Abstract
The Drosophila Necrotic protein is a serine proteinase inhibitor, which regulates the Toll-mediated innate immune response. Necrotic specifically inhibits an extracellular serine proteinase cascade leading to activation of the Toll ligand, Spätzle. Necrotic carries a polyglutamine extension amino-terminal to the core serpin structure. We show here that cleavage of this N-terminal extension occurs following immune challenge. This modification is blocked in PGRP-SAsemmelweiss mutants after Gram-positive bacterial challenge and in persephone mutants after fungal or Gram-positive bacterial challenge, indicating that activation of either of the Toll pathway upstream branches induces N-terminal cleavage of the serpin. The absolute requirement of persephone gene product for this cleavage indicates that Gram-positive bacteria activate a redundant set of proteinases upstream of Toll. Both full-length Necrotic and the core serpin are active inhibitors of a range of serine proteinases: the highest affinity being for cathepsin G and elastases. We found a 13-fold increase in the specificity of the core serpin over that of full-length Necrotic for one of the tested proteinases (porcine pancreatic elastase). This finding indicates that cleavage of the Necrotic amino-terminal extension might modulate Toll activation following the initial immune response.
Keywords: Insect immunity, Toll pathway, Proteinase inhibitor, Hemolymph proteins, Polyglutamine extension
Introduction
Serine proteinase inhibitors (serpins) regulate a wide range of processes such as blood coagulation, complement activation, and inflammation in mammals (Gettins, 2002; Silverman et al., 2001) and similar defense responses in invertebrates (Kanost, 1999). One of the best characterized of these invertebrate responses is the activation of the Drosophila Toll pathway, which is triggered by fungal or Gram-positive bacterial infections. A member of the family of peptidoglycan recognition proteins (PGRPs), PGRPSA, and the serine proteinase Persephone (Psh) have been shown genetically to delineate two separate signaling branches, upstream of Toll, and responsible, respectively, for the activation of the Toll pathway after Gram-positive bacterial challenge and natural fungal infection (Ligoxygakis et al., 2002a; Michel et al., 2001). Activation of the Toll receptor downstream of both these branches induces a signalling cascade leading to translocation of an NF-κB-like protein Dorsal-related immune factor (Dif) to the nucleus and synthesis of antimicrobial peptides (Hoffmann and Reichhart, 2002). The Toll receptor is activated by its ligand, a cysteine-knot growth factor called Spätzle (Spz), which is cleaved from its propeptide following infection (Levashina et al., 1999).
The Necrotic (Nec) serpin (previously called Spn43Ac) regulates Toll activation by inhibiting a proteinase involved in the cleavage of Spz (Levashina et al., 1999). Serpins are characterized by a highly conserved tertiary structure and a dynamic mechanism of inhibition. Native serpins have a folded core structure with an exposed reactive center loop (RCL), which is presented as an ideal substrate for the target proteinase. Cleavage of the RCL at the P1–P′1 position allows it to insert within a 5-strand β-sheet structure in the serpin core. During this process, the serpin relaxes and the proteinase is translocated by 70 Å from one pole of the serpin to the other. The proteinase molecule is distorted and trapped in a covalently linked serpin–proteinase complex, which is targeted for destruction (Gettins, 2002).
Nec has an alanine-rich hinge region and its active site is characterized by leucine and serine in the P1–P′1 positions. Nec is a highly unusual serpin in that the core structure carries an 80–100 amino acid N-terminal extension of unknown function. This extension has no obvious structure, but contains stretches of glutamines and prolines, including a 9-residue polyglutamine repeat. Following infection with a mixture of Gram-positive and Gram-negative bacteria, the Nec protein is cleaved (Levashina et al., 1999). We show here that, unexpectedly, this cleavage corresponds to the removal of the N-terminal polyglutamine-containing extension and is specifically linked to the induction of the Toll pathway following either Gram-positive bacterial or fungal infections. The cleavage of the N-terminal extension of Nec by both type of infections requires a wild-type psh gene, suggesting that Gram-positive bacteria activate a redundant proteinase upstream of Toll. We have been unable to express the full-length Nec protein (Nec-fl) in Escherichia coli cells, although the N-terminally truncated serpin (Nec-ΔN) is readily expressed and is a potent inhibitor of elastase- and chymotrypsin-like proteinases (Robertson et al., 2003). In this paper, we express Nec-fl in a baculovirus/insect cell system and show that it has similar stability and folding to the Nec-ΔN protein expressed in E. coli. The reaction kinetics and stoichiometry of inhibition (SI) ratios, for both forms of the serpin are compared with a range of serine proteinases. The observed change in specificity between Nec-fl and Nec-ΔN towards its target proteinase(s) could be a way to rapidly restore the initial conditions in the hemolymph after infection.
2. Materials and methods
2.1. Fly stocks and genetics
Flies were cultured at 25 °C. Oregon-R was used as the wild-type strain. To identify the faster-migrating Nec species (Levashina et al., 1999) we constructed a Nectag transgene. Myc and hemaglutinin (Ha) tags were introduced into the Nec-coding sequence by PCR. This construct carries the Myc tag between the signal peptide and the N-terminus of the secreted peptide (SP), while the Ha tag is at the C-terminus of the protein (Fig. 1A). The construct was cloned into the pUAST plasmid (Brand and Perrimon, 1993) by ligating a NotI-XhoI fragment from the pBluescript-SK vector into the corresponding sites of pUAST.
Fig. 1.
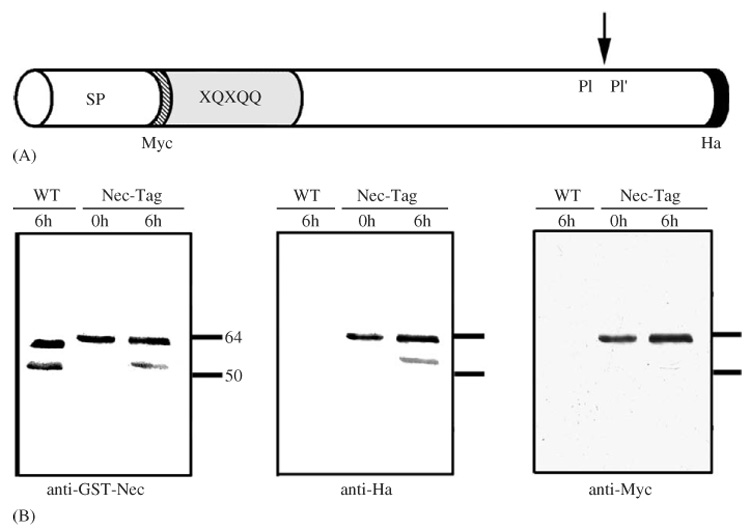
Immune-induced cleavage of Nec occurs at the N-terminus. (A) Schematic representation of the Nec-Tag transgene. The Myc tag is N-terminal, between the signal peptide (SP) and the polyglutamine rich N-terminal extension (XQXQQ). The Ha tag is at the C-terminal of the serpin molecule. (B) Western blot of hemolymph from wild type (WT) and Nec-tag (Df(2R)pk-78k/nec²; P[da-Gal4]/P[UAS-nectag]) flies. Samples were taken from unchallenged (0 h) flies or 6 h after challenge with Micrococcus luteus (6 h). The same membrane was used for probing with anti-GST-Nec, anti-Ha and anti-Myc antibodies.
