sporadic changes in rDNA copy number, whereas loss of both Trf4 and either the histone deacetylase Sir2 or the topoisomerase Top1 caused dramatic loss of rDNA repeats. Chromatin immunoprecipitation analyses showed that Trf4 is co-transcriptionally recruited to IGS1-R, consistent with a direct role in rDNA stability. Co-transcriptional RNA binding by Trf4 may link RNA and DNA metabolism and direct immediate IGS1-R degradation by the exosome following transcription termination.
Keywords: exosome, ncRNA, nucleolus, recombination, TRAMP
Introduction
The eukaryotic exosome is a complex of 10 core subunits, including a 3′–5′ exonuclease and RNA-binding proteins, that is involved in many aspects of RNA processing and surveillance. The purified exosome alone does not display strong RNA degradation activity (Mitchell et al, 1997) and is targeted to specific substrates by different cofactors (reviewed in Houseley et al, 2006). Two related complexes, TRAMP4 and TRAMP5, target aberrant tRNA (Kadaba et al, 2004; LaCava et al, 2005; Vanacova et al, 2005) and aberrant ribosomal RNA for exosome degradation (Dez et al, 2006; Houseley and Tollervey, 2006). These complexes also target a class of cryptic unstable transcripts (CUTs), resulting in their immediate degradation following transcription (Wyers et al, 2005; Arigo et al, 2006b; Thiebaut et al, 2006; Vasiljeva and Buratowski, 2006). An apparently similar class of RNA has been identified in mouse and human cells; like the CUTs, these superficially resemble mRNAs in being RNA polymerase II (Pol II) transcripts that carry 5′ caps and 3′ poly(A) tails, but lack evident protein coding capacity (reviewed in Willingham and Gingeras, 2006). Despite their apparent conservation, little is known about the function of these transcripts.
In many Eukaryotes, transcripts influence chromatin structure via the siRNA/RNAi system. In fission yeast, establishment of centromeric heterochromatin involves the poly(A) polymerases Cid12 (Motamedi et al, 2004) and Cid14 (Buhler et al, 2007), which are in the same family as budding yeast Trf4 and Trf5, and Cid14 resides in a TRAMP-like complex. Saccharomyces cerevisiae lacks homologs of key components of the machinery that processes siRNAs, but it seemed possible that other mechanisms might transfer information from the transcriptome back to the chromatin structure of the genome.
The TRAMP complexes are composed of a poly(A) polymerase, Trf4 or Trf5, a putative RNA-binding protein, Air1 or Air2, and the putative DEVH-box helicase Mtr4. Trf4 and Trf5 were originally isolated in a synthetic lethal screen with top1 mutations. Conditional double mutants of trf4 with top1-7 showed ribosomal DNA (rDNA) condensation phenotypes (Sadoff et al, 1995; Castano et al, 1996), and were reported to display defects in mitotic segregation (Wang et al, 2000; Edwards et al, 2003). In addition, strains lacking both Trf4 and Trf5 (which are synthetic lethal) fail to complete DNA replication (Wang et al, 2000). These reports suggested that Trf4 and Trf5 play some role in DNA metabolism, particularly in the rDNA, although the nature of this role remained unclear.
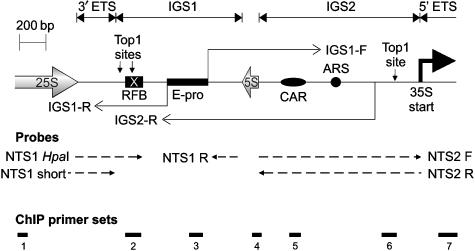
Wild-type yeast contain a tandem array of approximately 200 rDNA repeats on chromosome XII, the number of which is maintained by regulated recombination (Szostak and Wu, 1980). Each repeat contains the 35S and 5S ribosomal RNA genes separated by intergenic spacer regions IGS1 and IGS2 (see Figure 1). The IGS1 region of the rDNA repeat contains a replication fork barrier (RFB) (Voelkel-Meiman et al, 1987), where a protein complex including Fob1 imposes a unidirectional block to the progress of DNA replication forks. This region also contains a recombination hotspot that is necessary for double-strand break formation and subsequent recombination between sister chromatids to occur (Kobayashi and Horiuchi, 1996).
Figure 1.
Schematic of intergenic spacer region. Schematic representation of the rDNA intergenic spacer regions showing ncRNA transcripts. IGS, intergenic spacer; ARS, autonomously replicating sequence (origin of DNA replication); ETS, external transcribed sequence (the 35S has a 5′ ETS and a 3′ ETS); RFB, replication fork barrier. Dotted lines with arrows show strand-specific RNA probes; arrows point 5′–3′. Thick lines 1–7 indicate products of qPCR reactions used for ChIP analysis.
The repeat tracts in two sister chromatids are normally held together by the cohesin complex, which is concentrated over the cohesin-associated region (CAR) in IGS2 (see Figure 1) (Laloraya et al, 2000). This ensures that even if recombination is initiated, it occurs only with the aligned repeat in the sister chromatid, and does not result in a change of repeat number. Two divergent Pol II transcripts generated from the E-pro promoter within IGS1 (see Figure 1) (Santangelo et al, 1988) have been suggested to regulate recombination, as replacement of E-pro with a regulated, divergent GAL1-10 promoter resulted in cohesin displacement and extensive unequal recombination (Kobayashi and Ganley, 2005). Unequal recombination leads to a net change in repeat number for the chromatid that initiated recombination. However, these analyses did not address potentially distinct roles for the two Pol II transcripts, which we designate here as IGS1-F and IGS1-R. IGS1-F is transcribed through the CAR, consistent with its reported role in cohesin displacement, whereas IGS1-R is transcribed through the RFB.
The rDNA IGS regions, along with the telomeres and inactive mating loci, are silenced for Pol II transcription by the histone deactylase Sir2 (Smith and Boeke, 1997). Loss of Sir2 leads to elevated expression of the IGS1 and IGS2 transcripts (Li et al, 2006) and hyper-recombination within the rDNA array (Gottlieb and Esposito, 1989). This was proposed to be a consequence of cohesin displacement (Kobayashi et al, 2004).
Here we demonstrate that one of the non-coding RNA (ncRNA) transcripts generated from the intergenic spacer regions, IGS1-R, is targeted for degradation mediated by Trf4 and the exosome. Trf4 is recruited to the rDNA spacer region that includes the RFB, via the IGS1-R transcript, and is required for stability of the rDNA repeat copy number. These data provide evidence for novel links between RNA and DNA metabolism in budding yeast.