To give ubiquitous transgene expression in a nec null background, Df(2R)pk-78k; P[da-Gal4] flies were crossed to nec²; P[UAS-nectag] flies. The daughterless-Gal4 (da-Gal4) transgene gives ubiquitous expression of Gal4, the Df(2R)pk-78k chromosome is deleted for the nec transcript and the nec² allele carries a stop codon at the beginning of the coding sequence (Green et al., 2000). Crosses were performed at 25 °C, but moved to a 29 °C incubator after the third larval stage to increase transgenic protein expression. The nec² mutation was isolated by Heitzler et al. (1993); Df(2R)pk-78k by Gubb and Garcia-Bellido (1982); PGRP-SAsemmelweiss (PGRP-SAseml) by Michel et al. (2001) and psh by Ligoxygakis et al. (2002a).
2.2. Sample preparation and analysis
Infections, hemolymph collection, sample preparation and Western blot analysis were described in Ligoxygakis et al. (2002a, b). Western blots were incubated with one of three antibodies. Anti-GST-Nec antibody (Levashina et al., 1999) and anti-Ha-peroxidase antibody (Boehringer) were incubated overnight at 4 °C at a dilution of 1/5000 dilution and 1/2000, respectively. Anti-Myc-peroxidase antibody (Boehringer) was used at 1/4000 dilution and incubated for 1 h at RT. Filters were stripped and re-probed with each antibody following the manufacturer’s recommendation (Amersham).
2.3. Construction of Nec-fl expression baculovirus
The Nec cDNA was sub-cloned from pBluescript-SK and inserted into the SpeI-KpnI sites of the pFastBac1 vector. Selection of recombinant colonies and isolation of bacmid DNA followed manufacturer’s instructions (Invitrogen Life Technology). Spodoptera frugiperda Sf9 cells were transfected in the presence of CellFECTIN. The virus titer was maximized by two serial infections of the Sf9 cells (Wang et al., 2001).
2.4. Expression and purification of Nec-fl
Sf9 cells were inoculated at 2 × 106 cells/ml in 500 ml Sf-900 II serum-free medium (Invitrogen Life Technology). The culture was incubated at 27 °C for 96 h with gentle agitation (100 rpm). Cells were removed by centrifugation (500g for 20 min) and the conditioned medium stored at −70 °C in 0.5 mM benzamidine. Recombinant protein was isolated from 200 ml aliquots incubated with 6.0 ml dextran sulfate-Sepharose (Nakamura et al., 1985), equilibrated in 10 mM potassium phosphate, 0.5 mM benzamidine, 0.5 mM phenylmethylsulfonyl fluoride, pH 6.4. The mixture was gently stirred at 4 °C for 1 h and loaded onto a column (2.5 cm i.d. × 6.5 cm). Following washing, bound proteins were eluted from the column in the same buffer (at 0.5 ml/min for 80 min, with a linear gradient of 0–1.0 M NaCl). Fractions containing Nec-fl were pooled and made up to 1.0 M ammonium sulfate. After centrifugation (15,000g for 30 min), the supernatant was applied to a methyl HIC column (Econo-Pac, 5.0 ml, Bio-Rad) and eluted with a linear gradient of 1.0–0M ammonium sulfate at 1.0 ml/min (in 10 mM potassium phosphate, at pH 6.4 for 40 min). Purified Nec-fl was stored at −70 °C in 20 mM Tris–HCl, pH 8.5.
2.5. Characterization of Nec-fl
The molecular mass of Nec-fl was determined by MALDI mass spectrometry as described in Jiang et al. (2003). To determine the association-state, 20 µg of Nec-fl was separated on an HPLC gel filtration column (Bio-Silect SEC 250, Bio-Rad) equilibrated with 0.1 M sodium phosphate, 0.1 M NaCl, pH 6.9; absorbance was monitored at 280 nm. The column was calibrated with molecular weight standards (Bio-Rad) including thyroglobulin (670 kDa), bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa), equine myoglobin (17 kDa) and vitamin B12 (1.35 kDa).
2.6. Determination of secondary structure and thermal unfolding
Far UV circular dichroism (CD) spectra were measured using a J-810 spectropolarimeter (Jasco). Nec-fl protein was dialyzed against 100 mM Tris, pH 8.0, 150 mM NaCl and diluted to a concentration of 0.25 mg/ml. CD spectra were scanned between 200 and 260 nm using a 0.05 cm path-length cuvette, at 25 °C. Each scan was repeated 25 times.
Thermal unfolding of Nec-fl was measured by the change in ellipticity at 222 nm. Samples were prepared as for CD spectrum measurement and heated from 25 to 90 °C, at 1 °C/min (The 222 nm signal originates from ordered α-helices and its intensity reflects the proportion of folded material accurately). Data was fitted to a two-state transition model (Lawrence et al., 1994).
Eight M urea gel electrophoresis was performed as described by Mahadeva et al. (2002). Purified, transgenic Nec protein was visualized by Coomassie brilliant blue staining. Endogenous Nec, in whole-fly extracts, was visualized using anti-GST-Nec antibody (Levashina et al., 1999; Green et al., 2003).
2.7. Complex-formation assays and SDS-PAGE analysis of serpin-proteinase complexes
Nec-fl was incubated at room temperature for 15 min with human neutrophil cathepsin G (Athens Research and Technology, Inc.), human neutrophil elastase (HNE) (Athens Research and Technology, Inc.), bovine pancreatic α-chymotrypsin (Sigma), porcine pancreatic elastase (PPE) (Worthington), human thrombin (Sigma), or bovine pancreatic trypsin (Sigma) at molar ratios of 1:1 or 4:1 in 50 mM HEPES buffer, 150 mM NaCl, 0.01% dodecylmaltoside, pH 7.4. Reaction mixtures and Nec and proteinase control samples were boiled for 3 min in SDS sample buffer, before separation on 10% SDS-gels. Proteins were visualized with Coomassie blue staining.