Results
Cryptic transcripts from regions of repressed chromatin accumulate in strains with defects in the TRAMP complex or exosome
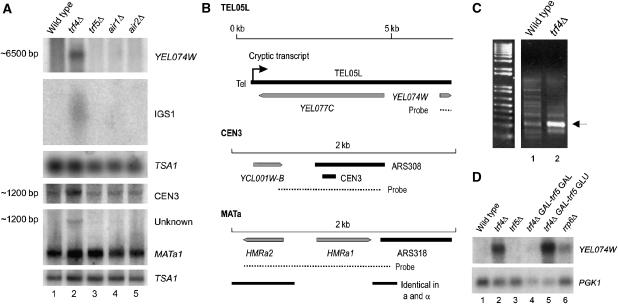
We speculated that transcripts may be generated from regions of repressed chromatin in the yeast genome but degraded by the TRAMP and exosome complexes. We therefore tested a telomeric region (TEL05L), the intergenic spacer (IGS) region of the rDNA repeat, a centromeric region (CEN3) and the silenced mating-type cassettes (MATa/α). In each case, we saw increased levels of a transcript in strains lacking Trf4, but not in single mutant strains lacking Trf5, Air1 or Air2 (Figure 2A, compare lane 2 with lanes 3–5).
Figure 2.
Cryptic transcripts can be detected from regions of repressed chromatin in strains lacking TRAMP activity. (A) Northern analysis of TRAMP mutants grown in YPD at 25°C. RNA is resolved on a 1.2% agarose glyoxal gel (three upper panels) or a 6% acrylamide 7 M urea gel (three lower panels). Probes are (top to bottom) YEL074W, NTS1 HpaI, TSA1, CEN3, MAT and TSA1. (B) Schematic representations of probed regions. (C) 5′ RACE PCR was performed on wild-type and trf4Δ RNA, with a primer located 138 bp from the 3′ end of the annotated YEL077C ORF. DNA products were stained with SYBR Safe. The indicated band was excised, cloned and sequenced. (D) Northern analysis of TRAMP and exosome mutants probed with YEL074W and PGK1. Strains were grown at 25°C on YPD except trf4Δ GAL-trf5, which was grown on YPGal and shifted to YPD for 18 h prior to RNA extraction.
At many yeast telomeres, the terminal repeats are flanked by the ‘Y′ region', which is conserved in whole or in part at 16 yeast telomeres. At some telomeres, this region encodes a putative DNA helicase (Yamada et al, 1998), designated Yel077c in the case of TEL05L (Figure 2B). Strand-specific probes demonstrated that the transcript elevated in the trf4Δ strain is an ncRNA expressed antisense to YEL077C. The TEL05L ncRNA is ∼6.5 kb in length and was detected by a probe located within YEL074W (Figures, 2A and B) and by a second probe located 2.8 kb further toward the chromosome end (data not shown), showing it to extend across the Y′ region. 5′ RACE generated a product that was enriched in trf4Δ (arrow in Figure 2C), which was cloned and sequenced. Three 5′ ends were identified, located 106–128 bp beyond the 3′ end of the open reading frame (ORF) of YEL077C. The ncRNA therefore starts close to the chromosome end and runs antisense through the entire putative helicase ORF. Polymorphisms in sequenced products demonstrate that the ncRNA is transcribed from at least two telomeres (data not shown).
The TEL05L ncRNA was also elevated in a strain lacking Rrp6 (Figure 2D, lane 6), indicating that it is a target for exosome degradation. Depletion of Trf5 by growth of a trf4Δ GAL-trf5 strain on glucose medium (Figure 2D, lane 5) increased accumulation of the TEL05L transcript relative to the trf4Δ single mutant. Overexpression of Trf5, in GAL-trf5 strains grown in galactose medium, can suppress the phenotype of trf4Δ strains on some TRAMP substrates (Houseley and Tollervey, 2006), and this was the case for the TEL05L transcript (Figure 2D, lane 4). We conclude that TRAMP4 and TRAMP5 both participate in degradation, with TRAMP4 probably playing the major role, and function with the exosome to degrade large ncRNAs generated from telomeric regions.
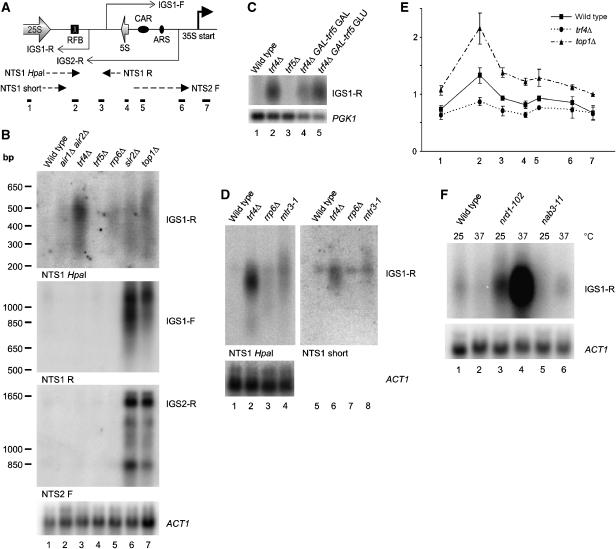
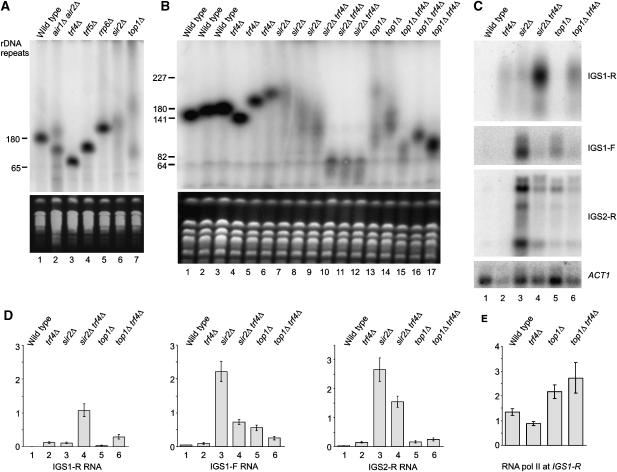
We have not further characterized the ∼1.2 kb CEN3 transcript, and the structure of the MAT loci prevents unambiguous assignment of the observed ∼1.2 kb transcript to a silent cassette. However, connections between Trf4 and Top1 mutations and rDNA structure (see Introduction) suggested a functional link to IGS transcription, which was therefore further investigated. The ncRNA products of transcription from E-pro, IGS1-F and IGS1-R (Figure 1) are normally present at very low levels (Santangelo et al, 1988) and we speculated that this might reflect rapid degradation involving the TRAMP and exosome complexes. To visualize these ncRNAs, northern blots of RNA from TRAMP and exosome mutants were probed with strand-specific probes to both rDNA intergenic spacer regions (Figure 3B). An sir2Δ strain, which overexpresses IGS transcripts, was used as a positive control. A top1Δ strain was also analyzed, as top1 trf4 double mutants were reported to show synthetic lethality and rDNA condensation phenotypes (Sadoff et al, 1995; Castano et al, 1996). Strains carrying sir2Δ and top1Δ showed elevated levels of three ncRNAs IGS1-R, IGS1-F and IGS2-R (Figure 3B, lanes 6 and 7).
Figure 3.