2.8. Inhibitory-activity assays
Proteinase samples were mixed with different concentrations of Nec-fl and Nec-ΔN (in 0.1 M Tris–HCl, 0.1 M NaCl, 5 mM CaCl2, pH 7.8). After incubation at room temperature for 10 min, a chromogenic substrate (150 µl) was added to measure residual enzyme activity. Absorbance at 405 nm was measured using a VERSAmax microplate reader (Molecular Devices). Enzymes and substrates were: 0.2 µg human neutrophil cathepsin G, with 100 µM N-succinyl-Ala-Ala-Pro-Phe-pNA (Sigma); 0.02 µg bovine pancreatic α-chymotrypsin, with 50 µM N-succinyl-Ala-Ala-Pro-Phe-pNA (Sigma); 0.2 µg of HNE with 100 µM N-succinyl-Ala-Ala-Pro-Leu-pNA (Sigma); 0.2 µg of PPE, with 50 µM N-succinyl-Ala-Ala-Pro-Leu-pNA; 0.02 µg bovine pancreatic trypsin with 50 µM H-d-Phe-pipecolyl-Arg-pNA (S-2238, Amersham Pharmacia Biotech) and 5 ng human thrombin, with 50 µM S-2238.
2.9. Determination of reaction parameters describing proteinase inhibition
The inhibition rate constant (ka) for Nec was determined for all serine proteinases except thrombin under pseudo-first order conditions (i.e., [I] ⩾ 10[E]0) using the progresss-curve method (Bieth, 1995; Morrison and Walsh, 1988). Rate constants of inhibition were measured at 25 °C in 50 mM Hepes, 150 mM NaCl, 0.01% w/v dodecyl-malto-side, pH 7.4 by adding the enzyme (20 nM final) to a mixture of Nec protein (from 200 to 800 nM) and the appropriate substrate (1 mM final) and recording the release of product as a function of time. The progress curves were then analyzed according to Eq. (1):
| (1) |
where υz is the initial velocity, υs is the steady-state velocity at completion of the reaction and kobs is the pseudo-first rate order rate constant for the approach towards steady state. The values of each variable were obtained by fitting the progress curve to Eq. (1) using nonlinear regression analysis, and were then used to calculate ka according to Eq. (2) under the assumption that the inhibition takes place through a simple bi-molecular reaction:
| (2) |
The association rate constant (ka) for thrombin was determined by reacting thrombin and Nec for different periods of time before addition of substrate that slows down the association process enough to allow measurement of residual enzyme activity. Pseudo-first order conditions were used and the data fitted to the following exponential equation (Bieth, 1984; Belorgey et al., 1996):
| (3) |
where [E] is the concentration of free enzyme at any time, t, and [E]0 is the concentration at t = 0. [E]0 and [E] are proportional to the rate of substrate hydrolysis at t = 0 and at any time t, respectively. ka was calculated from the pseudo-first order rate constant ka[I]0.
3. Results
3.1. Immune-induced cleavage of Nec released an N-terminal fragment
The double-tagged construct, P[UAS-nectag] (Fig. 1A, Materials and Methods), suppressed the nec phenotype of mutant flies when expressed under control of the ubiquitous da promoter (in nec−; P[da-Gal4]/P[UAS-nectag] flies, data not shown). This result shows that the Nectag protein remains biologically active. As previously reported, the anti-GST-Nec antibody recognized a single band (of about 60 kDa) in hemolymph from unchallenged flies and double bands (of about 60 and 52 kDa) after immune-challenge (Fig. 1B, anti-GST-Nec). The anti-Ha antibody detected both species of Nec in hemolymph from nec−; P[da-Gal4]/P[UAS-nectag] flies 6 h after Gram-positive bacterial challenge (Fig. 1B, anti-Ha). In contrast, the anti-Myc antibody identified only the higher band (Fig. 1B, anti-Myc) in either challenged or unchallenged flies. Note that the same membranes were stripped and reprobed with each antibody and that in neither case did the anti-tag antibodies detect bands in hemolymph from wild-type flies. These results establish that the lower band, which is only detected after immune-challenge, corresponds to the N-terminally cleaved Nec serpin. We never detected the N-terminal released fragment, neither with the anti-Myc antibody nor with anti-GST-Nec antibody, most likely because it is rapidly degraded.
3.2. Nec was cleaved by stimuli that activate the Toll pathway
The Toll pathway in Drosophila responds to fungal and Gram-positive bacterial infection, while the immune-deficient (Imd) pathway responds to Gram-negative bacterial infection (Hoffmann and Reichhart, 2002). Both fungal and Gram-positive responses require the Toll receptor, although their extracellular signalling pathways show distinct branches (Michel et al., 2001; Ligoxygakis et al., 2002a). We found that either fungal or Gram-positive bacterial infection can induce N-terminal cleavage of Nec. In contrast, only the 60 kDa Nec band was present following Gram-negative infection, or aseptic injury (Fig. 2A). N-terminal cleavage in response to Gram-positive infection was blocked in PGRP-SAseml mutants (Fig. 2B), which lack functional receptors required for the Gram-positive response (Michel et al., 2001). Similarly, following fungal infection, Nec N-terminal cleavage is blocked by mutations in the serine proteinase psh (Fig. 2B), which is required for the fungal response (Ligoxygakis et al., 2002a). Surprisingly, psh mutation also abolished the cleavage of the serpin induced by Gram-positive bacteria (Fig. 2B), but not the expression of Drosomycin (data not shown, Ligoxygakis et al., 2002a). Finally, we observed that the cleavage is not blocked in any mutant affecting genes of the Toll intracellular pathway (data not shown). These results establish that both upstream Toll pathway activating branches (Fig. 2B) can induce N-terminal Nec cleavage but that this cleavage is not a consequence of the Toll pathway activation.
Fig. 2.
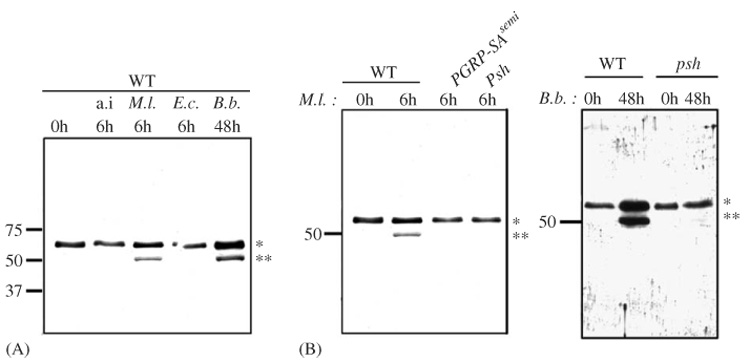
Cleavage of Nec is specifically induced by stimuli that activate the Toll pathway. Western blots of hemolymph from A, wild-type flies (WT) with aseptic injury (a.i.), Micrococcus luteus (M.l.) or Escherichia coli (E.c.) inoculation, or infected naturally with Beauveria bassiana (B.b.) and B, wild-type flies (WT), PGRP-SASemmelweiss (PGRP-SAseml) or persephone (psh) mutants challenged with M. luteus (M.l.) and wild-type flies (WT) or persephone (psh) mutants challenged with B. bassiana (B.b.). Hemolymph was collected before infection (0 h) and either 6 or 48 h post-infection. Membranes were probed with anti-GST-Nec antibody. The band corresponding to full-length and cleaved Nec are indicated by * and **, respectively.
3.3. Expression and purification of full-length Nec
Nec-fl was expressed in a baculovirus/insect cell system and purified as described in Experimental Procedures. The purified protein migrated as a doublet band on SDS-PAGE (Fig. 3A and B). MALDI-TOF mass spectrometry revealed two corresponding major species (Fig. 3C), at 51,792 and 52,838 Da. These values are significantly larger than the predicted molecular mass of 51,417 Da, suggesting post-translational modifications, such as glycosylation. The mass difference between the two species (1043 Da) corresponds exactly to the molecular mass of the first eight N-terminal residues indicating that the protein may be cleaved at these residues. This form is unlikely the N-terminally truncated form observed in vivo as the N-terminal cleavage releases an 8 kDa fragment. The apparent molecular size of the native protein, as assayed by size-exclusion chromatography (Fig. 3D), confirms that the serpin is present in monomeric form.
Fig. 3.
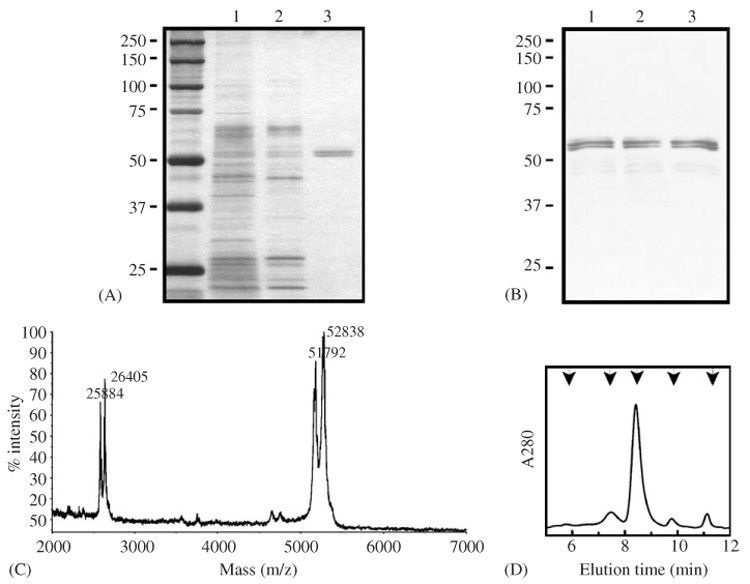
Purification of Nec-fl. Analysis of protein samples at different stages of purification. (A) silver-stained and (B) antibody-stained. Lane 1, conditioned medium; lane 2, elutate from dextran-sulfate-Sepharose column; lane 3, concentrated elutate from methyl-HIC column. Molecular mass standards are shown in left-hand. (C) MALDI-TOF mass spectrum of purified Nec-fl. Calibrated with rabbit muscle aldolase (Mm: 39,212). (D) Gel filtration chromatography of Nec-fl, on Bio-Silect SEC 250 column. The single major peak at 8.42 min indicates Nec-fl monomer. Molecular mass standards (arrows): thyroglobulin; bovine γ-globulin; chicken ovalbumin; equine myoglobin; vitamin B12, in order of decreasing mass.
3.4. Characterization of the conformation of purified Nec-fl
The inhibitory mechanism of serpins depends on the transition from a stressed (S), metastable conformation, characteristic of an active serpin, to a relaxed (R) conformation. This transition alters the electrophoretic mobility of the serpin. In addition, there are a number of routes by which an S → R transition can occur in the absence of proteinase inhibition (Gettins, 2002). Serpins can adopt a ‘latent’ conformation (in which RCL insertion occurs without cleavage), can polymerize (with the RCL of one serpin inserting within a second serpin protein), or can be cleaved and released (without the proteinase being trapped in a complex). Each of these conformational changes produces an inactive inhibitor with an altered electrophoretic mobility and thermal stability (Dafforn et al., 2004). For this reason, we undertook extensive characterization of the conformation state of Nec-fl. Far UV CD (Fig. 4A) shows a flat-bottomed negative minimum extending between 222 and 208 nm, characteristic of the serpin α/β fold. Thermal denaturation (Fig. 4B) shows a single sharp transition, corresponding to a loss of secondary structure of the native serpin around 51 ± 0.4 °C. A characteristic serpin denaturation was also shown following trypsin treatment of the Nec-fl protein analyzed by 8 M urea gel electrophoresis. The native protein shows a lower mobility compared to Nec-fl cleaved with trypsin (Fig. 4C). Tryptic digestion gave a dramatic increase of stability of the serpin (without complex formation), characteristic of an S → R transition. These data indicate that Nec-fl is in its native, metastable state.
Fig. 4.