IGS1-R is degraded by TRAMP and the exosome. (A) Locations of hybridization probes and ChIP primer sets used. (B) Northern analysis of TRAMP and exosome mutants and control strains grown in YPD at 25°C. Probes are (top to bottom) NTS1 HpaI, NTS2 R, NTS2 F and ACT1. (C) Accumulation of IGS1-R in trf4Δ GAL-trf5 strain. Strains were grown at 25°C on YPD (lanes 1–3) or YPGal (lane 4) and/or pre-grown in YPGal and shifted to YPD for 18 h. Northern blot of poly(A)+ selected RNA was probed with random primed NTS1 and PGK1. (D) Northern analysis of TRAMP and exosome mutant strains grown in YPD at 25°C, probed with NTS1 HpaI, NTS1 short and ACT1. (E) ChIP analysis of RNA polymerase II density in intergenic spacer regions, with primer sets shown in Figure 1. Error bars=±1 standard error, n=3. (F) Northern analysis of nrd1 and nab3 mutant strains grown in YPD at 23°C and shifted to 37°C for 1 h, probed with NTS1 HpaI and ACT1.
The levels of IGS1-F and IGS2-R were unaltered in TRAMP or exosome mutants, whereas the level of IGS1-R was substantially increased in trf4Δ and to a lesser extent in air1Δ air2Δ and rrp6Δ strains. No stabilization of IGS1-R was seen in trf5Δ strains, and overexpression of Trf5 under GAL regulation did not suppress the trf4Δ phenotype (Figure 3C, lanes 3 and 4). This suggests that Trf5 does not efficiently target IGS1-R, even in the absence of Trf4. IGS1-R stabilization in the rrp6Δ exosome mutant strain was weaker than that in trf4Δ (Figures 3B (lane 5) and D (lane 3)) and we therefore also examined the core exosome mutant mtr3-1 (Figure 3D). Even at permissive temperature (25°C), accumulation of IGS1-R was visible in the mtr3-1 strain. We conclude that TRAMP4 functions with both Rrp6 and the core exosome to degrade IGS1-R, whereas degradation of IGS1-F and IGS2-R presumably involves other activities.
Two major forms of IGS2-R of approximately 1.6 kb and 850 nt in length were detected (see also Li et al, 2006). The longer IGS2-R species also hybridized to a downstream probe to IGS1, whereas the short species was not detected with this probe, indicating that it is 3′ truncated (data not shown). This would be consistent with termination of some IGS2-R transcripts around the location of the CAR, even in top1Δ and sir2Δ strains. A shorter probe directed against the 3′ region of IGS1-R (NTS1 short; see Figure 3A) was also used. This region is extremely AT rich and the probe hybridized poorly; however, only the longer transcripts were detected by NTS1 short probe (Figure 3D, compare lanes 1–4 with 5–8). This indicates that the IGS1-R transcripts show 3′ heterogeneity.
To assess whether increased IGS1-R RNA reflects increased transcription or post-transcriptional stabilization, chromatin immunoprecipitation (ChIP) was performed to determine RNA Pol II occupancy in wild-type, trf4Δ and top1Δ cells (Figure 3E) and sir2Δ (data not shown). This revealed a clear peak of Pol II within the IGS1-R region in the wild-type strain. This peak was elevated in the top1Δ strain, consistent with constitutive derepression (Bryk et al, 1997). In contrast, the Pol II signal in the trf4Δ strain was lower than that in the wild-type strain, despite elevated levels of the transcript.
We next wanted to determine how the TRAMP complex is recruited to the IGS1-R transcripts. The Nrd1–Nab3 heterodimer of RNA-binding proteins was reported to recruit the exosome to substrate RNAs (Arigo et al, 2006b; Thiebaut et al, 2006). The longest observed IGS1-R transcript contains 10 potential binding sites for Nab3 (UCUU) including two prominent clusters, and seven binding sites for Nrd1 (GUAA/G). The level of IGS1-R was therefore assessed in ts-lethal nab3-11 and nrd1-102 mutant strains (Figure 3F). IGS1-R was strongly stabilized in the nrd1-102 mutant, demonstrating that it is targeted by the Nrd1–Nab3 pathway. In contrast, the nab3-11 mutant conferred little or no stabilization (Figure 3F). Allele specificity has, however, previously been reported for mutations in these proteins (Conrad et al, 2000) and the data do not demonstrate that Nrd1 is primarily responsible for targeting IGS1-R for degradation. On other transcripts, Nrd1–Nab3 are responsible for transcription termination (Steinmetz et al, 2001; Arigo et al, 2006a, 2006b; Kim et al, 2006; Thiebaut et al, 2006). However, we saw no clear differences in the migration of the IGS1-R transcripts in the nrd1 or nab3 mutants relative to the wild type (Figure 3F), suggesting that termination is not strongly impaired.
From the ChIP and northern data, we conclude that transcription of IGS2-R and IGS1-F is strongly repressed by a chromatin structure that requires Sir2 and Top1 for its maintenance. IGS1-R is more actively transcribed than IGS2-R or IGS1-F, but the resulting transcripts are rapidly degraded by a mechanism that requires the Nrd1/Nab3 complex, TRAMP4 and both the core exosome and Rrp6.
IGS1-R is polyadenylated and shows extensive 3′ heterogeneity
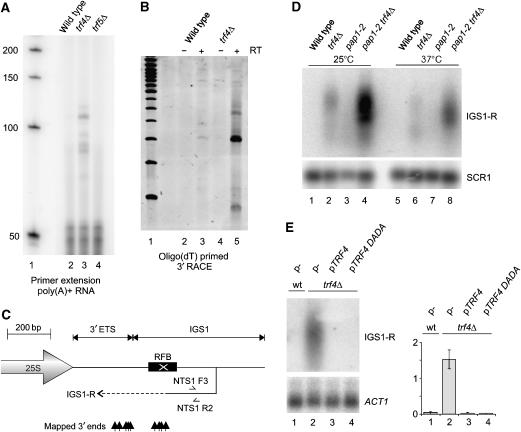
Primer extension was used to define the 5′ end of the IGS1-R transcript (Figure 4A). The 5′ end was mapped by running the primer extension products alongside a sequencing ladder on a 40 cm denaturing polyacrylamide gel (data not shown). The major 5′ end lies at +599 nt from the end of the 25S rRNA, with some secondary 5′ ends spanning about 20 bp. This position is 26 bp 3′ to that originally described (Santangelo et al, 1988) and around +175 nt from the RFB.
Figure 4.
IGS1-R is a 3′-heterogeneous transcript polyadenylated by Pap1. (A) Primer extension analysis of IGS1-R performed on poly(A)+ RNA from oligonucleotide NTS1 F3 (see C). (B) Ethidium-stained 6% acrylamide gel of 35-cycle 3′ RACE reactions using NTS1 R2 (see C). (C) Schematic of IGS1-R showing mapped 3′ ends and primer locations for 3′ RACE and primer extension. (D) Northern analysis of TRAMP and Pap1 mutants grown at 25°C in YPD and shifted to 37°C for 1 h, probed with NTS1 HpaI and SCR1. (E) trf4Δ strains were transformed with Trf4 plasmids (Vanacova et al, 2005), grown to mid-log in synthetic media and probed with NTS1 HpaI and ACT1. Error bars=±1 standard error, n=3.
Conventional analyses of polyadenylation using RNase H and oligo(dT) were complicated by the presence of two genome-encoded poly(A) tracts within IGS1-R (Supplementary Figures 1A and B). However, the results were consistent with 3′ heterogeneity, as were the northern analyses shown in Figure 3D. Oligo-dT-directed 3′ RACE detected multiple transcripts (Figure 4B), showing that IGS1-R is polyadenylated even in trf4Δ strains, and of 16 3′ RACE clones sequenced, 6 terminated in genomic encoded poly(A) tracts and the remaining 10 clones contained 9 different 3′ ends, demonstrating substantial 3′ heterogeneity (Figure 4C).
Depletion of Trf5 from the trf4Δ strain had little effect on transcript length (Figure 3C, lane 5), whereas transfer of a ts-lethal pap1-2 trf4Δ strain to 37°C resulted in shorter IGS1-R transcripts (Figure 4D, compare lanes 4 and 8). At permissive temperature, the abundance of IGS1-R transcripts was greater in the pap1-2 trf4Δ double mutant than in either single mutant (Figure 4D, compare lanes 2, 3 and 4). These results indicate that IGS1-R is polyadenylated by Pap1 and suggest that efficient polyadenylation promotes IGS1-R degradation.
We tested whether the polyadenylation activity of Trf4 is required for degradation of IGS1-R (Figure 4E). Plasmids expressing either Trf4 or the catalytically inactive Trf4-DADA (Vanacova et al, 2005) fully suppressed the IGS1-R stabilization phenotype in the trf4Δ strain. Recruitment of the exosome and IGS1-R degradation does not therefore require polyadenylation by Trf4 although the presence of the protein is necessary.
Trf4 functions in rDNA copy number control
TRAMP and exosome mutants were tested for alterations in rDNA copy number using pulsed field gel electrophoresis (PFGE) (Figures 5A and B). In the wild-type strain, chromosome XII (∼60% of which is composed of rDNA repeats) showed a well-defined length corresponding to an rDNA array of ∼200 repeats. In contrast, sir2Δ and top1Δ strains showed hyper-recombination phenotypes that caused chromosome XII to appear as smears (compare lane 1 with lanes 6 and 7 in Figure 5A and lanes 1–3 with lanes 7–9 and 13 and 14 in Figure 5B) (Christman et al, 1988; Gottlieb and Esposito, 1989; Bryk et al, 1997). Strains lacking the TRAMP components, trf4Δ or air1Δ air2Δ, showed sporadic deviations from the wild-type rDNA copy number, with the greatest effect in trf4Δ strains. These deviations from wild-type repeat number had only limited penetrance, with three out of eight trf4Δ clones analyzed showing a clear repeat number reduction and two strains showing repeat expansion. We observed no evidence of an rDNA hyper-recombination phenotype, which would be indicated by smearing of the rDNA band, in any trf4Δ single mutant analyzed, in contrast to a previous report (Sadoff et al, 1995).
Figure 5.
Loss of Trf4 alters rDNA copy number. (A, B) Pulsed field analysis of rDNA copy number in TRAMP mutants and controls. Strains were grown to stationary phase in YPD, resolved on a 0.8% pulsed field gel and probed with 18S to highlight chromosome XII. Ethidium bromide staining of other chromosomes shows integrity of preparation; the top chromosome visible is chromosome IV. Repeat numbers were calculated from an S. cerevisiae PFGE marker (A) and an H. wingii PFGE marker (B). (C) Northern analysis of strains grown to mid-log in YPD and probed with (top to bottom) NTS1 HpaI, NTS1 R, NTS2 F and ACT1. (D) Quantification of transcripts in (C). Results have been normalized to average repeat number in the culture determined by Southern blot analysis of BglII-digested genomic DNA (data not shown). (E) ChIP analysis of RNA polymerase II levels at the IGS1-R locus. The experiment is performed in Smc1-13Myc strains; however, northern analysis shows that this does not alter transcript levels (data not shown). Error bars=±1 standard error, n=3.
A MET∷trf4 top1Δ strain was viable on restrictive high methionine medium, and multiple top1Δ trf4Δ transformants isolated in four independent experiments were all growth-impaired but viable. This is in contrast to their reported synthetic lethality (Sadoff et al, 1995; Castano et al, 1996; Pan et al, 2006), and may reflect differences in alleles or strain background. We also combined trf4Δ with sir2Δ, and growth of the double mutant strain was similar to that of the trf4Δ single mutant. The double mutants of trf4Δ with either sir2Δ or top1Δ showed large losses in rDNA repeat number (Figure 5B, lanes 10–12 and 15–17), and this phenotype was observed in all clones tested (at least four of each combination). In contrast, the combination of trf5Δ with either sir2Δ or top1Δ had no apparent effect on rDNA repeat number (Supplementary Figure 2A). We also tested the exosome mutant rrp6Δ (Supplementary Figure 2B). Loss of Rrp6 alone had no clear effect on repeat number (lane 2), but an rrp6Δ top1Δ double mutant showed reduced heterogeneity relative to top1Δ single mutant strains (lanes 3–7).
To assess potential links between stability of the rDNA and transcription of IGS1-R, IGS1-F and IGS2-R, the trf4Δ sir2Δ and trf4Δ top1Δ double and single mutant strains were analyzed by northern hybridization (Figures 5C and D) and Pol II ChIP (Figure 5E).
Levels of IGS1-R appeared to correlate with rDNA copy number instability, being higher in sir2Δ trf4Δ and top1Δ trf4Δ double-mutant strains than in the sir2Δ and top1Δ single mutants. In Figure 5D, the quantification of the northern data is normalized to rDNA repeat number. The levels of IGS1-F were highest in the sir2Δ and top1Δ single mutants, which showed the greatest repeat heterogeneity. In contrast, the levels of IGS2-R appeared principally dependent on the presence or absence of Sir2. The ratio of abundance of the ncRNAs was altered between top1Δ and sir2Δ strains expressing or lacking Trf4. It is possible that this has an effect on repeat number, although we have no direct evidence for this. Loss of Trf5 from either the sir2Δ or top1Δ strains had no clear effect on the levels of any of the IGS ncRNAs (Supplementary Figure 2C).
ChIP for RNA Pol II at the IGS1-R locus (Figure 5E) shows that Pol II occupancy is not significantly altered in top1Δ trf4Δ strains compared to top1Δ. Hence, the reduced heterogeneity of the rDNA repeats in the trf4Δ top1Δ double mutant relative to the top1Δ single mutant is not due to reduced transcription. This also confirms that the high accumulation of transcript is due to increased RNA stability in the absence of Trf4.