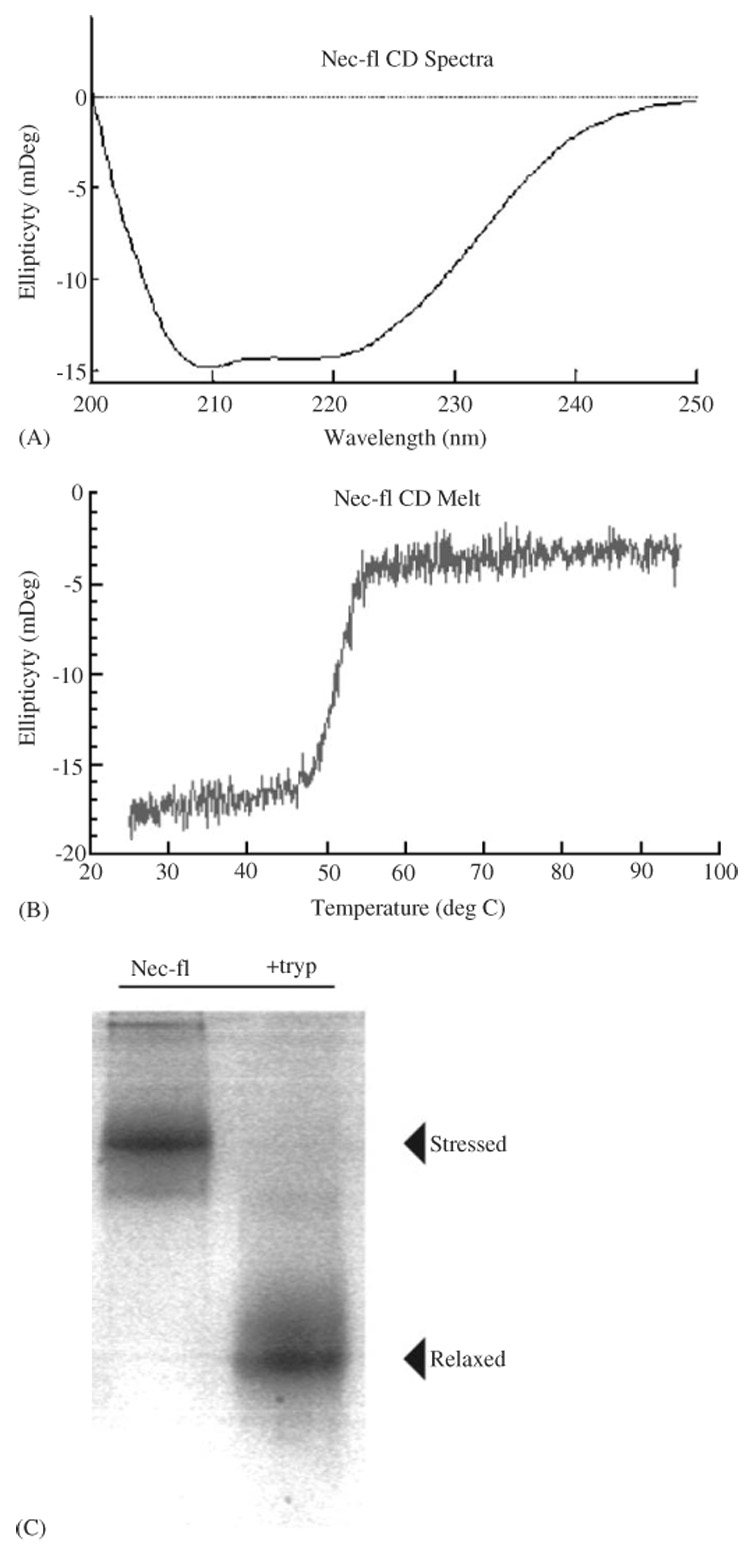
Conformational analysis of Nec-fl. (A) CD spectra of Nec-fl showing the presence of a mixed α-helix and α-sheet secondary structure, consistent with the known serpin tertiary structure. This figure is representative of three independent experiments carried out in a 100 mM Tris–HCl, 150 mM NaCl, pH 8.0 at 25 °C. (B) Thermal denaturation of Nec-fl shows a single transition at 51 ± 0.4 °C, indicating that Nec-fl is in the native conformation. Thermal unfolding was measured by monitoring the change in protein secondary structure assessed by the CD signal at 222 nm, with respect to temperature. (C) Trypsin-cleaved Nec-fl (right) had a higher motility on a 8 M Urea PAGE than intact serpin, indicating that cleaved serpin underwent the transition and remained in its folded state. Serpin was incubated with bovine-trypsin at a 500:1 serpin:proteinase molar ratio for 10 min at room temperature. Reactions were stopped with 0.1 mM PMSF and run on an 8 M Urea gel.
3.5. Characterization of Nec conformation in vivo
Total proteins from control flies and flies challenged with Gram-positive bacteria were analyzed on transverse urea gradient (TUG) gels. Both gels revealed the classic “S” curve with a shift around 3 M urea, characteristic of the gradual unfolding of a native serpin (Fig. 5). This result shows that Nec-fl and N-terminally cleaved Nec, which were both detected in fly hemolymph after infection, are in the active conformation.
Fig. 5.
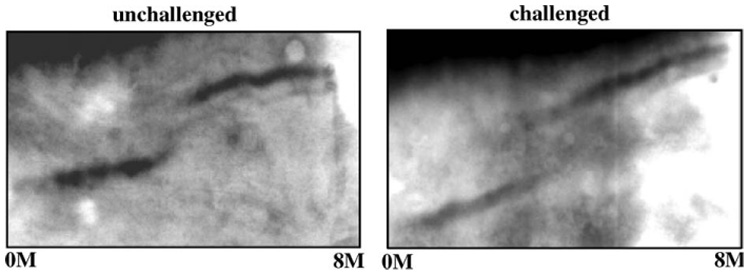
TUG gel analysis (0–8 M urea gradient). Whole fly extracts from unchallenged and immune-challenged wild-type flies. Both samples show a S-shaped curve with increasing urea concentration, characteristic of a serpin S → R transition. Note that inactive (R) serpin was not detected in the absence of urea, in either unchallenged or challenged flies.
3.6. Inhibition of a panel of well-characterized proteinases
Nec formed SDS-stable complexes with HNE, cathepsin G, chymotrypsin, PPE and thrombin (Fig. 6), but failed to form complexes with trypsin or kallikrein (not shown), for which Nec-fl is not an active inhibitor. With cathepsin G, Porcin Pancreatic Elastase Human Neutrophile Elastase and α-chymotrypsin, increasing the ratio of serpin to proteinase increased complex-formation. The Nec-thrombin complex could barely be detected, even with a large excess of serpin (Fig. 6). The association rate constant (ka) and SI values for each proteinase are compared between Nec-fl and Nec-ΔN in Table 1. Nec-fl is a potent inhibitor of cathepsin G, α-chymotrypsin and the elastase-like proteinases HNE and PPE. The interaction of Nec-fl is the most rapid with cathepsin G and HNE with ka values ranging from 5.3 × 105 to 1.8 × 105 M−1 s−1, and an SI values of 1.1 (Table 1). Nec-fl inhibited α-chymotrypsin and PPE moderately, but thrombin only weakly (the SI value for thrombin was not measurable with a serpin excess of 100:1, retaining greater than 50% proteinase activity). Again, Nec-fl gave no inhibition of trypsin, or kallikrein (data not shown).