We conclude that deletion of TRF4, but not TRF5, from the sir2Δ or top1Δ strains leads to greatly increased levels of IGS1-R, due to increased RNA stability, and a drastic loss of rDNA repeats. Deletion of RRP6 from the top1Δ strain appeared to reduce the heterogeneity in rDNA repeat number without clearly decreasing average repeat length.
Trf4 catalytic activity is not required for repeat regulation
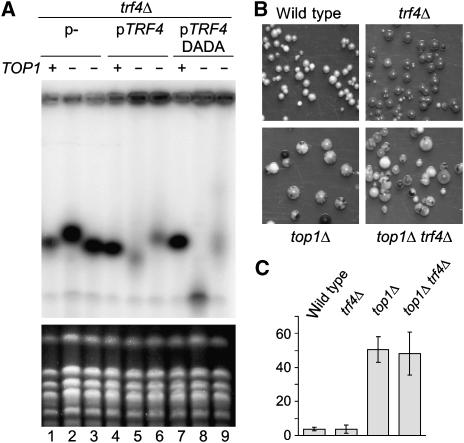
One explanation for the loss of rDNA repeats in trf4Δ mutants would be that Trf4 is a DNA polymerase involved in rDNA replication, as previously proposed (Wang et al, 2000). Were this the case, the polymerase activity of Trf4 would be required to allow the hyper-recombination observed in top1Δ strains.
To test this, we made use of the suppression of the rDNA repeat heterogeneity seen in top1Δ strains by loss of Trf4 (Figure 6A). We compared trf4Δ TOP1 (lanes 1, 4 and 7) and trf4Δ top1Δ (lanes 2–3, 5–6 and 8–9). These strains also carried plasmids lacking an insert (lanes 1–3), expressing intact Trf4 (lanes 4–6) or expressing the catalytically inactive Trf4-DADA (lanes 7–9). The top1Δ strains expressing wild-type Trf4 showed a hyper-recombination phenotype (lanes 5 and 6). Hyper-recombination was suppressed in the absence of Trf4 (lanes 2 and 3), but was clearly present when only Trf4-DADA was expressed (lanes 8 and 9). Western blotting confirmed that the mutant and wild-type Trf4 were expressed at similar levels (data not shown). In this experiment, strains were grown on minimal media to select for plasmid maintenance and recombination was less active than on complete YPD media used in the experiments shown in Figure 5. Thus, the poly(A) polymerase activity of Trf4 is not required for IGS1-R degradation and is also not required for hyper-recombination.
Figure 6.
The role of Trf4 in rDNA recombination. (A) Pulsed field analysis of trf4Δ top1Δ strains with Trf4 plasmids. One TOP1 and two top1Δ samples are shown in each case. Cells were grown in synthetic media and harvested at stationary phase approximately 50 generations after TOP1 deletion. Gel was run and probed as in Figure 5B. (B) An rDNA MET25 reporter was randomly integrated in each strain, and cells were grown to mid-log before plating on modified lead acetate plates at 25°C. Once colonies were ∼1 mm diameter, they were imaged and scored for the presence or absence of sectoring. (C) Recombination frequency assessed by colony sectoring. Three separate MET25 insertion clones were analyzed twice for each strain. Error bars=±1 standard error, n=6.
rDNA recombination frequency is unaffected by loss of TRF4
These analyses do not resolve differences in recombination frequency and stability of the rDNA repeat tract. Recombination rates within the rDNA tract can be assessed by integrating a single-copy marker gene and scoring for its loss. The presence or absence of a functional MET25 gene can be scored by a colony color test on medium containing Pb2+, on which met25Δ colony sectors turn dark brown (Figure 6B) (Smith and Boeke, 1997). A MET25-GFP construct was integrated into the IGS2 region of a single rDNA repeat in one strain of each genotype used in Figure 5. Three independent MET25 insertion clones from each strain were then scored for colony sectoring phenotypes (Figure 6C). Comparison of sectoring levels in top1Δ and top1Δ trf4Δ shows that loss of TRF4 has little or no effect on recombination frequency. A transgene inserted at this location was previously shown to be repressed by Sir2 (Smith and Boeke, 1997), and we confirmed this for our insert by western blotting (data not shown). Met25-GFP expression was lower in western blots from trf4Δ strains than trf4Δ sir2Δ strains (data not shown), showing that Sir2-dependent silencing was maintained. This indicates that hypoacetylation of H3 and H4 by Sir2 is not lost, since Sir2-dependent silencing requires its deacetylation activity (Li et al, 2006).
In trf4Δ strains, transcription of the MET25-GFP reporter was frequently reduced, resulting in dark pigmentation (Figure 6B). This is consistent with the reduction of IGS1-F expression seen in sir2Δ trf4Δ strains compared to sir2Δ, but the effect was variable between fresh transformants and old cells. The sir2Δ trf4Δ strain proved hypersensitive to Pb2+ ions and could not be assessed in the sectoring assay.
Trf4 is recruited co-transcriptionally to IGS1
We hypothesized that the altered rDNA stability in trf4Δ strains is due to direct effects of Trf4 on the rDNA or chromatin rather than indirect consequences of defects in RNA processing. In this case, the role of IGS1-R transcription might be to recruit Trf4 to the rDNA in the vicinity of the RFB region. Were this model correct, we would detect an association of Trf4 with the rDNA IGS1-R region that is dependent on RNA Pol II transcription.
ChIP analysis of Trf4-Myc over the rDNA IGS regions (Figure 7B) showed clear enrichment over IGS1-R. To confirm that this association was dependent on RNA Pol II transcription, we analyzed an rpb1-1 strain, which carries a fast-acting temperature-sensitive mutation in RNA Pol II (Figure 7A). The ChIP signal for Trf4 was substantially reduced across the IGS1-R region at the restrictive temperature, consistent with its recruitment by IGS1-R nascent transcripts, although the ChIP signal for Pol II showed greater reduction than the Trf4 signal (Figure 7B). To ensure that the protein context of the rDNA is not generally altered in this strain, lysates from this experiment were checked by ChIP for the presence of Sir2, which was only mildly reduced at 37°C (Supplementary Figure 3).
Figure 7.
Trf4 is recruited co-transcriptionally to the IGS1-R region of the rDNA. (A, B) ChIP analysis of Trf4 recruitment to the IGS in an rpb1-1 strain. rpb1-1 TRF4-13Myc cells were grown to mid-log at 25°C in YPD and half were shifted to 37°C for 30 min. Chromatin was precipitated from 25°C and 37°C cultures using anti-RNA polymerase II 8WG16 (A) and anti-Myc 9E10 (B). (C) ChIP analysis of Smc1-13Myc distribution in cells growing at 25°C in YPD. Error bars=±1 standard error, n=4 (A, B), n=3 (C).
These data indicate that Trf4 is recruited co-transcriptionally to the nascent IGS1-R transcripts, probably via Nrd1/Nab3. This is likely to be an important factor in allowing very rapid degradation of IGS1-R as soon as transcription termination generates a free 3′ end for exosome activity. This may also be the case for other cryptic ncRNA transcripts.
Alterations in rDNA copy number in trf4Δ strains do not reflect differences in cohesin recruitment
In order to address whether cohesin is displaced by IGS1-R accumulation in the trf4Δ strains, we performed ChIP analysis using a 13-Myc-tagged cohesin subunit Smc1. The Smc1-Myc fusion is the only form of Smc1 present in the cells, which showed no growth impairment. In addition, rDNA recombination rates were unaltered, as judged by the Pb2+ plate method described above (data not shown), indicating that the fusion protein is fully functional.
In the wild-type background, Smc1-Myc showed the expected distribution with a clear peak over the CAR (Figure 7C). Neither the distribution nor the intensity of the Smc1 ChIP signal was altered by the loss of Trf4. In contrast, the Smc1 ChIP signal was strongly reduced by loss of Top1, either in the presence or absence of Trf4. This makes it unlikely that the effects of Trf4 on rDNA copy number regulation are related to removal of cohesin from the IGS region.
Discussion
Cryptic transcripts are generated from repressed chromatin regions in S. cerevisiae
Genome-wide analyses of RNA Pol II density in yeast using ChIP and tiling microarrays showed that almost the entire genome is transcribed (David et al, 2006; Steinmetz et al, 2006). A striking feature of these analyses was that regions of repressed chromatin, notably in the rDNA IGS1 region and telomeres, showed Pol II occupancy that was significantly below the background of ‘non-transcribed' regions. Paradoxically, these repressed regions can be actively transcribed under some conditions, leading to the suggestion that initial rounds of transcription might be needed to establish subsequent silencing (Steinmetz et al, 2006). In strains lacking the poly(A) polymerase Trf4, we detected cryptic transcripts derived from CEN3 and from regions of repressed chromatin: a telomeric region (TEL05L) and the rDNA intergenic spacer region IGS1-R.
The TEL05L ncRNA transcript stabilized in TRAMP and exosome mutants is ∼6.5 kb in length and is derived from the Y′ region, which is fully or partially conserved at most yeast telomeres (Louis and Haber, 1992). The RNA initiates close to the terminal telomeric repeats and runs antisense to an ORF (YEL077C), which potentially encodes a 143 kDa protein proposed to function as a DNA helicase involved in telomere maintenance (Yamada et al, 1998). Sequence analyses showed that ncRNAs are generated from at least two telomeres, and we speculate that they may be transcribed from many or all telomeres. The identification of these ncRNAs might be consistent with the model that cryptic transcripts play a role in establishing silenced chromatin regions (Steinmetz et al, 2006). In S. cerevisiae, centromeric regions are very small, lack clear heterochromatic regions and were not previously reported to be transcribed. In contrast, centromeric regions in Schizosaccharomyces pombe are transcribed, but the RNAs are rapidly degraded by the TRAMP and exosome complexes (Buhler et al, 2007). We detected an RNA of ∼1.2 kb apparently derived from CEN3, which was elevated in the trf4Δ strain, suggesting that cryptic centromeric transcripts may also be present in S. cerevisiae.
Previous reports functionally linked Trf4 with Top1 and cohesion and condensation in the rDNA repeats (Castano et al, 1996), and we therefore analyzed the IGS1 transcripts in more detail. Two transcripts, IGS1-R and IGS1-F, are generated by divergent transcription from the E-pro promoter. We showed that IGS1-R, but not IGS1-F, is recognized and degraded by TRAMP4 and the exosome. Expression of a catalytically inactive form of Trf4 showed that its poly(A) polymerase activity is not required for degradation of IGS1-R. The catalytic activity of Trf4 was also dispensable for degradation of HSP104 mRNA in THO complex mutants (Rougemaille et al, 2007), but was required for degradation of hypomethylated tRNAiMet (Vanacova et al, 2005). This suggests that polyadenylation aids degradation of structured substrates, but is dispensable on less structured RNAs. The 3′ ends of the IGS1-R transcripts are polyadenylated by the canonical poly(A) polymerase Pap1, and are very heterogeneous. We speculate that this heterogeneity may result in part from termination by collision with oncoming RNA polymerase I molecules.
ChIP analyses showed that Trf4 could be crosslinked to the rDNA over the IGS1-R region, but not over IGS1-F. This association is largely dependent on functional RNA Pol II, since it was substantially reduced in an rpb1-1 mutant strain at non-permissive temperature. We therefore propose that Trf4 is recruited co-transcriptionally to the nascent IGS1-R transcripts. The IGS1-R region contains multiple predicted binding sites for the RNA-binding proteins Nab3 and Nrd1, which were previously reported to act as cofactors for the exosome in RNA degradation (Vasiljeva and Buratowski, 2006). Consistent with this, degradation of IGS1-R was strongly inhibited by a mutation in Nrd1, indicating the involvement of the Nrd1/Nab3 heterodimer in recruiting the TRAMP and/or exosome complexes. Since Nrd1/Nab3 also function in transcription termination by RNA Pol II (Arigo et al, 2006b; Thiebaut et al, 2006), they clearly have the potential to bind co-transcriptionally to nascent transcripts. We saw no evidence for effects of mutations in Nrd1 or Nab3 on termination of the IGS1-R transcript, but we predict that they act to recruit the TRAMP complex to nascent IGS1-R transcripts.
Co-transcriptional binding of Nrd1–Nab3 and Trf4 may be an important factor in the very rapid degradation that makes many transcripts from yeast and other eukaryotes ‘cryptic'. The exosome cannot degrade co-transcriptionally, since it requires a free 3′ end, but it appears that the IGS1-R transcripts are already targeted during transcription, potentially allowing their immediate degradation when the transcript is released from the polymerase.
Links between Trf4 and rDNA copy number regulation
Strains carrying trf4Δ displayed sporadic alterations in copy number. In contrast, strains carrying either top1Δ or sir2Δ showed hyper-recombination, manifested as extensive rDNA repeat length heterogeneity. The combination of trf4Δ with either top1Δ or sir2Δ resulted in a synergistic phenotype with drastic loss of rDNA repeats. Analysis of an integrated MET25 marker indicated that this does not reflect altered recombination rates, indicating that rDNA instability is responsible for repeat loss. Deletion of exosome component Rrp6 in top1Δ reduced the hyper-recombination phenotype, indicating that Rrp6 is also required for normal rDNA stability.
The cohesin complex is believed to hold sister chromatids together in the rDNA, so that recombination does not lead to alterations in copy number (Huang et al, 2006). However, loss of Trf4 did not detectably affect cohesin binding over IGS1 and IGS2. Changes in rDNA recombination rate without alteration of cohesin association were previously observed in mutants of Lrs4/Csm1 (Huang et al, 2006).