Fig. 6.
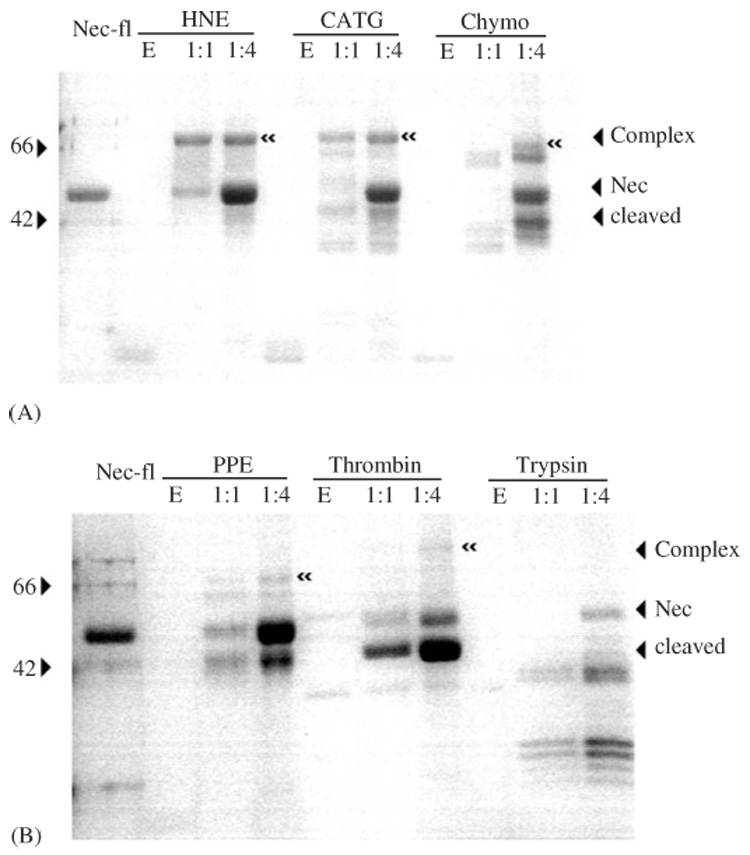
Detection of SDS-stable serpin-proteinase complexes by SDS-PAGE analysis. Tracks contain Nec-fl, proteinase, or a mixture of serpin and proteinase at Molar ratios of 1:1 or 4:1. Tested proteinases are human neutrophil cathepsin G (CATG), human neutrophil elastase (HNE), bovine pancreatic α-chymotrypsin (Chym), porcine pancreatic elastase (PPE), human thrombin (Throm) and bovine pancreatic trypsin (Tryp). “≪” indicates SDS-stable serpin-proteinase complex band.
Table 1.
Kinetic constants for inhibition of a panel of serine proteinase by Nec-fl and Nec-ΔN*
| Proteinase | Nec-fl |
Nec-ΔN |
||
|---|---|---|---|---|
| ka (M−1 s−1) | SI (inhibitor/proteinase) | ka (M−1 s−1) | SI (inhibitor/proteinase) | |
| Human neutrophile elastase | 1.8 × 105 | 1.1 | 1.5 × 105 | 1.6 |
| Porcine pancreatic elastase | 1.7 × 104 | 9.0 | 2.2 × 105 | 1.8 |
| Chymotrypsin | 4.6 × 104 | 3.9 | 4.0 × 104 | 3.1 |
| Thrombin | 1.9 × 10² | >100 | 1.2 × 10² | >100 |
| Trypsin | Undetectable | Infinity | Undetectable | Infinity |
| Cathepsin G | 5.3 × 105 | 1.1 | 3.2 × 105 | 1.1 |
Errors for the analysis of the kinetic parameters are 10%. The results represent the mean of, at least, three independent experiments.
In general, the inhibitory profiles of Nec-fl and Nec-ΔN are very similar (Table 1). The association rate of Nec-fl with PPE, however, is 13-fold lower, and the SI value 5-fold higher, than with Nec-ΔN (Table 1 and Fig. 7), suggesting that the N-terminally cleaved protein is a much better inhibitor of the elastase-like proteinase PPE. All values for Nec-ΔN were within 10% of those reported in Robertson et al. (2003), with the exception of thrombin and cathepsin G (which was not tested in Robertson et al. 2003).
Fig. 7.
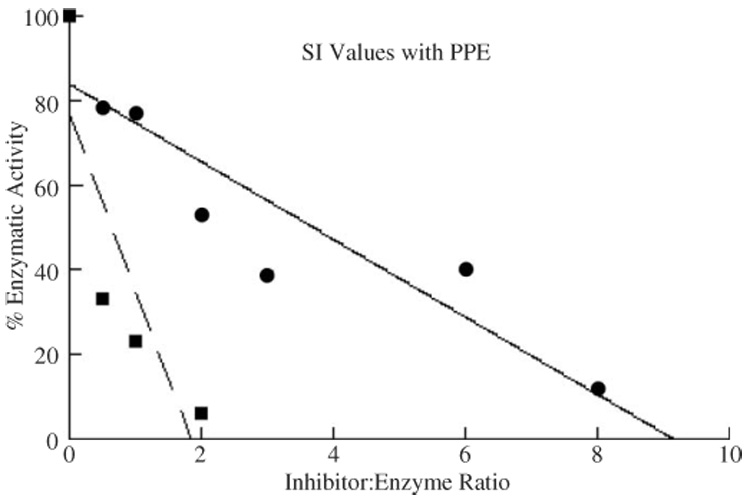
Concentration-dependent inhibition of Porcine Pancreatic Elastase by Nec proteins. Residual proteinase activities were assayed after 10 min incubations at different serpin:proteinase molar ratios. Residual activities (%) are plotted against the molar ratios of Nec-fl and proteinases. Values indicated with circles are Nec-fl and with squares are Nec-ΔN.