Together the data are consistent with the model in Figure 8. Transcription of IGS1-F through the CAR located in IGS2, or the balance between transcription of IGS1-F and IGS2-R may play important roles in cohesin displacement (Kobayashi and Ganley, 2005; Li et al, 2006). However, we predict that IGS1-R has a distinct function, which is important for rDNA stability, although its exact nature remains unclear. Trf4 and other factors binding to the nascent IGS1-R transcript may enhance rDNA stability at the site of transcription, possibly by promoting the repair of DNA damage. Alternatively, IGS1-R transcripts that escape degradation might exert a dominant negative effect on rDNA repair or recombination at other sites, and these models are not mutually exclusive. The ChIP data showing co-transcriptional recruitment of Trf4 would be consistent with association of the TRAMP complex with the rDNA at the site of transcription via IGS1-R. Rrp6 retains mRNAs with aberrant 3′ ends close to the site of transcription (Hilleren et al, 2001), and it is conceivable that IGS1-R, which shows high 3′ heterogeneity, could also be linked to the transcription site by Rrp6.
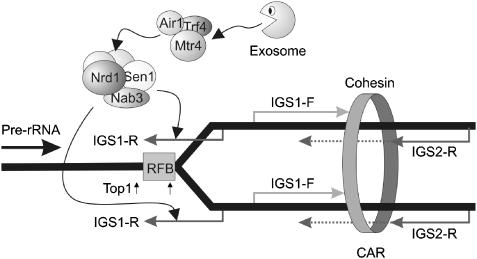
Figure 8.
Model for the roles of ncRNAs in the rDNA spacer regions. A DNA replication fork is shown blocked at the RFB. Transcription of IGS1-F and IGS2-R may concentrate cohesin at the CAR. This would potentially resemble the role of RNA Pol II in concentrating cohesin to other sites of convergent transcription (Lengronne et al, 2004). Alterations in the level or balance of IGS1-F and IGS2-R may also be responsible for regulated cohesin displacement, allowing misalignment of sister chromatids and alterations in repeat number. The high rate of pre-rRNA transcription and attendant DNA supercoiling may lead to DNA damage, particularly at the DNA replication fork stalled at the RFB, when Top1 is absent. The IGS1-R transcript is proposed to recruit Nrd1 and Nab3, which generally function together with the Sen1 RNA helicase. These factors may stimulate co-transcriptional association of the TRAMP complex and the exosome with the IGS1-R transcript. Interactions between TRAMP, the exosome and the RFB region of the rDNA, via the IGS1-R transcript, are envisaged to alter the chromatin structure and/or promote DNA repair (see Discussion).
The IGS1-R transcript passes through the RFB, a key region for rDNA copy number regulation. Replication forks emanate from replication origins (termed ARS; Figure 1) in each rDNA repeat, but forks moving against the direction of pre-rRNA transcription are stalled at the RFB, presumably to reduce collision with RNA Pol I. Top1 binding sites flank the RFB and are required to relieve DNA supercoiling generated by the high Pol I transcriptional rate. We speculate that the lack of Top1 activity leads to DNA damage over RFB regions carrying stalled DNA replication forks. Strains lacking Sir2 should not have excessive supercoiling at the RFB, but will have more stalled replication forks in this region due to increased activation of rDNA replication origins when Sir2 is absent (Pasero et al, 2002).
There are a number of potential links between the exosome and its cofactors and the repair of DNA damage in yeast. Strains lacking Trf4 are hypersensitive to DNA cleavage induced by the Top I inhibitor camptothecin (Walowsky et al, 1999). This was also the case for air1Δ air2Δ double-mutant strains (our unpublished observations), showing this phenotype to be TRAMP-related. Strains lacking Trf4 were also reported to be hypersensitive to DNA damage caused by treatment with the alkylating agent MMS (Walowsky et al, 1999). In addition, the nuclear exosome components Rrp6 and cofactor Rrp47/Lrp1/C1D are implicated in the repair of UV-induced DNA damage (Hieronymus et al, 2004), and rrp47Δ strains are defective in both non-homologous end joining and homologous recombination (Erdemir et al, 2002). We speculate that the TRAMP and exosome complexes also play a role in the repair of DNA damage at the RFB. Increased histone mRNA levels leading to defects in DNA metabolism have recently been reported (Reis and Campbell, 2006); however, this was not found to be the case for a trf4Δ single mutant, and we have also not observed this in our strains (data not shown).
Recombination events are frequent in top1Δ mutants but rare in trf4Δ. We predict that recombination-based repair is infrequent in trf4Δ strains due to the action of Top1 in removing supercoils and preventing DNA damage. This may explain the low penetrance of the repeat number change phenotype in trf4Δ, with events altering repeat number occurring only once in many generations. Notably, however, some trf4Δ and air1Δ air2Δ samples contained two populations with discrete rDNA lengths (Figure 5A and data not shown), suggesting that a recombination event had occurred during early growth of the culture. The top1Δ trf4Δ strains are predicted to undergo frequent rDNA damage repair by recombination. However, the repeat tract collapses as these events are biased toward contraction, being based on strand invasion rather than homologous recombination.
Degradation of the IGS1-R transcript resembles that of the CUTs, which account for a significant proportion of the genome of S. cerevisiae. A crude estimate based on the microarray analyses of Wyers et al (2005) suggests a minimum of 600 CUTs, or just under 10% of the number of annotated genes (see Supplementary Figure 4 for calculations). Strains lacking Trf4 are reported to show defects in chromosome arm cohesion (Wang et al, 2000; Edwards et al, 2003). Whether this is related to the recruitment of TRAMP to the numerous and widely dispersed CUTs remains to be determined.
Materials and methods
Strains and plasmids
Yeast transformation was performed by standard methods. Yeast strains are described in Supplementary Table 1. Cells were grown in YPD (2% peptone, 2% glucose, 1% yeast extract) or synthetic media (0.5% (NH4)SO4, 1.7% yeast nitrogen base, 2% glucose, amino acids) at 25°C; temperature shifts to 37°C were performed in a shaking water bath. Plasmids are described in Supplementary Table 2 and oligonucleotides used for cloning are listed in Supplementary Table 5.
Recombination assays were performed as described by Cost and Boeke (1996).
RNA analysis
Yeast RNA extraction and northern analysis were performed as described (Tollervey, 1987); high molecular weight RNA was separated on 1.2% glyoxal gels. Experimental details and probes are described in Supplementary data and Supplementary Table 3.
PFGE
PFGE was performed as described (Kobayashi, 2003); see Supplementary Materials and Methods for a detailed protocol.
ChIP
ChIP was carried out as described (Kotovic et al, 2003) with modifications (see Supplementary Materials and Methods and Supplementary Table 4).
Supplementary Material
Supplementary Information
Acknowledgments
We thank M Grunstein, T Kobayashi, S Vanacova, W Keller, R Grainger and D Ciais for reagents, L Aragon for helpful discussions, C Schneider for critical reading, S Innocente for help with ChIP and M Vogelauer, B Ward and D Leach for advice and equipment. This work was supported by the Wellcome Trust. JH is supported by the Leverhulme Trust and AEH by EMBO.
References
- Arigo JT, Carroll KL, Ames JM, Corden JL (2006a) Regulation of yeast NRD1 expression by premature transcription termination. Mol Cell 21: 641–651 [DOI] [PubMed] [Google Scholar]
- Arigo JT, Eyler DE, Carroll KL, Corden JL (2006b) Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell 23: 841–851 [DOI] [PubMed] [Google Scholar]
- Bryk M, Banerjee M, Murphy M, Knudsen KE, Garfinkel DJ, Curcio MJ (1997) Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev 11: 255–269 [DOI] [PubMed] [Google Scholar]
- Buhler M, Haas W, Gygi SP, Moazed D (2007) RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell 129: 707–721 [DOI] [PubMed] [Google Scholar]
- Castano IB, Brzoska PM, Sadoff BU, Chen H, Christman MF (1996) Mitotic chromosome condensation in the rDNA requires TRF4 and DNA topoisomerase I in Saccharomyces cerevisiae. Genes Dev 10: 2564–2576 [DOI] [PubMed] [Google Scholar]
- Christman MF, Dietrich FS, Fink GR (1988) Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell 55: 413–425 [DOI] [PubMed] [Google Scholar]
- Conrad NK, Wilson SM, Steinmetz EJ, Patturajan M, Brow DA, Swanson MS, Corden JL (2000) A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics 154: 557–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cost GJ, Boeke JD (1996) A useful colony colour phenotype associated with the yeast selectable/counter-selectable marker MET15. Yeast 12: 939–941 [DOI] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM (2006) A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci USA 103: 5320–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C, Houseley J, Tollervey D (2006) Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J 25: 1534–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S, Li CM, Levy DL, Brown J, Snow PM, Campbell JL (2003) Saccharomyces cerevisiae DNA polymerase epsilon and polymerase sigma interact physically and functionally, suggesting a role for polymerase epsilon in sister chromatid cohesion. Mol Cell Biol 23: 2733–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdemir T, Bilican B, Cagatay T, Goding CR, Yavuzer U (2002) Saccharomyces cerevisiae C1D is implicated in both non-homologous DNA end joining and homologous recombination. Mol Microbiol 46: 947–957 [DOI] [PubMed] [Google Scholar]
- Gottlieb S, Esposito RE (1989) A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56: 771–776 [DOI] [PubMed] [Google Scholar]
- Hieronymus H, Yu MC, Silver PA (2004) Genome-wide mRNA surveillance is coupled to mRNA export. Genes Dev 18: 2652–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH (2001) Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413: 538–542 [DOI] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D (2006) RNA-quality control by the exosome. Nat Rev Mol Cell Biol 7: 529–539 [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D (2006) Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep 7: 205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Brito IL, Villen J, Gygi SP, Amon A, Moazed D (2006) Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev 20: 2887–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J (2004) Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Vasiljeva L, Rando OJ, Zhelkovsky A, Moore C, Buratowski S (2006) Distinct pathways for snoRNA and mRNA termination. Mol Cell 24: 723–734 [DOI] [PubMed] [Google Scholar]
- Kobayashi T (2003) The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork. Mol Cell Biol 23: 9178–9188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Ganley AR (2005) Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 309: 1581–1584 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Horiuchi T (1996) A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells 1: 465–574 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Horiuchi T, Tongaonkar P, Vu L, Nomura M (2004) SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 117: 441–453 [DOI] [PubMed] [Google Scholar]
- Kotovic KM, Lockshon D, Boric L, Neugebauer KM (2003) Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol Cell Biol 23: 5768–5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D (2005) RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724 [DOI] [PubMed] [Google Scholar]
- Laloraya S, Guacci V, Koshland D (2000) Chromosomal addresses of the cohesin component Mcd1p. J Cell Biol 151: 1047–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F (2004) Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430: 573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Mueller JE, Bryk M (2006) Sir2 represses endogenous polymerase II transcription units in the ribosomal DNA nontranscribed spacer. Mol Biol Cell 17: 3848–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis EJ, Haber JE (1992) The structure and evolution of subtelomeric Y′ repeats in Saccharomyces cerevisiae. Genetics 131: 559–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell 91: 457–466 [DOI] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D (2004) Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119: 789–802 [DOI] [PubMed] [Google Scholar]
- Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD (2006) A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124: 1069–1081 [DOI] [PubMed] [Google Scholar]
- Pasero P, Bensimon A, Schwob E (2002) Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev 16: 2479–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis CC, Campbell JL (2006) Contribution of Trf4/5 and the nuclear exosome to genome stability through regulation of histone mRNA levels in Saccharomyces cerevisiae. Genetics 175: 993–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougemaille M, Gudipati RK, Olesen JR, Thomsen R, Seraphin B, Libri D, Jensen TH (2007) Dissecting mechanisms of nuclear mRNA surveillance in THO/sub2 complex mutants. EMBO J 26: 2317–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoff BU, Heath-Pagliuso S, Castano IB, Zhu Y, Kieff FS, Christman MF (1995) Isolation of mutants of Saccharomyces cerevisiae requiring DNA topoisomerase I. Genetics 141: 465–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo GM, Tornow J, McLaughlin CS, Moldave K (1988) Properties of promoters cloned randomly from the Saccharomyces cerevisiae genome. Mol Cell Biol 8: 4217–4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Boeke JD (1997) An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev 11: 241–254 [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ, Conrad NK, Brow DA, Corden JL (2001) RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413: 327–331 [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA (2006) Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell 24: 735–746 [DOI] [PubMed] [Google Scholar]
- Szostak JW, Wu R (1980) Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature 284: 426–430 [DOI] [PubMed] [Google Scholar]
- Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D (2006) Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1–nab3 pathway in genome surveillance. Mol Cell 23: 853–864 [DOI] [PubMed] [Google Scholar]
- Tollervey D (1987) A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J 6: 4169–4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W (2005) A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3: e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L, Buratowski S (2006) Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol Cell 21: 239–248 [DOI] [PubMed] [Google Scholar]
- Voelkel-Meiman K, Keil RL, Roeder GS (1987) Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell 48: 1071–1079 [DOI] [PubMed] [Google Scholar]
- Walowsky C, Fitzhugh DJ, Castano IB, Ju JY, Levin NA, Christman MF (1999) The topoisomerase-related function gene TRF4 affects cellular sensitivity to the antitumor agent camptothecin. J Biol Chem 274: 7302–7308 [DOI] [PubMed] [Google Scholar]
- Wang Z, Castano IB, De Las Penas A, Adams C, Christman MF (2000) Pol kappa: A DNA polymerase required for sister chromatid cohesion. Science 289: 774–779 [DOI] [PubMed] [Google Scholar]
- Willingham AT, Gingeras TR (2006) TUF love for ‘junk' DNA. Cell 125: 1215–1220 [DOI] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, Libri D, Jacquier A (2005) Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737 [DOI] [PubMed] [Google Scholar]
- Yamada M, Hayatsu N, Matsuura A, Ishikawa F (1998) Y′-Help1, a DNA helicase encoded by the yeast subtelomeric Y′ element, is induced in survivors defective for telomerase. J Biol Chem 273: 33360–33366 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information