4. Discussion
The mammalian and invertebrate innate immune system share highly conserved signalling pathways. In both cases, a central receptor, Toll or one of the mammalian homologous Toll-like receptors (TLRs), signals downstream to a Rel family transcription factor that in turn activates hundreds of effector genes (Hoffmann, 2003). However, while mammalian TLRs bind to and directly recognize microbial molecules, Drosophila Toll receptor is activated by its endogenous ligand Spz (Weber et al., 2003). In the fly, Gram-positive bacteria are sensed by soluble, blood-borne pattern recognition receptors of the PGRP and Gram-negative binding protein (GNBP) families (Michel et al., 2001; Gobert et al., 2003). As the results on Toll activation became gradually apparent, the recognition events were pushed further upstream. Hence, it became central to understand how the Toll ligand Spz is activated by cleavage from its precursor. The discovery of the Nec serpin and its genetically interacting proteinase Psh gave the first clues that proteinase cascade(s) could link microbial pattern binding and Toll activation (Levashina et al., 1999; Ligoxygakis et al., 2002a). Nec mutations result in constitutive cleavage of Spz and concomitant expression of the Toll target gene Drosomycin. Subsequently, mutations in the catalytic triad of the serine proteinase Psh were discovered to suppress the nec mutant phenotype, but also the activation of the Toll pathway by fungal infection, confirming that the processing of the Toll ligand requires at least one serine proteinase regulated by nec.
Nec differs from most other serpins in carrying a long N-terminal extension, rich in glutamine, lysine and praline residues. Prior to immune challenge, Nec is present in the fly as a single molecular species of about 60 kDa. We show here that any Toll pathway activating immune challenge results in N-terminal cleavage of Nec protein to give a truncated 52 kDa form. Cleavage resulting from either fungal or Gram-positive infections is blocked by mutations in psh, showing that the serine proteinase Psh is absolutely required for this cleavage. It is uncertain at present if Nec is the direct target of Psh itself or if Psh activates a downstream proteinase that is responsible for the N-terminal cleavage of Nec.
Cleavage of Nec resulting from Gram-positive infection is suppressed in a psh mutant background. Psh is therefore required for N-terminal cleavage of Nec after a Gram-positive infection but, as was shown previously, it is not necessary for subsequent Toll pathway activation (Ligoxygakis et al., 2002a). The best explanation for this apparent paradox is that Gram-positive infection activates at least two redundant proteinases or proteinase cascades, both targeting the cleavage of Spz and the activation of the Toll pathway. The first proteinase (or proteinase cascade) is directly upstream of the enzyme that is responsible for the cleavage of Spz. The second proteinase is aimed at Psh, resulting in the N-terminal cleavage of Nec, and hence activates indirectly Spz and the Toll pathway via Psh. This redundancy may also explain why several genetic screens have failed to isolate mutations in the serine proteinases acting upstream of Toll during the immune response of the fly.
Nec-fl protein expressed in a baculovirus system is an active serpin inhibitor, which forms covalent complexes with different serine proteinases. Nec-fl is a potent inhibitor of cathepsin G and elastases (Table 1) and a moderate inhibitor of chymotrypsin. Thrombin is only weakly inhibited, while trypsin is not inhibited by Nec-fl. The inhibitory profile of Nec-ΔN is very similar to that of Nec-fl, with the exception of PPE: the association rate of Nec-fl with PPE is 13-fold slower, and the SI value 5-fold higher, than with Nec-ΔN (Table 1 and Robertson et al., 2003). Other serpins that carry N-terminal extensions are Heparin cofactor II and antithrombin, which carry proteinase-binding (Baglin et al., 2002) and heparin-binding (Jin et al., 1997) sites, respectively. Although these N-terminal extensions are shorter than that of Nec and show no sequence similarity, they give examples mechanisms that can modify the specificity of a core serpin.
The finding that PPE is more strongly inhibited by Nec-ΔN than by Nec-fl indicates a mechanism for feedback inhibition following immune challenge. The generalized activation of serine proteinases in the fly hemolymph would be extremely deleterious; indeed nec mutant flies die within a few days of hatching as adult flies (Heitzler et al., 1993). In this situation, keeping the activation of the Toll pathway localized or rapidly down-regulating the different proteinase cascades becomes essential and increasing the inhibitory activity of Nec would fulfil this function.
The psh mutation is a genetic suppressor of the nec mutation and the S1 specificity pocket of Psh matches well with the RCL sequence of Nec, but it is currently unclear whether the Psh proteinase is the direct and only target of Nec. The Drosophila genome contains 10 elastase-like serine proteinases with a predicted signal sequence, eight of which are strongly up-regulated after an immune challenge (Ross et al., 2003). The analysis of these eight candidates should allow the isolation of the adult equivalent of easter, the elusive proteinase responsible for the cleavage of Spz in the hemolymph and the activation of the Toll pathway during the immune response.
Acknowledgments
This work was supported by the French Centre National de la Recherche Scientifique and the National Institutes of Health. We thank Drs. M. Kanost and V. Leclerc for critical reading of the manuscript, Dr. Steve Hartson at Oklahoma State University Recombinant DNA/Protein Resource Facility for assistance in MALDI-mass spectrometry and A. Meunier for technical help. TRD is an MRC Career Development Fellow.
Abbreviations
- PGRP
Peptidoglycan recognition protein
- Psh
Persephone
- Dif
Dorsal-related immune factor
- Spz
Spätzle
- Nec
Necrotic
- RCL
Reactive center loop
- Nec-fl
Full-length Nec
- Nec-ΔN
Nec with the N-terminal deletion
- Ha
Hemaglutinin
- CD
Circular dichroism
- HNE
Human neutrophil elastase
- PPE
Porcine pancreatic elastase
- TUG
Transverse urea gradient
- SI
Stoichiometry of inhibition
- TLR
Toll-like receptor
- GNBP
Gram-negative binding protein
- SP
Signal peptide
References
- Baglin TP, Carrell RW, Church FC, Esmon CT, Huntington JA. Crystal structures of native and thrombin-complexed heparin cofactor II reveal a multistep allosteric mechanism. Proc. Natl. Acad. Sci. USA. 2002;99:11079–11084. doi: 10.1073/pnas.162232399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belorgey D, Dirrig S, Amouric M, Figarella C, Bieth JG. Inhibition of human pancreatic proteinases by mucus proteinase inhibitor, eglin c and aprotinin. Biochem. J. 1996;313:555–560. doi: 10.1042/bj3130555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieth JG. In vivo significance of kinetic constants of macromolecular proteinase inhibitors. Adv. Exp. Med. Biol. 1984;167:97–109. doi: 10.1007/978-1-4615-9355-3_7. [DOI] [PubMed] [Google Scholar]
- Bieth JG. Theoretical and practical aspects of proteinase inhibition kinetics. Meth. Enzymol. 1995;248:59–84. doi: 10.1016/0076-6879(95)48007-2. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Dafforn TR, Pike RN, Bottomley SP. Physical characterization of serpin conformations. Methods. 2004;32:150–158. doi: 10.1016/s1046-2023(03)00206-8. [DOI] [PubMed] [Google Scholar]
- Gettins PG. Serpin structure, mechanism, and function. Chem. Rev. 2002;102:4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- Gobert V, Gottar M, Matskevich A, Rutschmann S, Royet J, Belvin M, Hoffmann JA, Ferrandon D. Dual activation of the Drosophila toll pathway by two pattern recognition receptors. Science. 2003;302:2126–2130. doi: 10.1126/science.1085432. [DOI] [PubMed] [Google Scholar]
- Green C, Levashina E, McKimmie C, Dafforn T, Reichhart JM, Gubb D. The necrotic gene in Drosophila corresponds to one of a cluster of three serpin transcripts mapping at 43A1.2. Genetics. 2000;156:1117–1127. doi: 10.1093/genetics/156.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C, Brown G, Dafforn TR, Reichhart JM, Morley T, Lomas DA, Gubb D. Drosophila necrotic mutations mirror disease-associated variants of human serpins. Development. 2003;130:1473–1478. doi: 10.1242/dev.00350. [DOI] [PubMed] [Google Scholar]
- Gubb D, Garcia-Bellido A. A genetic analysis of the determination of cuticular polarity during development in Drosophila melanogaster. J. Embryol. Exp. Morphol. 1982;68:37–57. [PubMed] [Google Scholar]
- Heitzler P, Coulson D, Saenz-Robles MT, Ashburner M, Roote J, Simpson P, Gubb D. Genetic and cytogenetic analysis of the 43A-E region containing the segment polarity gene costa and the cellular polarity genes prickle and spiny-legs in Drosophila melanogaster. Genetics. 1993;135:105–115. doi: 10.1093/genetics/135.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann JA. The immune response of Drosophila. Nature. 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- Hoffmann JA, Reichhart JM. Drosophila innate immunity: an evolutionary perspective. Nature Immunol. 2002;3:121–126. doi: 10.1038/ni0202-121. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Yu XQ, Zhu Y, Kanost M. Prophenoloxidase-activating proteinase-3 (PAP-3) from Manduca sexta hemolymph: a clip-domain serine proteinase regulated by serpin-1J and serine proteinase homologs. Insect Biochem. Mol. Biol. 2003;33:1049–1060. doi: 10.1016/s0965-1748(03)00123-1. [DOI] [PubMed] [Google Scholar]
- Jin L, Abrahams JP, Skinner R, Petitou M, Pike RN, Carrell RW. The anticoagulant activation of antithrombin by heparin. Proc. Natl. Acad. Sci. USA. 1997;94:14683–14688. doi: 10.1073/pnas.94.26.14683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanost MR. Serine proteinase inhibitors in arthropod immunity. Dev. Comp. Immunol. 1999;23:291–301. doi: 10.1016/s0145-305x(99)00012-9. [DOI] [PubMed] [Google Scholar]
- Lawrence DA, Olson ST, Palaniappan S, Ginsburg D. Engineering plasminogen activator inhibitor 1 mutants with increased functional stability. Biochemistry. 1994;33:3643–3648. doi: 10.1021/bi00178a022. [DOI] [PubMed] [Google Scholar]
- Levashina EA, Langley E, Green C, Gubb D, Ashburner M, Hoffmann JA, Reichhart JM. Constitutive activation of toll-mediated antifungal defense in serpin-deficient Drosophila. Science. 1999;285:1917–1919. doi: 10.1126/science.285.5435.1917. [DOI] [PubMed] [Google Scholar]
- Ligoxygakis P, Pelte N, Hoffmann JA, Reichhart JM. Activation of Drosophila Toll during fungal infection by a blood serine protease. Science. 2002a;297:114–116. doi: 10.1126/science.1072391. [DOI] [PubMed] [Google Scholar]
- Ligoxygakis P, Pelte N, Ji C, Leclerc V, Duvic B, Belvin M, Jiang H, Hoffmann JA, Reichhart JM. A serpin mutant links Toll activation to melanization in the host defence of Drosophila. EMBO J. 2002b;21:6330–6337. doi: 10.1093/emboj/cdf661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadeva R, Dafforn TR, Carrell RW, Lomas DA. 6-mer peptide selectively anneals to a pathogenic serpin conformation and blocks polymerization. Implications for the prevention of Z α1-antitrypsin-related cirrhosis. J. Biol. Chem. 2002;277:6771–6774. doi: 10.1074/jbc.C100722200. [DOI] [PubMed] [Google Scholar]
- Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756–759. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- Morrison JF, Walsh CT. The behavior and significance of slow-binding enzyme inhibitors. Adv. Enzymol. Relat. Areas Mol. Biol. 1988;61:201–301. doi: 10.1002/9780470123072.ch5. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Morita T, Iwanaga S. Intracellular proclotting enzyme in limulus (Tachypleus tridentatus) hemocytes: its purification and properties. J. Biochem. 1985;97:1561–1574. doi: 10.1093/oxfordjournals.jbchem.a135213. [DOI] [PubMed] [Google Scholar]
- Robertson AS, Belorgey D, Lilley KS, Lomas DA, Gubb D, Dafforn TR. Characterization of the necrotic protein that regulates the Toll-mediated immune response in Drosophila. J. Biol. Chem. 2003;278:6175–6180. doi: 10.1074/jbc.M209277200. [DOI] [PubMed] [Google Scholar]
- Ross J, Jiang H, Kanost MR, Wang Y. Serine proteases and their homologs in the Drosophila melanogaster genome: an initial analysis of sequence conservation and phylogenetic relationship. Gene. 2003;304:117–131. doi: 10.1016/s0378-1119(02)01187-3. [DOI] [PubMed] [Google Scholar]
- Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PG, Irving JA, Lomas DA, Luke CJ, Moyer RW, Pemberton PA, Remold-O’Donnell E, Salvesen GS, Travis J, Whisstock JC. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins: evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 2001;276:33293–33296. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Jiang H, Kanost MR. Expression and purification of Manduca sexta prophenoloxidase-activating proteinase precursor (proPAP) from baculovirus-infected insect cells. Protein Exp. Purif. 2001;23:328–337. doi: 10.1006/prep.2001.1517. [DOI] [PubMed] [Google Scholar]
- Weber AN, Tauszig-Delamasure S, Hoffmann JA, Lelievre E, Gascan H, Ray KP, Morse MA, Imler JL, Gay NJ. Binding of the Drosophila cytokine Spatzle to Toll is direct and establishes signaling. Nature Immunol. 2003;4:794–800. doi: 10.1038/ni955. [DOI] [PubMed] [Google Scholar]


